Abstract
The POK family of proteins plays an important role in not only embryonic development and cell differentiation, but also in oncogenesis. Leukemia/lymphoma-related factor (LRF) belongs to the POK family of transcriptional repressors and is also known as POK erythroid myeloid ontogenic factor (POKEMON), which binds to short transcripts of HIV-1 (FBI-1) and TTF-1 interacting peptide (TIP21). Its oncogenic role is known only in lymphoma, non-small cell lung carcinoma, and malignant gliomas. The functional expression of LRF in human breast carcinoma has not yet been confirmed. The aim of this study was to investigate and compare the expression of LRF in human breast cancer tissues and other human tumors. The expression of LRF mRNA transcripts and protein was observed in twenty human benign and malignant breast biopsy tissues. Expression of LRF was observed in several formalin-fixed tissues by immunohistochemistry and immunofluorescence. All malignant breast tissues expressed mRNA transcripts and protein for LRF. However, 40% and 15% benign breast biopsy tissues expressed LRF mRNA transcripts and protein, respectively. The overall expression of LRF mRNA transcripts and total protein was significantly more in malignant breast tissues than the benign breast tissues. LRF expression was also observed in the nuclei of human colon, renal, lung, hepatocellular carcinomas and thymoma tumor cells. In general, a significantly higher expression of LRF was seen in malignant tissues than in the corresponding benign or normal tissue. Further studies are warranted to determine the malignant role of LRF in human breast carcinoma.
Keywords: Breast Cancer, Pokemon, LRF, ZBTB7, POK
INTRODUCTION
Leukemia/lymphoma-related factor (LRF) can impair cellular differentiation and promote oncogenesis (Schubot and Tropea, 2006). A 72kDa protein and the product of the ZBTB7A gene (located on chromosome 19p13.3), LRF is characterized by a zinc finger at the carboxy-terminal domain and a BTB-POZ (Broad Complex, Tramtrack and Bric-a-brac - Pox-virus and Zinc-finger/DNA-binding Zinc finger) domain at the amino-terminal domain. It associates and co-localizes with another BTB/POZ domain protein, LAZ-3/BCL-6 (Lymphoma Associated Zinc finger on chromosome 3/B-cell Lymphoma 6), in the nucleus but not with the other POZ domain proteins such as promyelocytic leukemia zinc finger (PLZF) and the Drosophila transcriptional activator known as GAGA factor (Davie et al, 1999; Morrison et al, 1999). Structural and sequential analyses suggest some similarity between BCL-6 and LRF, but not with PLZF or diffuse large cell lymphomas (Stogios et al, 2007).
Comparative genomic hybridization analysis indicated that LRF is expressed in adult human malignant glioma (Rovin and Winn, 2005). It is also involved in the differentiation of preadipocytes (Laudes et al, 2004; Laudes et al, 2008). Osteoclast derived zinc finger (OCZF), the rat homologue of LRF, is involved in osteoclast differentiation (Kukita et al, 1999). LRF is involved in diverse aspects of development and differentiation and represses the function of extracellular matrix genes (Lee et al, 2002; Pessler and Hernandez, 2003). Modulation by sumoylation, a post-translational modification, and interaction with Sp1 transcription factor, BCL-6-interacting corepressor,, nuclear receptor corepressor, silencing mediator for retinoid and thyroid complex corepressor, and various histones regulate the activity of LRF as a transcription factor (Laudes et al, 2008; Lee et al, 2002; Jeon et al, 2008).
A study using mouse embryonic fibroblasts (MEFs) and oncogenes, c-Myc, H-ras, and T-antigen, indicated the potential oncogenic role of LRF in the pathogenesis of cancer. LRF overexpression in transgenic mice led to the oncogenic transformation of MEFs, significantly increasing its expression in more B-cell and T-cell lymphomas than in the normal thymic cells (Roh et al, 2007; Maeda et al, 2005a). LRF is known as the master regulator in determining the B-cell and T-cell lymphoid fate (Maeda et al, 2007).
Overexpressed LRF suppresses the transcription of p14ARF so that Mdm2 is no longer deactivated by p14ARF, leaving p53 to continue the cellular growth process (Agrawal et al, 2006). It is inferred that LRF induces tumorigenesis by suppressing the p53 pathway in cancers of the bladder, lung, colon, and breast (Maeda et al, 2005b). LRF also aberrantly opposes the Notch signaling pathway by targeting upstream components that have yet to be determined (Maeda et al, 2007).
Despite the known role of LRF in lymphoma, non-small cell lung carcinoma, malignant gliomas, and speculation of its function in various physiological processes, the functional expression of LRF has not been observed in human solid tumors. In this study, we examined the basal expression of LRF in various malignant and benign human tissues.
MATERIALS AND METHODS
Human breast tissue biopsy samples and tumor tissue arrays
Twenty fresh biopsy samples (9 malignant and 11 benign) from female patients were obtained from Creighton University Medical Center (CUMC), Omaha, NE (IRB #0513792). Patient identifiers were coded to protect confidentiality. The fresh tissue was flash-frozen by placing on dry ice and then cut into two sections for RNA and protein analysis. Paraffinized blocks of breast biopsy patients and their subsequent surgically removed tissue and other breast, kidney, colon, mesothelioma, and lymphoma tissues were provided by the Department of Pathology at CUMC. The accompanying pathology reports indicated the stage of carcinoma in patients according to the AJCC classification. An array (#MNT241, Pantomics, Inc, San Francisco, CA) of multi-normal and tumor tissue sections, consisting of seven common tumor tissues and corresponding normal tissue of brain, breast, colon, liver, lung, prostate, and tonsil on one slide was purchased. A breast disease array (#BRD181, Pantomics, Inc.) consisting of 18 tissues including normal breast tissue, hyperplasia, fibroadenoma, invasive ductal carcinoma, invasive lobular carcinoma, and Paget’s disease on one slide was purchased. A third assay (#UNC241, Pantomics, Inc.) consisting of tonsil, colon, thyroid, thymoma, uterus, placenta, and melanoma tissues in duplicates on one slide was also purchased.
RNA Isolation and semi-quantitative reverse transcriptase-polymerase chain reaction (RT-PCR)
The flash frozen biopsy tissue was homogenized in Trizol Reagent (Sigma, St. Louis, MO) and the supplier’s instructions were followed for the remainder of the isolation. The quantity of the total RNA was determined using the GeneQuant 1300 spectrophotometer (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). RNA (1μg) was reverse-transcribed into cDNA using the ImProm-II Reverse Transcription System kit (#A3800, Promega, Madison, WI) and the supplier’s instructions were followed with modifications. Nuclease-free water (5.3μL) and MgCl2 (3.2μL) were used for the reverse-transcription reaction mixture which was placed on the heat block at 25°C for 5 minutes, 42°C for 60 minutes, 70°C for 5 minutes, and then 4°C for 5 minutes. Each PCR reaction mixture contained PCR Master Mix (25μL), cDNA (2μL), forward primer (5μL; 1μM), reverse primer (5μL; 1μM), and RNase/DNase Free Water (13μL). The PCR reaction mixtures were placed in the PCR machine heat block and the cDNA was initially denatured at 95°C for 15 minutes. PCR amplification was performed by denaturation for 60 seconds at 42°C, annealing for 60 seconds at 53°C, primer extension for 60 seconds at 72°C. The samples were amplified for an optimized number of cycles and annealing temperatures. For all experiments, 18S was used as an internal control. The PCR product of LRF (NM_015898.2) and 18S were 365bp and 247bp, respectively as deduced from the following sequences:
LRF (forward): 5′ GTCGCAGAAGGTGGAGAAGAAGAT 3′
LRF (reverse): 5′ AGCCGTCTTTCTTGAGGTGTCTCT 3′
18S (forward): 5′ TTGCCATCACTGCCATTAAGGGTG 3′
18S (reverse): 5′ TCTTCACGGAGCTTGTTGTCCAGA 3′
The PCR product was loaded onto an 1.5% agarose gel containing ethidium bromide. After electrophoresis, the gels were photographed under UV light using the UVP Bioimaging System (Upland, CA). Each experiment was performed three times.
Western immunoblotting
The protein concentration of harvested cell lysates from the breast biopsy tissues and cultured breast cells was measured using the Bio-Rad Bradford Protein Assay. The assay was performed in a flat-bottom 96-well microplate according to the supplier’s instructions. The concentrations of the bovine serum albumin solutions that were used as standards for the assay were as follows: 0.05, 0.125, 0.25 and 0.5mg/mL. The samples were mixed thoroughly for three seconds in the microplate mixer and the absorbance was measured at 595nm using a Bio-Rad microplate reader.
Protein solutions were diluted (1:1 v/v) with Laemmli loading buffer with 10% β-mercaptoethanol and heated at 95°C for five minutes before application to the SDS-PAGE gels. A solution containing reference proteins (Precision plus protein dual color) was similarly applied. Electrophoresis was performed at 100V for 1 hour at 25mM Tris buffer containing 192mM glycine and 0.1% (w/v) SDS pH 8.3. The protein was transferred from the gels onto an Immuno-blot PVDF membrane and sandwiched between filter papers. The protein transfer was performed at 400mA for one hour in cold 25mM Tris buffer containing 192mM glycine pH 8.3, methanol (20% v/v), and distilled water (70% v/v). The membranes were incubated overnight in blocking solution [100mL PBS, 5% (w/v) nonfat dried milk, and 0.05% Tween-20] to minimize non-specific protein binding.
The membranes were incubated for one hour at room temperature in a solution of the anti-LRF rabbit polyclonal antibody (1:500) and PBS containing 0.05% Tween-20. The anti-LRF rabbit polyclonal antibody was raised against a peptide derived from the C-terminal region of LRF (NP_056982, amino acids 533–547, Cys-GQEKHFKDEDEDEDV) prepared by Pacific Immunology (Ramona, CA). For all experiments, anti-GAPDH monoclonal mouse antibody (1:2000; NB-1300, Novus Biologicals, Littleton, CO) was used as an internal control. The membranes were incubated in HRP-conjugated secondary antibody solutions (1:1000, anti-rabbit (NB-730H) or anti-mouse (NB-720H), Novus Biologicals) for one hour at room temperature. The antibody-binding was detected using the SuperSignal West Dura Extended Duration Substrates (Pierce, Rockford, IL) and recorded in the UVP Bioimaging System.
Immunohistochemistry
Expression of LRF was analyzed using four antibodies specific to four different epitopes of LRF. The LRF antibodies (Pacific Immunology, Novus Biologicals, Abcam, and Santa Cruz Biotechnology) were applied to the tissue sections at dilutions of 1:800 for the brain, tonsil, liver, uterus, placenta, thymus, thymoma, melanoma, and 1:3000 for lung, breast, and colon tissues. Absence of staining (negative control) was demonstrated in control experiments using pre-immunization sera from rabbits immunized with LRF and also when the primary antibody was omitted. hematoxylin and eosin (H&E) staining of sections was used to confirm the clinicopathological status of tissues.
Paraffin sections (4–6μM) were cut from the paraffin-embedded blocks and placed onto Superfrost plus slides (#631-0108, VWR International, Lutterworth, Leicestershire, UK). The sections were deparaffinized and hydrated in xylene and solutions of ethanol gradients [100, 95, 80, and 70 (v/v)]. For immunostaining, antigen was exposed using Target Retrieval Solution (#S1699, DAKO, Carpenteria, CA) by boiling the sections in a steam cooker for 30 minutes. Endogenous peroxidase was blocked by incubating the sections in 3% hydrogen peroxide in methanol for 15 minutes. Blocking of non-specific binding sites, incubation with primary antibody (one hour), secondary antibodies, and avidin-biotin complex (ABC) was performed according to the supplier’s instructions from the Vectastain ABC elite kit (PK-6101, Vector Laboratories, Burlingame, CA). Sections were stained with 3,3′-Diaminobenzidine (DAB; SK-4100, Vector Laboratories) and counterstained using hematoxylin for five seconds, dehydrated in solutions of ethanol gradients [70, 80, 95, and 100 (v/v)] and immersed in xylene. Sections were mounted using Permount (#SP-15-100, Fisher Scientific, Pittsburgh, PA) and photographed by an Olympus DP71 camera at 200x or 400x magnification.
Immunofluorescence
Sections were incubated in blocking solution containing phosphate-buffered saline (PBS), 0.25% Triton X-100, 10mg/mL bovine serum albumin (BSA), and 5% normal goat serum (#015-000-120, Jackson Laboratories, West Grove, PA) at room temperature. The cells were incubated in primary antibody solution [anti-LRF (1:1,000) antibody, PBS, 0.1% Triton X-100, 10mg/mL BSA, and 1% normal goat serum] at room temperature. After washing with PBS containing 0.1% BSA three times for five minutes each, a secondary antibody (affinity purified goat anti-rabbit Cy2 antibody, 1:200) was applied to the sections for one hour in the dark to visualize Pokemon-labeled cells (Jackson Immunolabs, West Grove, PA). Negative controls were run in parallel using rabbit pre-immune serum PAC-767 (Pacific Immunology, Ramona, CA) instead of primary antibody or complete omission of primary antibody. Sections were washed with PBS with 0.1% BSA three times for five minutes and dipped into distilled water for two seconds. Fluorescence was preserved by sealing specimens with a solution of equal parts of PBS and glycerol containing 10mg/mL n-propyl gallate, and 1.5mg/mL 4′,6-diamidino-2-phenylindole (DAPI). To prevent the escape of the mounting medium from the coverslips, a single layer of nail polish was placed around the edges. Pictures were taken within 1 hr of the mounting using the Olympus DP71 camera.
Statistical Analysis
Values of all measurements were expressed as the mean ± SE. The Graph Pad Prism 4.0 biochemical statistical package (GraphPad Software, Inc., San Diego, CA, USA) software was used to draw graphs. Values of p<0.05 were considered statistically significant. Nuclear staining for IHC was scored by two independent observers, including a pathologist, in a blinded manner. There was <5% interobserver variation. The semi-quantitative scoring of the nuclear immunostaining was performed as follows: 0 - undetectable, 1+ - weakly positive, 2+ - moderately positive and 3+ - strongly positive.
RESULTS
Expression of LRF in breast cancer tissues
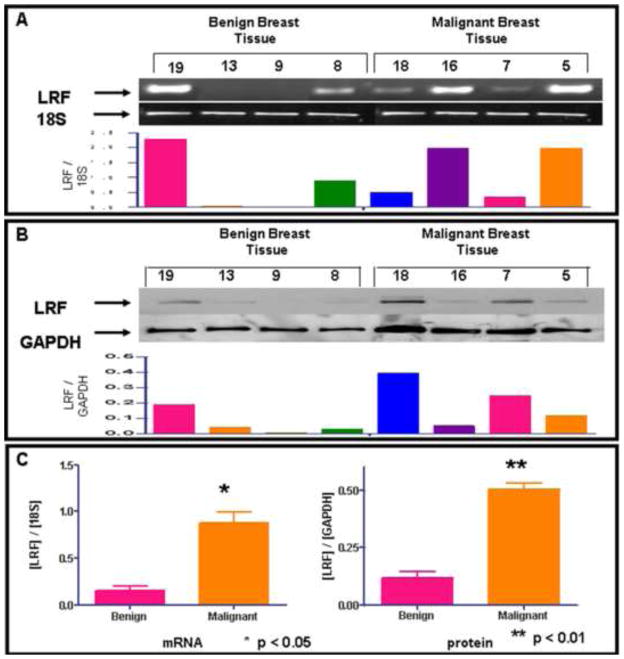
Expression of LRF mRNA transcripts and protein was observed in 11 benign and 9 malignant breast biopsy tissues. The representative data from 4 tissues in each group is shown in Figure 1A and 1B. The clinicopathological characteristics of the patients along with AJCC classification of the tumor stage were taken from the pathology reports and are presented in Table 1. Of the 20 samples, all 9 malignant breast tissues expressed mRNA transcripts and protein for LRF. However, 40% and 15% benign breast biopsy tissues expressed LRF mRNA transcripts and LRF protein, respectively. Densitometric analysis revealed significantly higher expression of LRF mRNA transcripts (p<0.05) and LRF protein (p<0.01) in 9 malignant breast tissues than in 11 normal/benign breast tissues (Figure 1C).
Figure 1. Expression of LRF and its mRNA transcripts in human benign and malignant breast biopsy tissues.
A - Representative samples of LRF mRNA transcripts expression by RT-PCR with densitometric analysis. B - Representative samples of LRF protein expression by western blotting with densitometric analysis. C - Densitometric analysis of all twenty samples indicating significantly higher expression of LRF mRNA and protein in malignant tissues than in the benign tissues. (*p<0.05; **p<0.01)
Table 1.
Clinicopathological characteristics of 20 breast biopsy tissues.
| Sample |
Age |
Breast (L/R) |
Cancer (Y/N - Stage) |
ER/PR% |
Ki-67 |
|---|---|---|---|---|---|
| 1 | 46 | L | Y (I) | 99/99 | 24 |
| 2 | 46 | R | N | - | - |
| 3 | 32 | R | N | - | - |
| 4 | 44 | L | N | - | - |
| 5 | 73 | R | Y (II/III) | 99/98 | 19 |
| 6 | 70 | R | N | - | - |
| 7 | 58 | R | Y (I) | 99/16 | 32 |
| 8 | 82 | L | N | - | - |
| 9 | 82 | L | N | - | - |
| 10 | 50 | R | Y (I) | 97/43 | 38 |
| 11 | 60 | R | Y (I) | 98/94 | 36 |
| 12 | 40 | R | N | - | - |
| 13 | 58 | R | N | - | - |
| 14 | 47 | R | N | - | - |
| 15 | 44 | L | Y (II) | 77/88 | 16 |
| 16 | 58 | R | Y (II) | 100/98 | 28 |
| 17 | 38 | L | N | - | - |
| 18 | 48 | R | Y (II) | 85/86 | 40 |
| 19 | 43 | L | N | - | - |
| 20 | 55 | L | Y (II/III) | 100/86 | 41 |
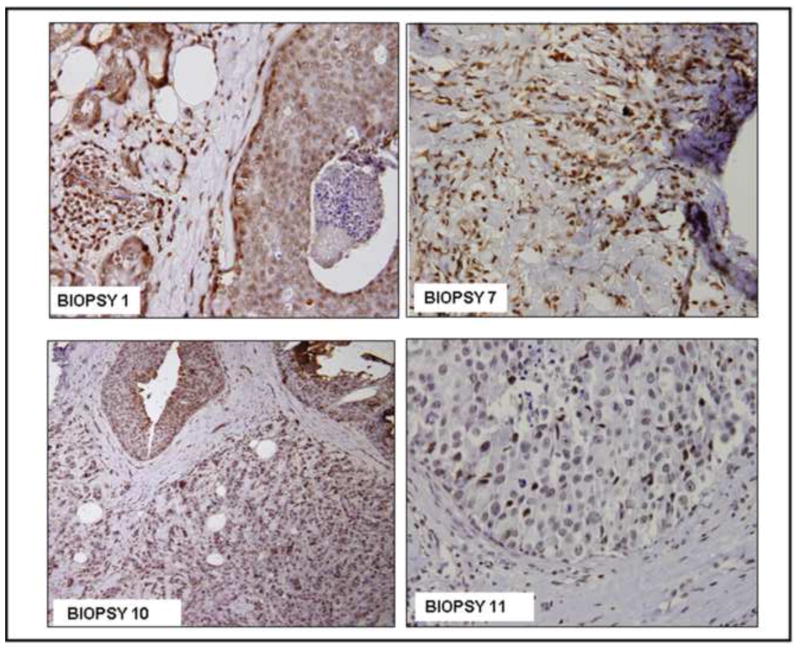
Immunohistochemical analysis validated the nuclear expression of LRF in the breast tissues (Figures 2–5). Biopsy samples #1, 7, 10, and 11 were characterized as invasive ductal carcinoma and they strongly expressed LRF (score: 3+; Figure 2). Biospy samples #1 and 10 were from patients with DCIS in addition to invasive ductal carcinoma. Staining for LRF in the surrounding stroma, which was infiltrated with inflammatory cells, was weakly positive and negative in the necrotic area within the ducts (Figure 2). Nuclear expression of LRF was clearly evident in malignant breast tissues and surrounding fibroblasts, lymphocytes, and stromal cells were also positive for expression of LRF, but not as vividly as the tumor (Figure 2). Staining for LRF with similar intensity was observed in the normal adjacent and malignant components of a core-needle biopsy sample (Figure 3A). This expression was confirmed by immunofluorescence and by the overlay with the DAPI nuclear stain (Figure 3B).
Figure 2. Immunohistochemical expression of LRF in representative human breast biopsy tissues.
DAB was used as a chromogen and hematoxylin as counterstain (200x).
Figure 5. Immunofluorescence of LRF expression in breast diseases.
Left Panels - Immunofluoresence using goat anti-rabbit Cy2 as the secondary antibody showing LRF expression, Right Panels - DAPI (blue) was used to a nuclear counterstain (200x – 400x).
Figure 3. LRF expression in malignant and normal adjacent tissues of a patient with breast cancer.
A - Immunohistochemical expression of LRF. Sections were stained with DAB and counterstained using hematoxylin. B - Immunofluoresence of LRF expression using goat anti-rabbit Cy2 as the secondary antibody, C - DAPI (blue) was used to stain nuclei (400x).
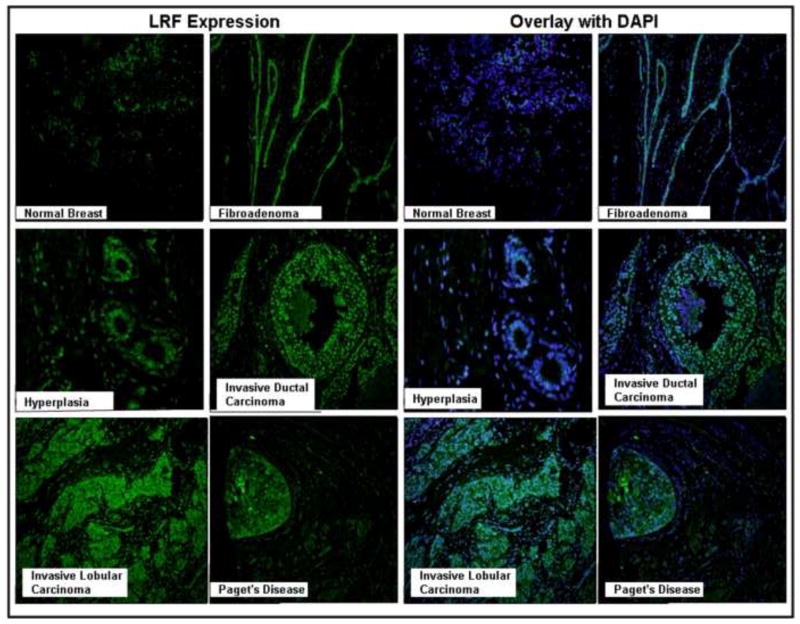
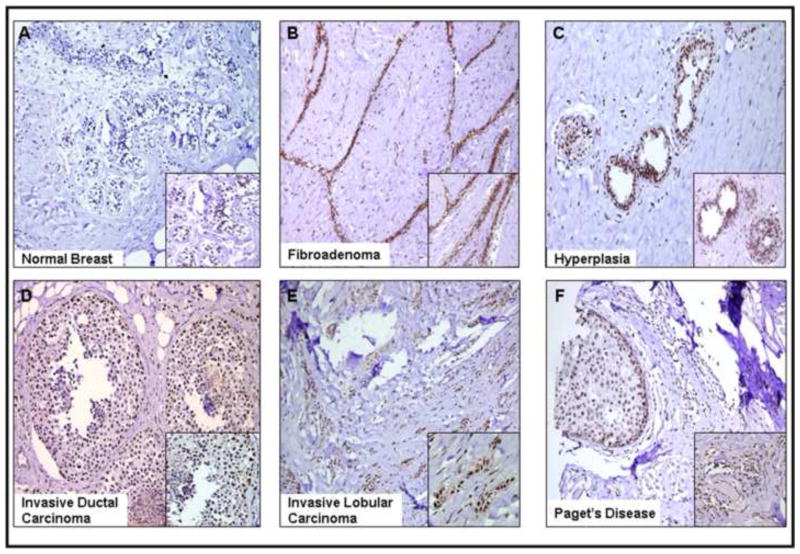
Little to no expression was observed in normal breast tissues (Figure 4A) while tissues of fibroadenoma consistently and strongly expressed LRF within the tissue fibers (Figure 4B). The intracanalicular fibroadenoma showed elongation, distortion and thinning of the epithelial elements of the breast tissue where LRF expression was strongest (Figure 4B). LRF was weakly stained in the ductal hyperplasia (Figure 4C). Strong nuclear expression of LRF was observed in invasive ductal carcinoma and invasive lobular carcinoma (Figure 4D and 4E). The breast tissue from a patient with Paget’s disease also expressed LRF within the invasive ductal carcinoma and DCIS components (Figure 4F). In general, little-to-no expression was observed in the adipocytes, inflammatory cells, and other stromal cells of the breast tissues and expression of LRF was confined to the tumor. LRF immunofluorescence was also shown in breast tissues and results were similar to those obtained by immunohistochemistry (Figure 5). The pathology scores for expression of LRF in the tissues of various breast diseases are presented in Table 2.
Figure 4. Immunohistochemical expression of LRF in various breast diseases.
DAB was used as a chromogen and counterstained with hematoxylin. Representative tissues from each disease are shown (200x – 400x).
Table 2.
Semi-quantified Expression of LRF in Breast Diseases Tissues from Figure 4.
| Sample |
Expression Level |
|---|---|
| Normal | 0 |
| Fibroadenoma | 3+ |
| Hyperplasia | 2+ |
| Invasive Lobular | |
| Carcinoma | 3+ |
| Invasive Ductal | |
| Carcinoma | 3+ |
| Paget’s disease | 2+ |
Expression of LRF in various human tumors and their corresponding normal tissues
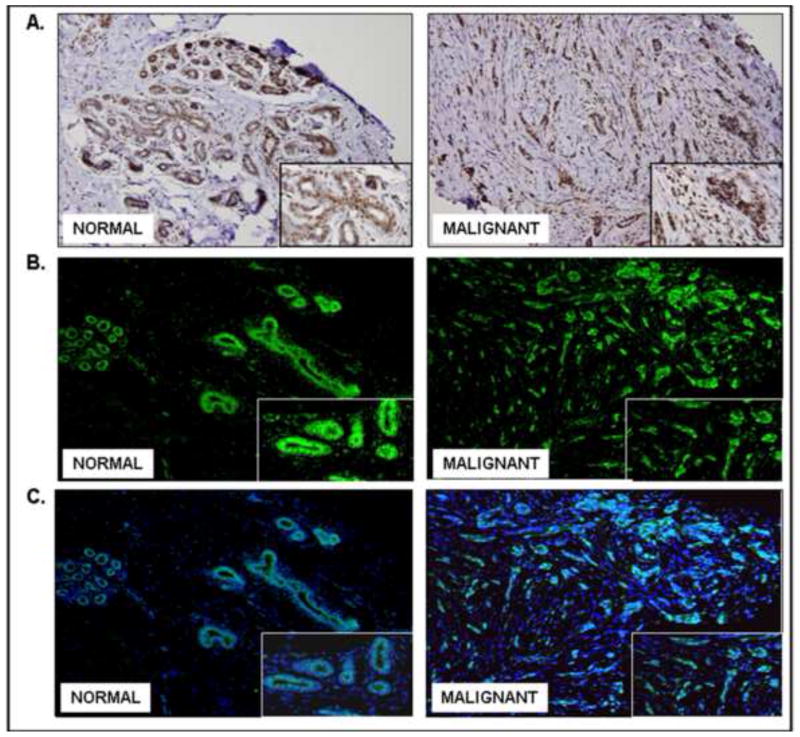
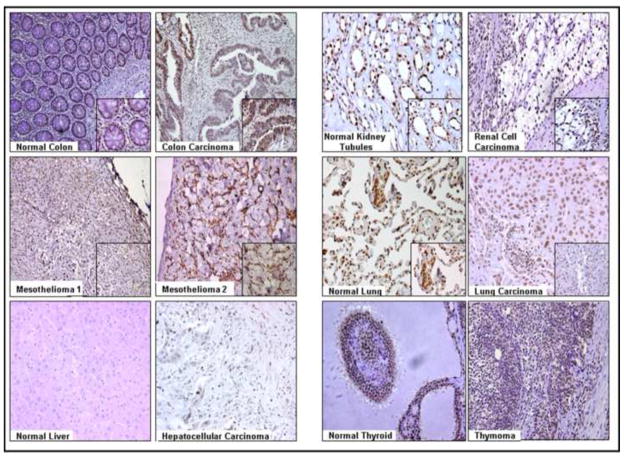
The clinicopathological characteristics of the patients along with the AJCC classification of the tumor stage were taken from the pathology reports and are presented in Table 3. The multi-normal and tumor tissue section array consisting of 24 tissues needed to be analyzed several times due to varying optimal dilutions (as listed in the Methods) of the anti-LRF antibody for different tumors. Each tissue (n=2) was present in duplicates in the array. Cerebral cortex was present in duplicates the array (unpublished observations) and did not show any expression of LRF. Tonsil tissue showed an increased expression of LRF in the germinal centers of the lymphatic tissue (unpublished observations). Normal colon showed weak expression of LRF while colon cancer strongly expressed LRF in the epithelia (Figure 6). Expression of LRF was observed in all tubules of the normal kidney as well as in renal cell carcinoma (Figure 6).
Table 3.
Clinicopathological characteristics of 13 carcinoma tissues.
| Tissue |
Cancer (Y/N - Stage) |
ER/PR% |
Ki-67 |
|---|---|---|---|
| Colon | Y | - | |
| Colon | Y | - | |
| Mesothelioma | Y | - | |
| Mesothelioma | Y | - | |
| Kidney | Y (II) | - | |
| Kidney | Y (II) | - | |
| Lymphoma | Y | - | |
| Breast | Y (II) | 100/5 | 70 |
| Breast | Y (II) | 99/15 | 70 |
| Breast | Y (III) | - | 18 |
| Breast | Y (III) | 90/0 | 94 |
| Breast | Y (III) | 91/0 | 93 |
| Breast | Y (I) | - | - |
Figure 6. Immunohistochemical expression of LRF in various carcinomas and corresponding normal tissues.
DAB was used as a chromogen and hematoxylin as a counterstain. Representative samples from the array are shown (200x – 400x).
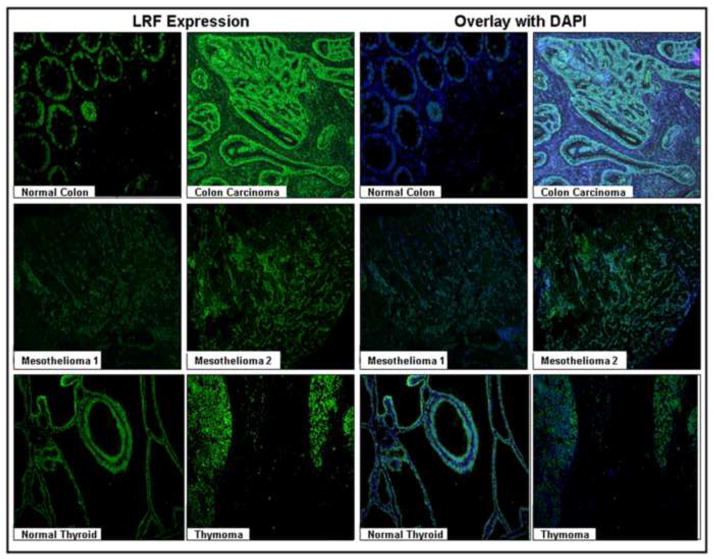
In the two cases of diffuse malignant mesothelioma, LRF was strongly expressed in the epithelial cells (Figure 6). LRF expression was slightly increased in lung carcinoma tissue (Figure 6). Normal liver tissue did not express LRF and hepatocellular carcinoma was stained weakly positive for LRF (Figure 6). Normal thyroid and thymoma tissues showed similar expression of LRF (Figure 6). The pathology scores for the expression of LRF in various tissues are presented in Table 4. The expression of LRF in the tissue sets (tumor and corresponding normal) was further examined and confirmed by immunofluorescence (Figure 7).
Table 4.
Semi-quantified expression of LRF in Tissues from Figure 6.
| Sample |
Normal |
Cancer |
|---|---|---|
| Colon | 1+ | 3+ |
| Kidney | 2+ | 2+ |
| Mesothelioma | N/A | 2+ |
| Lung | 1+ | 2+ |
| Liver | 0 | 1+ |
| Thyroid/Thymoma | 2+ | 2+ |
Figure 7. Immunofluorescence of LRF expression in various carcinomas and corresponding normal tissues.
Left Panels - Immunofluoresence using goat anti-rabbit Cy2 as the secondary antibody showing LRF expression, Right Panels - DAPI (blue) was used to a nuclear counterstain. Representative samples from the array are shown (200x).
DISCUSSION
The role of BCL-6 and PLZF POK family members has been extensively examined in carcinoma, but comparatively little is known about LRF. PLZF is involved in the progression of melanoma by coordinating the expression of many genes which are indicative of melanoma (Felicetti et al, 2004). BCL-6 is the third most common translocated region in non-Hodgkin lymphoma and functions in the germinal center of B-cells (Chang et al, 1996; Ye 2000; Bos et al, 2003). The functional role and expression of BCL-6 is acknowledged outside of the lymphoid system, such as in the skin and breast carcinomas (Kanazawa et al, 1997; Logarajah et al, 2003). The expression of LRF in various solid tumors has been examined, but little is known about its functional expression in human breast carcinomas.
The results in breast, colon, lung and liver tissues indicate that LRF has oncogenic properties and is involved in the malignant progression due to a strong difference in the expression of LRF mRNA and protein expression between the benign and malignant breast biopsy tissues. The few benign breast biopsy tissues that do express LRF could contain pre-malignant cells and adipocytes. A strong correlation could not be drawn from the clinicopathological characteristics of the breast biopsy tissues in this study, but Cui and associates (2007) found higher LRF expression in breast cancer tissues with axillary lymph node metastasis. LRF expression also correlated significantly with TNM staging in non-small cell lung carcinoma (Zhao et al, 2008).
Most transcription factors translocate from the cytoplasm to the nucleus. The members of the POZ family of transcription factors (i.e., BCL-6, Hypermethylated in Cancer-1, Kaiso, and PLZF) are predominantly located in the nuclei of mammalian cells (Dhordain et al, 1995; Kelly and Daniel, 2006). Consistent with observation by Lee and associates (2005) in HeLa cells, some cytoplasmic expression of LRF was seen in breast cells (unpublished observations) (Lee et al, 2005). Liu and associates (2004) showed that the POZ-domain of LRF interacted with various proteins and formed a heterodimer in both the nucleus and cytoplasm using the fluorescence-microscope.
The results that LRF expression was high in the breast tumor and the adjacent normal tissue agree with other findings in hepatocellular carcinomas (Cui et al, 2007; Ji et al, 2007). Frequently, cells with the non-cancerous breast tissue in patients with invasive carcinoma may have mutated and may express LRF as intensely as the malignant components of the tissue. Though the normal breast tissue does not express LRF, conditions that are associated with the development of carcinoma tend to express LRF. Expression of LRF was also observed in other neoplastic tissues (colon, lung, kidney, thymus, liver, and mesothelioma) indicating its involvement in carcinogenesis but not specificity for the type of tumor. Nuclear expression of LRF was not observed in the normal liver which is consistent with Ji and associates (2007).
The findings suggest that LRF has a oncogenic role in breast carcinoma because a strong correlation was observed in the epithelial cells of the proliferating benign breast lesions of the breast, fibroadenomas, and weak but consistent expression of LRF in the stromal cells and lymphocytes of carious carcinomas. Fibroadenomas are usually not associated with subsequent development of carcinoma, but exceptions have been noted (Dupont et al, 1994; Dorjgochoo et al, 2008). Continued studies in our lab using breast cell lines and MRC-9 fibroblast cells show expression of LRF and its mRNA transcripts. unpublished observations). LRF could be exerting its effects through these hormone receptors.
Potential mechanisms for the role of LRF in oncogenesis are not known. One possibility could be that LRF exerts its effect through hormone receptors, including estrogen and progesterone receptors. Alternatively or in addition, increased LRF expression and activity abrogates anti-oncogenic pathways involving 14ARF-Mdm2-p53 and Notch signaling, as recently reported (Maeda et al, 2005a; Maeda et al. 2007), But, upstream components causing aberrant regulation have yet to be identified. Obviously, additional studies on the underlying cellular and molecular mechanisms are warranted.
In summary, the results demonstrate that LRF is highly expressed in malignant tissues, but may not be a pan-marker for all tumors. Additional studies using more samples and clinical data could establish LRF as a marker for malignancy and develop therapeutic agents.
Acknowledgments
This work was supported by the Claire Booth Luce Fellowship at Creighton University (to AA). This work was also supported in part by research grants from the Department of Health and Human Services, Nebraska Cancer and Smoking – Related Diseases,, NIH grant HL073349, and the Carpenter Chair (to DKA) of Creighton University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal A, Yang J, Murphy RF, Agrawal DK. Regulation of the p14ARF-Mdm2-p53 pathway: an overview in breast cancer. Exp Mol Pathol. 2006;81:115–122. doi: 10.1016/j.yexmp.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Bos R, van Diest PJ, van der Groep P, Greijer AE, Hermsen MAJA, Heijnen I, Meijer GA, Baak JPA, Pinedo HM, Wall E, Shvarts A. Protein expression of B-cell lymphoma gene 6 (BCL-6) in invasive breast cancer is associated with cyclin D1 and hypoxia-inducible factor-1 alpha (HIF-1alpha) Oncogene. 2003;22:8948–8951. doi: 10.1038/sj.onc.1206995. [DOI] [PubMed] [Google Scholar]
- Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Xu H, Yang Y. Expression and clinical significance of Pokemon protein in breast cancer. J Clinical Surgery (Chinese) 2007;15:399–401. [Google Scholar]
- Davies JM, Hawe N, Kabaroswski J, Huan QH, Zhu J, Brand NJ, Leprince D, Dhordain P, Cook N, Morriss-Kay G, Zelenta A. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene. 1999;18:365–375. doi: 10.1038/sj.onc.1202332. [DOI] [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Ansieau S, Koken MHM, Deweindt C, Quief S, Lantoine D, Leutz A, Kerckaert J, Leprince D. The BTB/POZ domain targets the LAZ3/BCL6 oncoprotein to nuclear dots and mediates homodimerization in vivo. Oncogene. 1995;11:2689–2697. [PubMed] [Google Scholar]
- Dorjgochoo T, Deming SL, Gao YT, Lu W, Zhang Y, Ruan Z, Zheng W, Shu XO. History of benign breast disease and risk of breast cancer among women in China: a case-control study. Cancer Causes Control. 2008;19:819–828. doi: 10.1007/s10552-008-9145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont WD, Page DL, Parl FF, Vnencak-Jones CL, Plummer WD., Jr Long-term risk of breast cancer in women with fibroadenoma. N Engl J Med. 1994;331:10–15. doi: 10.1056/NEJM199407073310103. [DOI] [PubMed] [Google Scholar]
- Felicetti F, Bottero L, Felli N, Mattia G, Labbaye C, Alvino E, Peschle C, Colombo MP, Care A. Role of PLZF in melanoma progression. Oncogene. 2004;23:4567–4576. doi: 10.1038/sj.onc.1207597. [DOI] [PubMed] [Google Scholar]
- Hur MW. FBI-1 enhances transcription of the nuclear factor-kappaB (NF-kappaB)-responsive E-selectin gene by nuclear localization of the p65 subunit of NF-kappaB. J Biol Chem. 2005;280:27783–27791. doi: 10.1074/jbc.M504909200. [DOI] [PubMed] [Google Scholar]
- Jeon BN, Yoo JY, Choi WI, Lee CE, Yoong HG, Hur MW. Proto-oncogene FBI-1 (Pokemon/ZBTB7A) represses transcription of the tumor suppressor Rb gene via binding competition with Sp1 and recruitment of co-repressors. J Biol Chem. 2008;283:33199–33210. doi: 10.1074/jbc.M802935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, He XS, Zhu XF, Zhang R, Ou ZH. Expression of pokemon in hepatocellular carcinoma tissue, adjacent and far tissue, hepatocirrhosis and normal liver tissue. Chinese Journal of Experimental Surgery. 2007;24:187–189. [Google Scholar]
- Kanazawa N, Moriyama M, Onizuka T, Sugawara K, Mori S. Expression of Bcl-6 protein in normal skin and epidermal neoplasms. Pathol Int. 1997;47:600–607. doi: 10.1111/j.1440-1827.1997.tb04548.x. [DOI] [PubMed] [Google Scholar]
- Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–1997. [PubMed] [Google Scholar]
- Laudes M, Christodoulides C, Sewter C, Rochford JJ, Cosnidine RV, Sethi JK, Vidal-Puig A, O’Rahilly S. Role of the POZ zinc finger transcription factor FBI-1 in human and murine adipogenesis. J Biol Chem. 2004;279:11711–11718. doi: 10.1074/jbc.M310240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudes M, Bilkovski R, Oberhauser F, Droste A, Golmolka M, Leeser U, Udelhoven M, Krone W. Transcription factor FBI-1 acts as a dual regulator in adipogenesis by coordinated regulation of cyclin-A and E2F-4. J Mol Med. 2008;86:597–608. doi: 10.1007/s00109-008-0326-2. [DOI] [PubMed] [Google Scholar]
- Lee DK, Suh D, Edenberg HJ, Hur MW. POZ domain transcription factor, FBI-1, represses transcription of ADH5/FDH by interacting with the zinc finger and interfering with DNA binding activity of Sp1. J Biol Chem. 2002;277:26761–26768. doi: 10.1074/jbc.M202078200. [DOI] [PubMed] [Google Scholar]
- Lee DK, Kang JE, Park HJ, Kim MH, Yim TH, Kim JM, Heo MK, Kim KY, Kwon HJ, Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, Di Cesare PE. Leukemia/lymphoma related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J Biol Chem. 2004;279:47081–47091. doi: 10.1074/jbc.M405288200. [DOI] [PubMed] [Google Scholar]
- Logarajah S, Hunter P, Kramam M, Steele D, Lakhani S, Bobrow L, Venkitaramam A, Wagner S. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene. 2003;22:5572–5578. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005a;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- Maeda T, Hobbs RM, Pandolfi PP. The transcription factor Pokemon: a new key player in cancer pathogenesis. Cancer Res. 2005b;65:8575–8578. doi: 10.1158/0008-5472.CAN-05-1055. [DOI] [PubMed] [Google Scholar]
- Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van der Brink MR, Zelent A, Shigmatsu H, Akashi K, Teruya-Feldstein J, Cattoretti G, Pandolfi PP. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DJ, Pendergrast PS, Stavropoulos P, Colmenares SU, Kobayashi R, Hernandez N. FBI-1, a factor that binds to HIV-1 inducer of short transcripts (IST), is a POZ domain protein. Nucleic Acids Res. 1999;27:1251–1262. doi: 10.1093/nar/27.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessler F, Hernandez N. Flexible DNA binding of the BTB/POZ-domain protein FBI-1. J Biol Chem. 2003;278:29327–29335. doi: 10.1074/jbc.M302980200. [DOI] [PubMed] [Google Scholar]
- Roh HE, Lee MN, Jeon BN, Choi W, Kim YJ, Yu MY, Hur MW. Regulation of pokemon 1 activity by sumoylation. Cell Physiol Biochem. 2007;20:167–180. doi: 10.1159/000104164. [DOI] [PubMed] [Google Scholar]
- Rovin RA, Winn R. Pokemon expression in malignant glioma: an application of bioinformatics methods. Neurosurg Focus. 2005;19:E8. doi: 10.3171/foc.2005.19.4.9. [DOI] [PubMed] [Google Scholar]
- Schubot FD, Tropea JE, Waugh DS. Structure of the POZ domain of human LRF, a master regulator of oncogenesis. Biochem Biophys Res Commun. 2006;351:1–6. doi: 10.1016/j.bbrc.2006.09.167. [DOI] [PubMed] [Google Scholar]
- Stogios PJ, Chen L, Prive GG. Crystal stucture of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein Sci. 2007;16:336–342. doi: 10.1110/ps.062660907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH. BCL-6 in the pathogenesis of non-Hodgkin’s lymphoma. Cancer Invest. 2000;18:356–365. doi: 10.3109/07357900009012179. [DOI] [PubMed] [Google Scholar]
- Zhao ZH, Wang SF, Yu L, Wang J, Chang H, Yan W, Zhang J, Fu K. Overexpression of Pokemon in non-small cell lung cancer and foreshowing tumor biological behavior as well as clinical results. Lung Cancer. 2008;62:113–119. doi: 10.1016/j.lungcan.2008.02.014. [DOI] [PubMed] [Google Scholar]