Abstract
The integrase of the Saccharomyces cerevisiae retrotransposon Ty1 integrates Ty1 cDNA into genomic DNA likely via a transesterification reaction. Little is known about the mechanisms ensuring that integrase does not integrate non-Ty DNA fragments. In an effort to elucidate the conditions under which Ty1 integrase accepts non-Ty DNA as substrate, PCR fragments encompassing a selectable marker gene were transformed into yeast strains overexpressing Ty1 integrase. These fragments do not exhibit similarity to Ty1 cDNA except for the presence of the conserved terminal dinucleotide 5′-TG-CA-3′. The frequency of fragment insertion events increased upon integrase overexpression. Characterization of insertion events by genomic sequencing revealed that most insertion events exhibited clear hallmarks of integrase-mediated reactions, such as 5 bp target site duplication and target site preferences. Alteration of the terminal dinucleotide abolished the suitability of the PCR fragments to serve as substrates. We hypothesize that substrate specificity under normal conditions is mainly due to compartmentalization of integrase and Ty cDNA, which meet in virus-like particles. In contrast, recombinant integrase, which is not confined to virus-like particles, is able to accept non-Ty DNA, provided that it terminates in the proper dinucleotide sequence.
Keywords: DNA integration, Ty, Recombination, Target sites
Introduction
Ty elements are retrotransposons of the yeast Saccharomyces cerevisiae which contain long terminal repeats (LTRs) and resemble retroviruses in structure and function (for a recent review see Lesage and Todeschini 2005). In contrast to retroviruses, however, the life cycle of Ty elements lacks an extracellular phase. The genome of typical yeast laboratory strains contains about 45 full-length copies of the major class of Ty elements, Ty1, and the closely related class Ty2 (Kim et al. 1998). In addition, about 200 solo LTRs of Ty1 and Ty2 (called delta elements) are observed, which most likely arose by homologous recombination between direct repetitive LTRs and excision of the intervening sequence.
LTRs play a pivotal role in the life cycle of Ty1 elements. Transcription starts in the 5′ LTR and terminates in the 3′ LTR in a way that the resulting RNA molecule is terminally redundant. The redundant sequences define the R domain of the LTR, whereas U3 and U5 represent sequences unique to the 3′ and 5′ end, respectively. Ty1 elements contain two overlapping genes, GAG and POL. GAG encodes nucleocapsid proteins that form the structural components of the cytoplasmic virus-like particles (VLPs); POL encodes a polyprotein from which by ordered proteolytic processing the catalytic proteins protease, integrase, and reverse transcriptase/ribonuclease H are formed (Garfinkel et al. 1991; Merkulov et al. 2001).
Two Ty1 RNA molecules, a host initiator methionine tRNA (tRNAMeti), and the Ty1-encoded proteins are assembled in the VLPs, in which reverse transcription takes place. For priming of minus-strand cDNA synthesis, the 3′ end of the tRNA acceptor stem is bound to the complementary primer binding sequence (PBS) which is located adjacent to the 5′ LTR. Minus-strand cDNA is synthesized until the 5′ end of the genomic RNA is reached. A template switch is then necessary to allow elongation of the minus strand. Plus-strand synthesis is primed by short RNA sequences that are created by specific RNAse H cleavage at so-called polypurine tract (PPT) sequences. PPT1 lies adjacent to the 3′ LTR, and a second PPT site is found within the Ty1 coding region. Synthesis of the plus strand proceeds to the 5′ end of the minus strand. A second template switch is required to allow completion of plus-strand synthesis as well as completion of minus-strand synthesis. This generates the pre-integrative double-stranded full-length Ty cDNA, which bears complete (U3-R-U5) LTRs on both ends.
The LTRs also play an important role for the integration of the retroelements into genomic DNA. During the integration process, Ty1 integrase creates a 5-bp staggered cut in the target DNA and joins the ends of the blunt-ended cDNA molecules to the exposed 5′ phosphate groups of the target in a transesterification reaction (reviewed by Wilhelm et al. 2005). It is generally assumed that the conserved inverted dinucleotides 5′-TG-CA-3′ at the ends of the cDNA duplex are absolutely required for transposition in vivo, although only few sequence alterations have been investigated (Sharon et al. 1994). Anecdotal evidence for the importance of subterminal sequences has also been presented (Wilhelm et al. 1999). More detailed analyses of the requirement for cis-acting sequences in transposition were carried out using purified VLPs or recombinant integrase in model systems (Eichinger and Boeke 1990; Braiterman and Boeke 1994; Moore et al. 1995; Moore and Garfinkel 2000). These experiments hint at a relaxed integration specificity of recombinant integrase or purified VLPs in vitro. In addition, reverse transcriptase cooperates with integrase during reverse transcription and integration (Wilhelm and Wilhelm 2006).
Substrate specificity may, at least in part, be due to physical compartmentalization. Freshly synthesized cDNA and integrase meet in the VLPs, and both have to enter the nucleus for integration to occur. How this migration into the nucleus is achieved is not entirely clear. Since the yeast nuclear membrane remains intact throughout the cell cycle and since Ty1 VLPs are larger than the size limit for particle transport across the nuclear pore complex, a pre-integration complex (PIC) most presumably contains fewer constituents than the VLPs. The integrase protein contains a nuclear localization signal (NLS) at the C terminus which is essential and sufficient for translocation of integrase into the nucleus (Moore et al. 1998; Kenna et al. 1998). Thus, the Ty1 PIC may consist minimally of a cDNA element and integrase; the latter may be present as multimeric complex (Moore et al. 1998). While it is generally assumed that cDNA and integrase cross the nuclear membrane together, it cannot be excluded at present that cDNA and integrase are transported to the nucleus independently (Kenna et al. 1998). At least in certain situations, both factors have been found to act independently from each other. For example, in NLS-deficient integrase mutants retrotransposition is severely reduced, but insertion of Ty1 elements by cDNA recombination does take place frequently (Kenna et al. 1998), indicating that the cDNA is able to enter the nucleus even without integrase proteins. On the other hand, ectopically expressed integrase protein localizes to the nucleus even when highly overexpressed, strongly suggesting that this does not require a certain stochiometry between the number of integrase and of cDNA molecules (Moore et al. 1998; Kenna et al. 1998).
It has been suggested that host cell factors may contribute to substrate specificity in vivo (Wilhelm et al. 2005; Moore and Garfinkel 2000). Indeed, in a recent study of non-homologous integration (NHI; Schiestl and Petes 1991; Schiestl et al. 1993, 1994; Zhu and Schiestl 1996) in yku70 mutants we observed several examples where plasmid DNA was integrated into genomic DNA by a process apparently mediated by Ty1 integrase (Kiechle et al. 2000). In these cases, the plasmid DNA was processed to terminate in 5′-G/CT-C/GA-3′; the target sites exhibited a 5 bp duplication, and the target sites were compatible with known site preferences of Ty1 integrase. In addition, we observed several examples where composite elements, consisting of plasmid DNA and Ty-derived sequences, were integrated. These composite elements presumably arose by erroneous priming and strand switching events during reverse transcription. To further elucidate the conditions under which Ty1 integrase accepts non-Ty1-DNA in vivo as a substrate for integration, we studied the integration of PCR fragments encompassing the URA3 gene in yeast strains overexpressing Ty1 integrase. Our data strongly suggest that integrase readily accepts non-Ty1-DNA if expressed out of the context of regulated Ty1 life cycle.
Materials and methods
Strains and media
Saccharomyces cerevisiae strain RSY12 (MAT a leu2-3,112 his3-11,15 ura3Δ::HIS3) was used for all NHI experiments. In RSY12 the entire URA3 gene was replaced by the HIS3 gene (Schiestl and Petes 1991). Strain RSY12 Gal-Ty-IN was constructed as follows: plasmid pGTy1-IN, containing the Ty1 integrase ORF, was a generous gift of David Garfinkel, Department of Biochemistry and Molecular Biology, University of Georgia, Athens, GA, USA. This plasmid was used as template to amplify the coding region of Ty1-IN with Pfu Polymerase (Stratagene), using primers BamHI-TY-IN-2041-fw (5′-GCGGATCCAATGTCCATACAAGTGAAAG-3′) and BamHI-Ty-IN-3945-rc (5′-CGGGATCCTGCAATCAGGTGAATTCG-3′). After purification (Qiagen PCR purification kit) the PCR product was digested with BamHI. Plasmid pWY203 (Galli and Schiestl 1998) harbours the I-SceI-gene under the control of the Gal1-10 promoter, inserted within a 1.8 kb LYS2 fragment, and a hisG-URA3-hisG cassette. The I-SceI fragment was removed by BamHI digestion and the PCR-amplified integrase gene was inserted. The lys2::Gal1-10-Ty-IN-hisG-URA3-hisG::lys2 cassette was released from the backbone by AflII digest and transformed into RSY12. Transformants were selected on SD-Ura plates. They were checked for correct integration on SD-Lys plates and transferred to YPD media for 2–3 days to allow for removal of the URA3 gene due to recombination events between the hisG repeats. Loss of URA3 was selected by plating on 5-fluoro-orotic acid (5-FOA). Correct conformation of the locus was again confirmed by PCR.
PCR fragments and plasmids
Plasmid pM151 (Manivasakam and Schiestl 1998), a pUC18 derivative containing the URA3 gene, served as template for PCR amplification of URA3 cassettes. To create amplification products terminating either in 5′-TGA-3′ or in 5′-GTA-3′, two sets of primers were used (URA3-TGA-fw 5′-TGAACCTGCAGGCATGCAAGC-3′ and URA3-TGA-rc 5′-TGAGGATAACAATTTCACACAGG-3′; URA3-GTA-fw 5′-GTAACCTGCAGGCATGCAAGC-3′ and URA3-GTA-rc 5′-GTAGGATAACAATTTCACACAGG-3′). All PCR reactions for the generation of transformation cassettes were performed with Pfu polymerase to ensure high fidelity and to avoid nontemplated extensions at the 3′ ends. Plasmid YEplac195 contains the URA3 gene and the 2 μm origin of replication (Gietz and Sugino 1988); it is used to normalize transformation experiments with regard to transformation efficiency. Plasmids were maintained and amplified in Escherichia coli strain DH5α. Plasmid preparations were done using Qiagen plasmid purification kits.
Northern analysis
Preparation of mRNA and Northern analysis were done according to standard procedures, using a PCR-generated fragment of the integrase gene as probe.
Yeast transformation
Logarithmic cultures were centrifuged, washed twice with H2O, and split. Half of the culture was transferred to SC-glucose, the other half to SC-galactose. The cells were incubated at 30° for 6 h prior to the transformation with URA3 cassettes. Yeast transformations were performed using the lithium acetate-single-stranded (ss) DNA-polyethylene glycol transformation method described previously (Gietz et al. 1992), with the following modifications. For transformations with the PCR-derived URA3 cassettes, the amount of DNA was reduced from 7 to 2 μg per transformation. For parallel control transformations using the same batch of cells, 10 or 2 ng of circular YEplac195 were used. Transformants were grown on SD-Ura plates. To distinguish productive transformants and abortive transformants (Yap and Schiestl 1995) unambiguously the plates were replica plated after 2–3 days. Relative transformation frequencies were determined by normalizing the transformation rate (per μg of DNA) relative to the rate of transformation with control plasmid YEplac195.
Analysis of integration sites by direct genomic sequencing
Direct genomic sequencing (DGS; Horecka and Jigami 2000) was adapted for analysis of integration sites. Transformants were grown in 75 ml YPD media to early stationary phase (1 × 108 cells/ml) and DNA was extracted using the Qiagen Midi Genomic DNA kit. The incubation times for zymolyase (Zymolyase 100T, Seikagaku Corp., Tokyo, Japan) and proteinase K (Roche Molecular Biochemicals, Mannheim, Germany) treatment were extended to 1 and 1.5 h, respectively. DNA concentrations were determined with a conventional spectrometer (Eppendorf BioPhotometer). These modifications allowed a reduction of the amount of BigDye Terminator Ready Reaction Mix (Perkin Elmer Life and Analytical Sciences Inc., Boston, MA, USA) from 16 to 8 μl in the sequencing reactions. Primers URA3-out-5′-114 (5′-TCGTTCTTCCTTCTGCTCGGAGAT-3′) and URA3-out-3′-951 (5′-TGCAAAGGGAAGGGATGCTAAGGT-3′) were used, which were constructed using a web based primer design program (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Unincorporated terminators were removed with centri-sep spin columns (Princeton Separations, Adelphia, NJ, USA) that had been hydrated for at least 2 h prior to use. After elution from the columns the probes were dried in a vacuum centrifuge and resuspended in 30 μl water (chromatographic grade). Electrophoresis was carried out on an automated ABI Prism 3100 sequencer under the following conditions: Dye Set: E; Mobility file: DT3100POP6{BD}v2.mob; Run Module: RapidSeq36_POP6DefaultModule; Analysis Module: BC-3100POP6RR_SeqOffFtOff.saz. Sequencing data were analyzed by BLASTN homology search on SGD (http://seq.yeastgenome.org/cgi-bin/SGD/nph-blast2sgd).
Results
Construction of a strain with inducible Ty-IN expression
To generate S. cerevisiae strain RSY12 Gal-TyIN, a cassette containing the integrase gene under control of an inducible Gal1-10 promoter was inserted in the genomic LYS2 locus. The integrase ORF was obtained by PCR from a plasmid generously provided by D. Garfinkel. Basically, the resulting protein corresponds to the one used by Garfinkel et al. for in vitro integration assays (Moore et al. 1995, 1998; Moore and Garfinkel 1994, 2000; Wilhelm and Wilhelm 2006), except that several N-terminal amino acids are lacking which had been introduced by these authors to facilitate protein expression and purification. By switching the carbon source of the growth medium from glucose to galactose, induction of Ty integrase expression is achieved in this strain, as was confirmed by Northern blot analysis (Fig. 1). It should be noted that even under repressing conditions integrase mRNA is readily detectable in this strain (Fig. 1), while it is not detectable in the parental, non-transformed strain (data not shown), supporting the observation that the Gal1-10 promoter is leaky in some strain backgrounds (e.g., Bond et al. 2001; Minic et al. 2005).
Fig. 1.
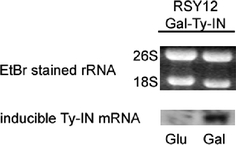
Northern Blot analysis of TyIN mRNA level after growth of strain RSY12 Gal-TyIN in SC-glucose (left) or SC-galactose (right). Ethidium bromide-staining of 18S and 26S rRNA served as a loading control
Growth characteristics
Yeast cells which have to adapt their metabolism to a new carbon source (e.g., switching from glucose to galactose) undergo a lag phase of 2–3 h before they continue to grow exponentially. After this lag phase the division times generally are similar for wild-type cells growing in SC-glucose or SC-galactose. In the case of RSY12, the doubling time is almost identical in both media, with 2.6 and 2.7 h, respectively (Fig. 2). The doubling time is slightly longer (3.0 h) for strain RSY12 Gal-TyIN grown in glucose medium. A severe effect is observed when strain RSY12 Gal-TyIN is grown in galactose medium, as the doubling time has almost doubled (5.5 h) as compared to wild type.
Fig. 2.
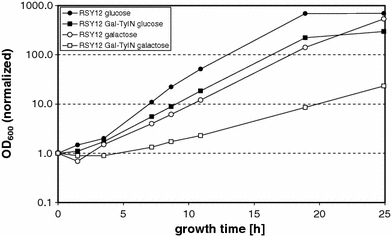
Growth in SC-glucose and SC-galactose of strains RSY12 and RSY12 Gal-TyIN after inoculation of logarithmic phase cells
Reduced growth of integrase-overexpressing strains is accompanied by an accumulation of cells without buds, which reflect G0 or G1 phase cells (Fig. 3). When analyzed 8.5 h after inoculation of stationary cells, the proportion of un-budded cells is less than 10% for RSY12 cells growing in glucose, as well as in galactose-containing medium (6.0 and 8.3%). The frequency of un-budded cells in strain RSY12 Gal-TyIN is higher (15.4%) under repressing conditions (glucose medium) and considerably higher (37.6%) after induction of the Ty1 integrase on galactose medium.
Fig. 3.
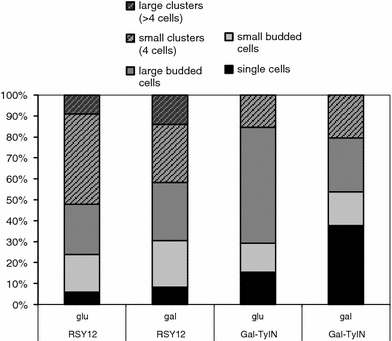
Cell morphology 8.5 h after transfer of stationary phase cells of the strains RSY12 and RSY12 Gal-TyIN to SC-glucose or SC-galactose, respectively
Frequency of non-homologous integration upon overexpression of TyIN
To investigate whether overexpression of Ty integrase affects the frequency of non-homologous integration (NHI) of DNA fragments, cells, which lack the entire URA3 ORF, were transformed with a PCR fragment encompassing the URA3 ORF. The integration frequency was determined as the number of Ura+ transformants, normalized with respect to the number of transformants obtained in a parallel control transformation with circular plasmid YEplac 195. This normalization step accounts for variations in transformation efficiency. In the first round of experiments, URA3 cassettes were used which terminated in 5′-CA-3′. This terminal dinucleotide is present on natural Ty1 elements and has been described as critical for Ty1 integrase-mediated integration of Ty1 elements in vivo (Sharon et al. 1994). In strain RSY12 the frequency of NHI is moderately reduced when the cells were cultivated in SC-galactose prior to transformation 60% of the value obtained in SC-glucose (Fig. 4). Strain RSY12 Gal-TyIN exhibits an almost sixfold increased frequency of NHI even when grown on SC-glucose, which probably is due to leakiness of the Gal promoter. Further induction of Ty-IN expression by growth in SC-galactose lead to a 23-fold increase of the NHI frequency, as compared to strain RSY12 grown on galactose. Thus, increased expression of Ty-IN correlates with increased frequency of NHI of fragments terminating in 5′-CA-3′.
Fig. 4.
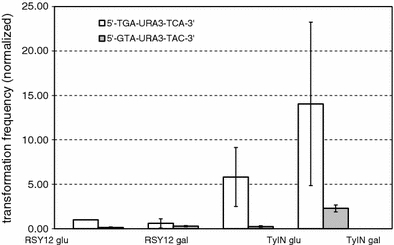
Frequency of non-homologous insertion of PCR fragments terminating in 5′-TG-CA-3′ (white bars) or 5′-GT-AC-3′ (grey bars) in strains RSY12 and RSY12 Gal-TyIN after 6 h incubation in SC-glucose or SC-galactose, respectively, prior to transformation. Transformation frequency per microgram DNA was normalized with respect to parallel transformations with a circular control plasmid to account for variations in transformation efficiency
To test the influence of the terminal dinucleotide, we performed transformation experiments with URA3 fragments that terminated in 5′-AC-3′ instead of 5′-CA-3′ (Fig. 4). This terminal sequence was found to reduce Ty1 integrase-mediated integration more than 100-fold in previous in vitro integration experiments (Moore and Garfinkel 2000). Generally, with the fragment terminating in 5′-AC-3′ the NHI frequencies were considerably reduced as compared to the fragment terminating in 5′-CA-3′. Overexpression of TyIN resulted, however, also in a strong enhancement of NHI events (15-fold, as compared to RSY12) which was still only about 16% of the frequency with the fragment terminating in 5′-CA-3′. A possible explanation for the enhancement with the wrong dinucleotide ending AC is given in the discussion.
Sequence analysis of NHI junctions
The mechanism leading to NHI can be inferred from sequence analysis of integration sites. For example, features such as 5 bp duplication of the sequence flanking the integration site would strongly suggest involvement of Ty1 integrase in the integration process, as would integration into known Ty1 integration hotspots. By direct genomic sequencing, we analyzed six integration events obtained by transforming strain RSY12 with a fragment terminating in 5′-CA-3′. Three events were obtained from cells grown prior to transformation in SC-glucose and SC-galactose, respectively (Fig. 5; Table 1). Characteristics from four integration events (MK03, MK68, MK69, MK70) were comparable to NHI events in wild-type cells as seen in earlier work (Schiestl and Petes 1991; Schiestl et al. 1993, 1994; Zhu and Schiestl 1996; Kiechle et al. 2002). For example, in MK68 and MK70, the URA3 fragments were found joined to non-contiguous stretches of mitochondrial DNA. In MK03 and MK69, insertion may have been facilitated by a 1 bp microhomology. Insertion of material of unknown origin was observed in MK68 (2 bp) and MK69 (7 bp). In contrast to these events, integration in clone MK04 exhibited clear foot prints of an event mediated by Ty1 integrase: a 5 bp duplication of the target site was seen, and the insertion occurred upstream of a tRNA gene (Table 1), in the vicinity of a solo LTR. This is the first time that hallmarks of an integrase-mediated event were observed in strain RSY12 after analysis of 104 events (mainly resulting from transformation of linearized plasmids and not having the blunt ends terminating in the integrase’s favourite dinucleotide of the currently used fragments) during the past 15 years. We found a few events that led to target site duplications of 1, 2 or 3 bp in those previous target sites, but never to 5 bp duplication (Schiestl and Petes 1991; Schiestl et al. 1993, 1994; Zhu and Schiestl 1996; Manivasakam and Schiestl 1998; Kiechle et al. 2002). Less clear is the situation in clone MK02, where the extent of target site duplication (5 or 7 bp) could not be determined unambiguously because of sequence homology. The sequence is compatible with a standard Ty integrase-mediated reaction, i.e., a staggered cut leaving a 5′ 5 bp overhang (GGATC) joined to a full-length blunt PCR fragment. Alternatively, in case of a 7 bp overhang, processing of the terminal dinucleotide of the PCR fragment would be required. The integration site lies in a region flanked by the promoters of two genes, which are transcribed divergently by RNA polymerase II, in a region not representing a clear Ty1 hotspot.
Fig. 5.
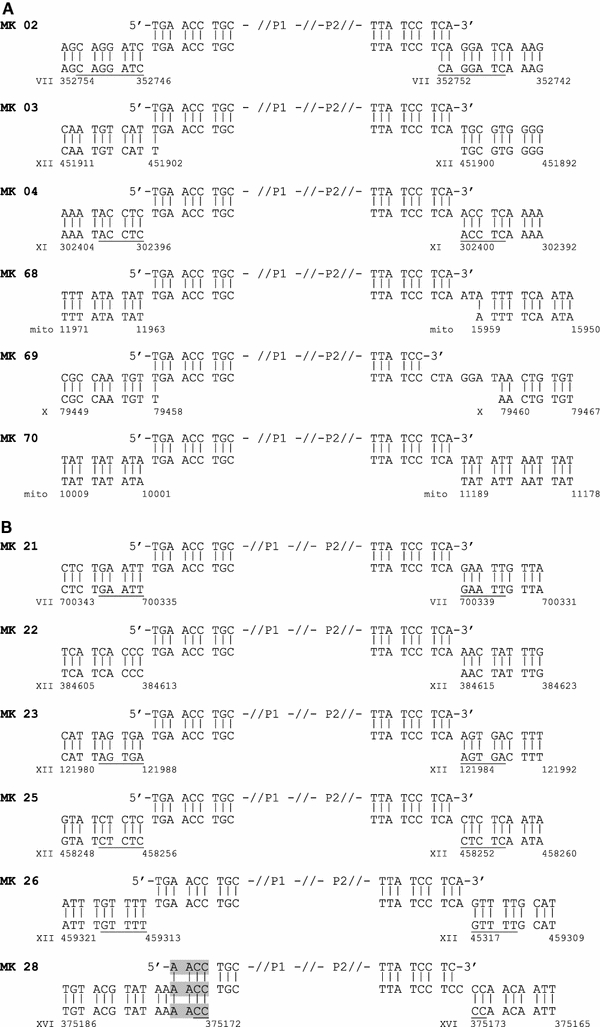
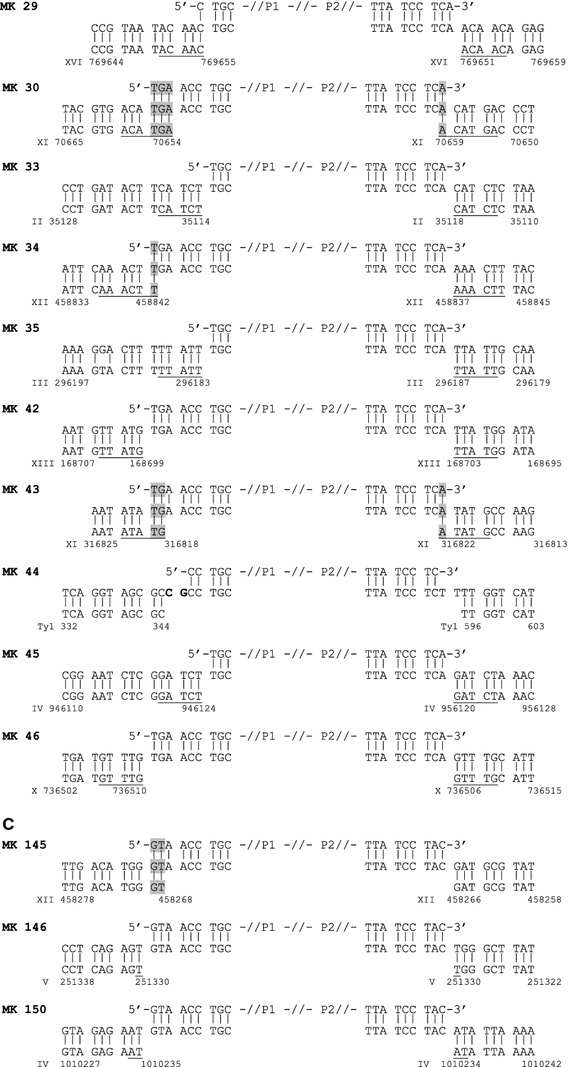
Sequence of insertion junctions after transformation of RSY12 with fragments terminating in 5′-TG-CA-3′ (a), RSY12 Gal-Ty-IN with fragments terminating in 5′-TG-CA-3′ (b), and RSY12 Gal-Ty-IN with fragments terminating in 5′-GT-AC-3′ (c). Sequences determined by direct genomic sequences (middle row) are depicted in 5′-to-3′ orientation and aligned with the sequence of the PCR fragment (top row) and the chromosomal sequence (bottom row). Sequence coordinates refer to SGD genomic coordinates, except for MK44, where coordinates refer to a standard Ty1 sequence. Target site duplications are underlined, microhomology regions are shaded. Bold nucleotides in MK44 depict the last two nucleotides of a 12 bp tRNA complementary sequence (see text for details)
Table 1.
Characterization of insertion events
| Clone | Strain/treatment/terminal bp | Insertion type | Location |
|---|---|---|---|
| MK02 | RSY12 glucose-CA | 5–7 bp overlap + MH | Upstream of Pol II-transcr. genes |
| MK03 | RSY12 glucose-CA | Other | |
| MK04 | RSY12 glucose-CA | 5 bp overlap | 165 bp upstream of tRNA gene |
| MK68 | RSY12 galactose-CA | Other | |
| MK69 | RSY12 galactose-CA | Other | |
| MK70 | RSY12 galactose-CA | Other | |
| MK42 | TyIN glucose-CA | 5 bp overlap | 94 bp upstream of tRNA gene |
| MK43 | TyIN glucose-CA | 5–7 bp overlap + MH | >3 kb upstream of tRNA gene |
| MK44 | TyIN glucose -CA | Composite element | |
| MK45 | TyIN glucose-CA | 5 bp overlap | 184 bp upstr. of tRNA gene, in Ty3 LTR |
| MK46 | TyIN glucose-CA | 5 bp overlap | Into Pol II-transcr. gene |
| MK21 | TyIN galactose-CA | 5 bp overlap | 342 bp upstream of tRNA gene |
| MK22 | TyIN galactose-CA | Other | |
| MK23 | TyIN galactose-CA | 5 bp overlap | 287 bp upstream of tRNA gene |
| MK25 | TyIN galactose-CA | 5 bp overlap | rDNA, 5′ ETS |
| MK26 | TyIN galactose-CA | 5 bp overlap | rDNA, 35 bp upstream of 5S rDNA gene |
| MK28 | TyIN galactose-CA | Other | |
| MK29 | TyIN galactose-CA | 5 bp overlap | 353 bp upstr. of tRNA gene, in Ty3 LTR |
| MK30 | TyIN galactose-CA | 5–7 bp overlap + MH | >3 kb upstream of tRNA gene |
| MK33 | TyIN galactose-CA | 5 bp overlap | 1,279 bp upstr. of tRNA gene, in Ty2 |
| MK34 | TyIN galactose-CA | 5-7 bp overlap + MH | rDNA, 836 bp upstr. of 5S rDNA gene |
| MK35 | TyIN galactose-CA | 5 bp overlap | in Ty1 LTR |
| MK145 | TyIN galactose-AC | Other | |
| MK146 | TyIN galactose-AC | Other | |
| MK150 | TyIN galactose-AC | Other |
For strain RSY12 Gal-TyIN the integration sites of 16 clones were sequenced after transformation with URA3 fragments terminating in 5′-CA-3′. Five transformants were derived from cells cultivated in SC-glucose (repressing conditions) and 11 transformants were grown in SC-galactose (inducing conditions) prior to transformation. Only two events (MK22 and MK 28) exhibited hallmarks of normal NHI events (Fig. 5). Among the remaining 14 transformants, unambiguous 5 bp duplication was detected in 10 integration events (MK21, MK23, MK25, MK26, MK29, MK33, MK35, MK42, MK45, MK46; Fig. 5). In six of these events, the ends of the transformed URA3 fragment remained intact, in three events, the 5′ ends were processed by removal of 6 bp; these processed ends terminate also in 5′-CA-3′. Most likely, the 5′ end of the remaining clone, MK29, was processed in the same manner, but due to sequence overlap this cannot be shown unequivocally.
In most of the events involving a 5 bp target site duplication, known Ty1 target site preferences were also evident: In five clones (MK21, MK23, MK29, MK42, MK45; see Table 1) integration occurred within a well-described 700 bp window upstream of tRNA genes (Ji et al. 1993; Devine and Boeke 1996; Bolton and Boeke 2003; Bachman et al. 2004). In two clones integration occurred within the rDNA cluster, with the integration site of MK25 falling into the non-coding 5′ external transcribed spacer (ETS) region and the integration site of MK26 falling into the non-transcribed spacer upstream of the 5S rDNA gene. Interestingly, the integration site of MK26 coincides with a site observed in earlier experiments after transformation of plasmid DNA in a yku70 mutant (19; clone MK19). In two further events, integration occurred within pre-existing Ty2 (MK33) or solo LTR (MK35) elements. Only in the case of clone MK46 integration did not occur at a known Ty1 hotspot. Taken together, the data strongly suggest that the ten events exhibiting 5 bp duplication were mediated by Ty1 integrase. The sequence composition of the target sites of all 11 events exhibiting clear-cut 5 bp duplication (including event MK04; Table 2) agrees well with the anti-consensus site proposed by Ji et al. (1993), further corroborating that integration of the URA3 fragment here followed the rules for Ty1-mediated events.
Table 2.
Distribution of nucleotides at positions 1–5 of unambiguous target sites (i.e. target sites with 5 bp duplication)
| Position | A | T | C | G | Consensus (Ji et al. 1993) |
|---|---|---|---|---|---|
| 1 | 3 | 2 | 2 | 4 | GAC |
| 2 | 3 | 5 | 2 | 1 | – |
| 3 | 4 | 5 | 2 | 0 | AT |
| 4 | 1 | 7 | 2 | 1 | – |
| 5 | 1 | 5 | 3 | 2 | CTG |
For comparison the consensus determined by Ji et al. (1993) is indicated in the right column
In another three events (MK30, MK34, MK43), the extent of the target site duplication could not be determined unequivocally because of sequence overlaps on one or both junctions. In the case of MK34, junctions and target site are compatible with a Ty1 integrase-mediated reaction (duplication of AAACT and ligation to full-length PCR fragment; target in non-transcribed spacer upstream of the 5S rDNA gene). In the case of MK30 and MK43, no known hotspot was found in the vicinity of the integration sites. If insertions were initiated by 5 bp staggered cuts, processing of the overhangs or of the PCR fragment 3′ ends would have been required for end joining to occur. It is possible that this kind of junction hints at an un-coupling of staggered cutting and joining in at least some reactions, in which case DNA fragments may be inserted by microhomology-mediated end joining.
In contrast, clear indications for an involvement of Ty metabolism in the integration reaction could be demonstrated for event MK44. Here, the inserted URA3 fragment is flanked by non-contiguous stretches of Ty1 sequence. Similar events were frequently observed in earlier work where the non-homologous integration of linearized plasmids was studied in yku70 mutants (Kiechle et al. 2000). In a large proportion of these former so-called composite elements, the structure of the junctions suggested that plasmid and Ty sequences were joined by template switching and/or erroneous priming mechanisms. In particular, the apparent involvement of pausing Ty1 replication intermediates (Lauermann and Boeke 1997; Mules et al. 1998) in several events corroborated this interpretation. This holds also for the insertion in MK44 presented here, where the P1 end of the URA3 fragment is joined, via a 2 bp (CG; bold in Fig. 5) insertion, to the 3′ end of a primer binding site (PBS). Replication intermediates terminating in the PBS sequence plus the dinucleotide CG (thus terminating in a 12 bp tRNA complementary sequence) are known to accumulate in the cell, presumably because they are dead-end products of the Ty1 replication process (Lauermann and Boeke 1997).
Since URA3 fragments terminating in 5′-CA-3′ apparently are readily used as Ty integrase substrates, the question arises in what manner insertion events are affected by alteration of the terminal dinucleotide to 5′-AC-3′. Unfortunately, the sequence of both junctions could only be determined in three clones (Fig. 5). In MK146 and MK150, a 1 bp, respectively, 2 bp target site duplication was observed, while in MK145 insertion may have been facilitated by a sequence overlap at one junction site. We did not observe processing of the fragment ends yielding integrase-compatible ends, although this kind of processing was frequently observed in yku70 mutants in our earlier study (Kiechle et al. 2000).
Discussion
In this work we provide evidence that Ty1 integrase under certain conditions readily accepts transformed DNA fragments that do not bear similarity to Ty cDNA except for the terminal dinucleotide, 5′-TG-CA-3′, as substrates for integration. This conclusion is mainly based on the observation that upon overexpression of Ty1 integrase the frequency of non-homologous insertion of these DNA fragments is enhanced and that the majority of integration events exhibit clear hallmarks of Ty integrase-mediated events such as 5 bp target site duplication and compatibility of observed target sites with both known target site preferences and target site sequence consensus of Ty1 integrase. Thus, upon overexpression of integrase alone the substrate specificity appears to be less stringent than it was described for overexpression of complete Ty1 elements (Sharon et al. 1994). It should be noted, however, that the sequence requirements for Ty integrase-mediated integration in vivo have not been studied in great detail, so far. Under in vitro conditions, using purified integrase or VLPs and purified substrate molecules, the substrate specificity generally appeared rather relaxed (Eichinger and Boeke 1990; Braiterman and Boeke 1994; Moore et al. 1995; Moore and Garfinkel 2000).
Overexpression of TyIN resulted, however, also in a strong enhancement of NHI events (15-fold with the dinucleotide ending in 5′-AC-3′, as compared to RSY12) which was still only about 16% of the frequency with the fragment terminating in 5′-CA-3′. A possible explanation for the events terminating in 5′-AC-3′ is as follows. We have previously shown that DNA double-strand breaks cause an increase of microhomology-mediated recombination events in trans (Chan et al. 2007). Since we were not able to construct the strain overexpressing Ty in the Ku mutant background, it seems likely that the expression of Ty integrase causes DNA damage that induces integration of DNA via the induced microhomology-mediated recombination pathway.
At first glance the data presented here remind of our earlier observations that in yku70 mutants transformed linearized plasmids are frequently inserted into the genome in a Ty-dependent manner (Kiechle et al. 2000). There are, however, clear differences in the frequency distribution of Ty-dependent insertion types in both studies. In yku70 mutants 11/14 Ty-mediated events led to insertion of so-called composite elements in which the transformed DNA was first joined to Ty1-derived sequences. This type of events, which most likely is explained by erroneous priming and strand-transfer events, did occur only once in cells overexpressing Ty1 integrase. In contrast, while insertion of complete (or slightly processed) fragments with Ty-specific target site duplication and preferences occurred in at least 10/16 events upon overexpression of integrase, it occurred less frequently (3/14) in yku70 mutants. This difference is highly significant (p ≤ 0.001).
There are two potential reasons for this difference: first, it may be due to the differences in substrate DNA used in the two studies: here, blunt-ended PCR fragments were used, which encompass a 1.1 kbp genomic HindIII-fragment including the URA3 locus flanked by some ten basepairs of pUC19-derived sequences. In the earlier study, however, BglII-linearized plasmid pM151 was used, which is about 3.8 kbp long and terminates in 5′ single-stranded overhangs. Second, it may be that integrase overexpression and Ku70 deficiency affect substrate specificity in different ways.
The Ku heterodimer, consisting of a Ku70 and a Ku80 subunit, is a DNA end-binding factor which plays a central role in the repair of DNA double-strand breaks by non-homologous end joining; in addition, it is involved in telomere stabilization (for reviews see Friedl 2002; Downs and Jackson 2004). In Ku-deficient yeast cells, increased 5′–3′ single-strand degradation is known to occur (Lee et al. 1998; Tomita et al. 2003), and indeed end-processing of the plasmid was frequently seen in Ku-deficient cells (Kiechle et al. 2000). In some of the composite elements, extensive microhomology was present at the plasmid-Ty junctions, and it seems reasonable to assume that exposure of single-stranded regions in the course of end-processing was instrumental in the formation of the junctions. The frequent occurrence of composite elements may also be taken to indicate that in the absence of Ku the VLPs are more accessible for non-Ty-DNA. With a VLP pore diameter of about 2.5 nm (Roth 2000), naked DNA fragments should be able to enter the VLPs, while DNA associated with proteins larger than about 30 kDa would be blocked. It is known that the yeast Ku heterodimers bind efficiently to DNA ends in vitro and in the nucleus (Feldmann and Winnacker 1993; Frank-Vaillant and Marcand 2002). While it has, to our knowledge, not been tested whether Ku heterodimer binds DNA ends in the cytoplasm, it is clear that heterodimer formation does occur, at least in part, in the cytoplasm (Koike and Koike 2005). Thus, it is possible that the presence of end-bound Ku is a major factor in preventing non-Ty-DNA from entering VLPs. In line with this model, it is interesting to note that Ku protein has been found associated with VLP particles (Downs and Jackson 1999). Composite elements, which require that the DNA fragments enter the VLPs, are thus expected to form easily in the absence of Ku, but rarely in the strains overexpressing integrase.
Combining the data on integrase overexpression and on Ku-deficiency, the following model emerges: In analogy to the situation in human immunodeficiency virus (HIV) it is assumed that proteolytic processing of the gag–pol polyprotein and thus liberation of integrase occurs in VLPs (Wilhelm et al. 2005); thus under normal conditions, integrase molecules should only be found within VLPs until they enter the nucleus. Integrase and freshly made Ty cDNA meet within the VLPs, where they form a complex, the PIC. This complex formation may depend on or strongly be favoured by the presence of the terminal dinucleotide 5′-GT-CA-3′. Integrase and associated cDNA then cross the nuclear membrane with the help of the integrase’s NLS sequence and presumably remain associated until eventual integration of the cDNA into the genomic DNA. Under these conditions, substrate specificity is mainly ensured by the fact that other DNA fragments do not come into contact with the integrase molecules. This specificity is, however, not absolute, as is seen in MK04 (and presumably MK02). The fact that integrase-mediated integration in wild-type cells was observed in this study, but not in any of the previous studies, may be explained by the present use of blunt-ended DNA terminating in integrase’s favourite dinucleotides.
The situation differs when integrase is overexpressed, since in this situation most likely a high proportion of integrase molecules will not be present within VLPs. Free integrase molecules may then either associate with DNA fragments they encounter in the cytoplasm and cross the nuclear membrane in a complex, or both integrases and DNA fragments may migrate to the nucleus independently and meet there. Again, association of integrase and DNA fragment may depend on the presence of the correct terminal dinucleotide, but apart from this constraint, any DNA fragment may be a suitable substrate. Composite elements, however, will not form easily, since the accessibility of VLPs is not affected by the integrase overexpression.
To test the above model, it would be interesting to investigate the distribution of event types in Ku-deficient cells overexpressing integrase. In spite of several attempts we did, however, not succeed in stably obtaining the appropriate strains. This may hint at a severe growth defect, which may be related to the moderate growth defect observed here in Ku-proficient cells upon overexpression of integrase. The reasons for the growth defect are not clear at present. While proper integrase-mediated integration is a concerted transesterification reaction that does not lead to an open break in the genomic DNA (reviewed by Wilhelm et al. 2005), unspecific endonucleolytic activity of integrase cannot be excluded. In this context it is interesting to note that expression of HIV or Moloney murine leukemia virus (M-MuLV) in yeast confers a lethal phenotype, in particular in strains compromised in the repair of DNA double-strand breaks (Caumont et al. 1996; Parissi et al. 2003; Vera et al. 2005). Detrimental effects of overexpression of Ty1 integrase have, to our knowledge, not been described before.
Our results raise the possibility of using integrase expression to stimulate DNA fragment insertion in biotechnical applications. Further research will be necessary to clarify the suitability of such an approach.
Acknowledgments
This research was supported by Grant KI 796/1-1 from the Deutsche Forschungsgemeinschaft to MK and Grant No 1 RO1 CA82473-01 from the National Cancer Institute, NIH, as well as a Stein-Oppenheimer Research award to RHS.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The contributions of A. A. Friedl and M. Kiechle are considered equal.
References
- Bachman N, Eby Y, Boeke JD. Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res. 2004;14:1232–1247. doi: 10.1101/gr.2052904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton EC, Boeke JD. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 2003;13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AT, Mangus DA, He F, Jacobson A. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol Cell Biol. 2001;21:7366–7379. doi: 10.1128/MCB.21.21.7366-7379.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braiterman LT, Boeke JD. Ty1 in vitro integration: effects of mutations in cis and trans. Mol Cell Biol. 1994;14:5731–5740. doi: 10.1128/mcb.14.9.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caumont AB, Jamieson GA, Pichuantes S, Nguyen AT, Litvak S, Dupont C. Expression of functional HIV-1 integrase in the yeast Saccharomyces cerevisiae leads to the emergence of a lethal phenotype: potential use for inhibitor screening. Curr Genet. 1996;29:503–510. doi: 10.1007/BF02426953. [DOI] [PubMed] [Google Scholar]
- Chan CY, Kiechle M, Manivasakam P, Schiestl RH. Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:5051–5059. doi: 10.1093/nar/gkm442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine SE, Boeke JD. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. Involvement of DNA end-binding protein Ku in Ty element retrotransposition. Mol Cell Biol. 1999;19:6260–6268. doi: 10.1128/mcb.19.9.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- Eichinger DJ, Boeke JD. A specific terminal structure is required for Ty1 transposition. Genes Dev. 1990;4:324–330. doi: 10.1101/gad.4.3.324. [DOI] [PubMed] [Google Scholar]
- Feldmann H, Winnacker EL. A putative homologue of the human autoantigen Ku from Saccharomyces cerevisiae. J Biol Chem. 1993;268:12895–12900. [PubMed] [Google Scholar]
- Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10:1189–1199. doi: 10.1016/S1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- Friedl AA. Ku and the Stability of the Genome. J Biomed Biotechnol. 2002;2:61–65. doi: 10.1155/S1110724302201035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli A, Schiestl RH. Effects of DNA double-strand and single-strand breaks on intrachromosomal recombination events in cell-cycle-arrested yeast cells. Genetics. 1998;149:1235–1250. doi: 10.1093/genetics/149.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel DJ, Hedge AM, Youngren SD, Copeland TD. Proteolytic processing of pol TYB proteins from the yeast retrotransposon Ty1. J Virol. 1991;65:4573–4581. doi: 10.1128/jvi.65.9.4573-4581.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation on intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horecka J, Jigami Y. Identifying tagged transposon insertion sites in yeast by direct genomic sequencing. Yeast. 2000;16:967–970. doi: 10.1002/1097-0061(200007)16:10<967::AID-YEA597>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ji H, Moore DP, Blomberg MA, Braiterman LT, Voytas DF, Natsoulis G, Boeke JD. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell. 1993;73:1007–1018. doi: 10.1016/0092-8674(93)90278-X. [DOI] [PubMed] [Google Scholar]
- Kenna MA, Brachmann CB, Devine SE, Boeke JD. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiechle M, Friedl AA, Manivasakam P, Eckardt-Schupp F, Schiestl RH. DNA integration by Ty integrase in yku70 mutant Saccharomyces cerevisiae cells. Mol Cell Biol. 2000;20:8836–8844. doi: 10.1128/MCB.20.23.8836-8844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiechle M, Manivasakam P, Eckardt-Schupp F, Schiestl RH, Friedl AA. Promoter-trapping in Saccharomyces cerevisiae by radiation-assisted fragment insertion. Nucleic Acids Res. 2002;30:e136. doi: 10.1093/nar/gnf136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Koike M, Koike A. The Ku70-binding site of Ku80 is required for the stabilization of Ku70 in the cytoplasm, for the nuclear translocation of Ku80, and for Ku80-dependent DNA repair. Exp Cell Res. 2005;305:266–276. doi: 10.1016/j.yexcr.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Lauermann V, Boeke JD. Plus-strand strong-stop DNA transfer in yeast Ty retrotransposons. EMBO J. 1997;16:6603–6612. doi: 10.1093/emboj/16.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/S0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lesage P, Todeschini AL. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- Manivasakam P, Schiestl RH. Nonhomologous end-joining during restriction enzyme-mediated DNA integration in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1736–1745. doi: 10.1128/mcb.18.3.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkulov GV, Lawler JF, Eby Y, Boeke JD. Ty1 proteolytic cleavage sites are required for transposition: all sites are not created equal. J Virol. 2001;75:638–644. doi: 10.1128/JVI.75.2.638-644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic J, Persuy MA, Godel E, Aioun J, Connerton I, Salesse R, Pajot-Augy E. Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J. 2005;272:524–537. doi: 10.1111/j.1742-4658.2004.04494.x. [DOI] [PubMed] [Google Scholar]
- Moore SP, Garfinkel DJ. Expression and partial purification of enzymatically active recombinant Ty1 integrase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:1843–1847. doi: 10.1073/pnas.91.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SP, Garfinkel DJ. Correct integration of model substrates by Ty1 integrase. J Virol. 2000;74:11522–11530. doi: 10.1128/JVI.74.24.11522-11530.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SP, Powers M, Garfinkel DJ. Substrate specificity of Ty1 integrase. J Virol. 1995;69:4683–4692. doi: 10.1128/jvi.69.8.4683-4692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SP, Rinckel LA, Garfinkel DJ. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mules EH, Uzun O, Gabriel A. In vivo Ty1 reverse transcription can generate replication intermediates with untidy ends. J Virol. 1998;72:6490–6503. doi: 10.1128/jvi.72.8.6490-6503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parissi V, Caumont A, de Soultrait VR, Desjobert C, Calmels C, Fournier M, Gourgue G, Bonneu M, Tarrago-Litvak L, Litvak S. The lethal phenotype observed after HIV-1 integrase expression in yeast cells is related to DNA repair and recombination events. Gene. 2003;322:157–168. doi: 10.1016/j.gene.2003.08.020. [DOI] [PubMed] [Google Scholar]
- Roth JF. The yeast Ty virus-like particles. Yeast. 2000;16:785–795. doi: 10.1002/1097-0061(20000630)16:9<785::AID-YEA550>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Petes TD. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Dominska M, Petes TD. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial sequences. Mol Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Zhu J, Petes TD. Effect of mutations in genes affecting homologous recombination on restriction-enzyme mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Burkett TJ, Garfinkel DJ. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14:6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T, Yoshinaga K, Ueno M. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Cell Biol. 2003;23:5186–5197. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera J, Parissi V, Garcia A, Zuniga R, Andreola ML, Caumont-Sarcos A, Tarrago-Litvak L, Leon O. Yeast system as a model to study Moloney murine leukemia virus integrase: expression, mutagenesis and search for eukaryotic partners. J Gen Virol. 2005;86:2481–2488. doi: 10.1099/vir.0.81006-0. [DOI] [PubMed] [Google Scholar]
- Wilhelm M, Wilhelm FX. Cooperation between reverse transcriptase and integrase during reverse transcription and formation of the preintegrative complex of Ty1. Eukaryotic Cell. 2006;5:1760–1769. doi: 10.1128/EC.00159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Heyman T, Boutabout M, Wilhelm FX. A sequence immediately upstream of the plus-strand primer is essential for plus-strand synthesis of the Saccharomyces cerevisiae Ty1 retrotransposon. Nucleic Acids Res. 1999;27:4547–4552. doi: 10.1093/nar/27.23.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm FX, Wilhelm M, Gabriel A. Reverse transcriptase and integrase of the Saccharomyces cerevisiae Ty1 element. Cytogenet Genome Res. 2005;110:269–287. doi: 10.1159/000084960. [DOI] [PubMed] [Google Scholar]
- Yap WY, Schiestl RH. Nature of abortive transformation in Saccharomyces cerevisiae. Curr Genet. 1995;28:517–520. doi: 10.1007/BF00518163. [DOI] [PubMed] [Google Scholar]
- Zhu J, Schiestl RH. Topoisomerase I involvement in illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]


