Abstract
The Saccharomyces cerevisiae protein Nhp6A is a model for the abundant and multifunctional high-mobility group B (HMGB) family of chromatin-associated proteins. Nhp6A binds DNA in vitro without sequence specificity and bends DNA sharply, but its role in chromosome biology is poorly understood. We show by whole-genome chromatin immunoprecipitation (ChIP) and high-resolution whole-genome tiling arrays (ChIP–chip) that Nhp6A is localized to specific regions of chromosomes that include ∼23% of RNA polymerase II promoters. Nhp6A binding functions to stabilize nucleosomes, particularly at the transcription start site of these genes. Both genomic binding and transcript expression studies point to functionally related groups of genes that are bound specifically by Nhp6A and whose transcription is altered by the absence of Nhp6. Genomic analyses of Nhp6A mutants specifically defective in DNA bending reveal a critical role of DNA bending for stabilizing chromatin and coregulation of transcription but not for targeted binding by Nhp6A. We conclude that the chromatin environment, not DNA sequence recognition, localizes Nhp6A binding, and that Nhp6A stabilizes chromatin structure and coregulates transcription.
Keywords: HMGB proteins, chromatin structure, nucleosome dynamics, transcription, DNA bending
Decades of research have established an integral role for chromatin structure in regulating many essential cellular processes such as genome replication, compaction, gene transcription, and DNA repair. The defining unit of eukaryotic chromatin is the nucleosome, composed of the histone octamer and ∼150 base pairs (bp) of DNA. Extensive studies have examined the removal, replacement, exclusion, or chemical modification of nucleosomes (Lee et al. 2004, 2007; Liu et al. 2005; Dion et al. 2007; Mavrich et al. 2008). Variant histones are critical in establishing and maintaining important chromatin regions. For example, CENP-A (CenH3) in centromeric chromatin or testes-specific histone H2B (TH2B) in sperm chromatin define discrete chromatin regions. In addition to histones, several other protein families are crucial to genome structure and function. For example, 96% of sperm chromatin consists of highly basic protamines, with canonical histones covering only 4% of the sperm DNA (Balhorn et al. 2000). The high-mobility group (HMG) family of proteins represents another important component of chromatin. In mitotic cells, HMG proteins are the most abundant component of chromatin after histones, and the HMGB family constitutes the most abundant of the three distinct families of HMG proteins (Bustin 1999; Bianchi and Agresti 2005).
The defining feature of HMGB proteins is the ∼75-residue HMG box, an L-shaped three-α-helical fold that binds to the minor groove face of DNA and sharply bends DNA (Thomas and Travers 2001). HMGB proteins can be divided into two classes based on whether they bind DNA in a sequence-specific or sequence-independent manner (Bustin 1999). Sequence-specific HMGB proteins, such as members of the SOX and LEF1/TCF families, play critical roles in stem cell biology and development, respectively (Eastman and Grosschedl 1999; Guth and Wegner 2008). These cell type-restricted, low-abundance molecules operate as transcription factors by binding to distinct DNA motifs within gene regulatory regions (Agresti et al. 2003).
HMGB proteins that bind DNA in a sequence-independent manner, such as mammalian HMGB1, have been shown to function in the nucleus as chromatin architectural factors and as extranuclear components of a Toll-like receptor (TLR)-mediated innate immunity response (Thomas and Travers 2001; Agresti et al. 2003; Yanai et al. 2009). Recruitment of sequence-independent HMGB proteins can be mediated by protein–protein interactions with sequence-specific transcription factors such as nuclear hormone receptors, HOXD9, and p53 (Zappavigna et al. 1996; Boonyaratanakornkit et al. 1998; Jayaraman et al. 1998). HMGB proteins can also function as DNA chaperones to facilitate assembly of specific protein–DNA complexes and are not necessarily associated with the final complex (McKinney and Prives 2002; Mitsouras et al. 2002).
The most abundant Saccharomyces cerevisiae HMGB protein is Nhp6A, at 50,000–70,000 molecules per haploid cell (about one molecule for every one to two nucleosomes) (Paull et al. 1996). Study of this protein offers a tractable system in which to dissect molecular functions of HMGB proteins in the nucleus. Nhp6A is one of seven S. cerevisiae proteins containing an HMG box conferring sequence-independent DNA binding. The C-terminal 75 amino acid residues of the Nhp6A protein fold into a typical HMG-box structure that wedges into the DNA minor groove, and the highly basic N-terminal 16 amino acids wrap around to the major groove to confer unusually high-affinity DNA binding for an HMGB protein (see Fig. 5C, below; Yen et al. 1998; Allain et al. 1999; Masse et al. 2002). Insertion of two hydrophobic residues into the base pair stack, together with the concave DNA-binding surface, results in highly distorted DNA upon Nhp6A binding. A close paralog, Nhp6B, exhibits 87% amino acid sequence identity with Nhp6A but is expressed in lower amounts. Strains containing knockout mutations of both nhp6A and nhp6B (Δnhp6) are viable but grow at ∼40% of the rate of single-knockout or NHP6A/B strains (Costigan et al. 1994; Yen et al. 1998).
Figure 5.
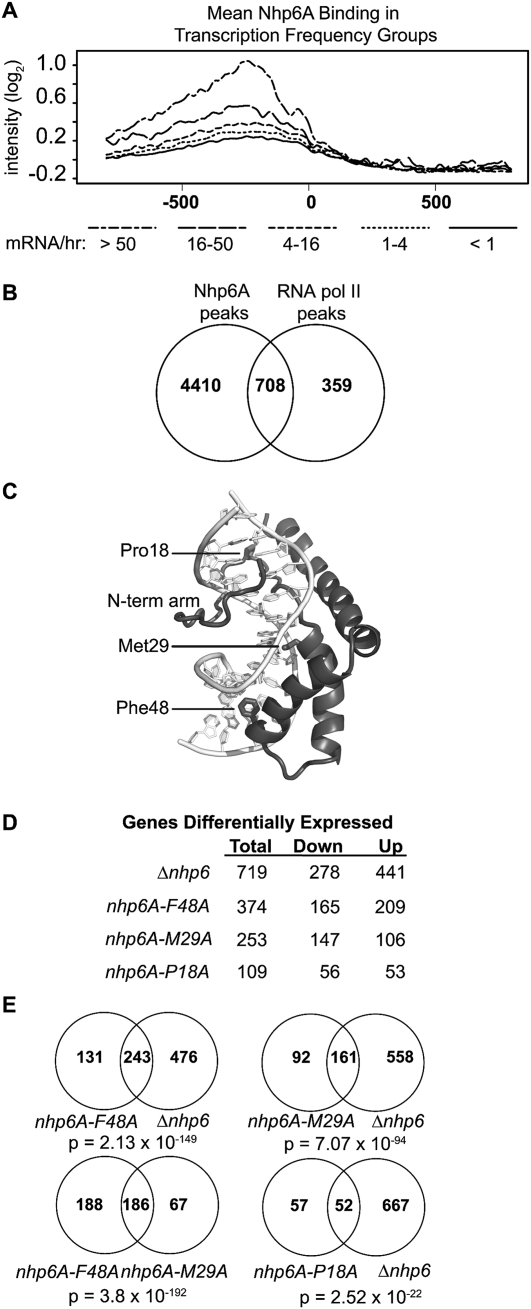
Transcription and Nhp6A DNA binding and bending. (A) Mean Nhp6A binding intensity in gene groups defined by transcriptional frequency (Holstege et al. 1998). The long/short dash line is Nhp6A binding in genes with >50 mRNAs per hour, the long dash represents 16–50 mRNAs per hour, the short dash indicates 4–16 mRNAs per hour the dotted line is one to four mRNAs per hour, and the solid line represents less than one mRNA per hour. (B) Venn diagram illustrating the overlap between Nhp6A-binding peaks and RNA Pol II-binding peaks (Bermejo et al. 2009). (C) Structure of the Nhp6A–DNA complex (PDB code 1J5N) highlighting the bending residues Met29 and Phe48 along with Pro18, which connects the N-terminal tail to the HMG box domain. (D) Number of genes differentially expressed by the loss of wild-type Nhp6A (Δnhp6) or by each of the Nhp6A mutants as compared with NHP6A Δnhp6B cells. (E) Venn diagrams of overlapping differentially expressed genes between genotypes. A hypergeometric test was used to calculate the significance of overlaps (Fury et al. 2006).
Previous studies demonstrating transcriptional induction defects in the absence of Nhp6A/B at RNA polymerase II (Pol II)-transcribed genes have been attributed to chromatin defects and direct or indirect interactions with activators, basal transcription factors, and chromatin remodeling complexes (Stillman 2010). Genetic studies have uncovered functional relationships between Nhp6 and the FACT, RSC, or Swi/Snf chromatin remodeling complexes (Brewster et al. 2001; Ruone et al. 2003; Biswas et al. 2004). In vitro experiments support direct collaborations between Nhp6 and Spt16/Pob3 (yFACT) and RSC (Formosa et al. 2001; Szerlong et al. 2003). Although a unifying mechanism for Nhp6A function remains elusive, a common theme within these studies is that Nhp6A interaction with chromatin or chromatin-modifying complexes leads to efficient transcription and optimal cell growth.
In this study, we use chromatin immunoprecipitation (ChIP) and high-resolution whole-genome tiling arrays (ChIP–chip) to identify which genomic regions Nhp6A binds in vivo, how Nhp6A interacts with chromatin in vivo, and how the loss of Nhp6 affects chromatin structure. We further relate the genomic binding patterns of Nhp6A with effects on global gene expression caused by loss of Nhp6. We show that Nhp6A binds to discrete genomic regions that are often promoters of functionally related gene clusters. Expression of genes within these clusters is often perturbed upon loss of Nhp6. Our in vivo and in vitro data indicate that Nhp6A targeting to specific loci is achieved through interaction with the chromatin environment and not through DNA sequence elements. We show that Nhp6A binding stabilizes nucleosomes in gene promoters and within 5′ gene-coding regions. Experiments with Nhp6A-bending mutants provide evidence that DNA bending is a key determinant for coregulation of transcription by Nhp6A. Furthermore, reduction of Nhp6A DNA-bending activity does not disrupt targeted binding but does disturb chromatin structure. The data provide strong support for an important role for Nhp6A in maintaining chromatin structure and as a coregulator of transcription through its strong DNA-bending activity. Our findings with the S. cerevisiae Nhp6A protein are likely to be broadly applicable to related HMGB chromatin proteins in metazoans.
Results
Nhp6A binds to discrete genomic regions
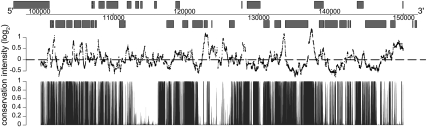
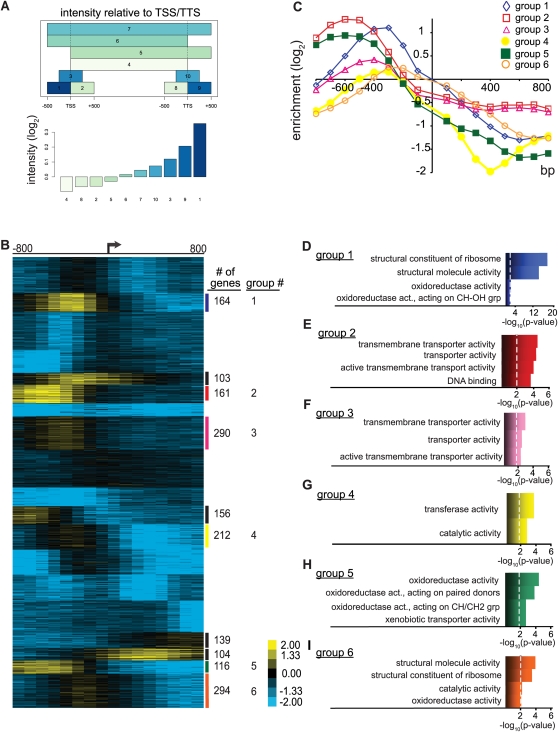
Previous studies demonstrated that Nhp6A binds linear naked DNA with relatively high affinity (KD 1–10 nM) in a sequence-independent manner, but it had not been determined how Nhp6A associates with DNA in the in vivo chromatin environment (Paull and Johnson 1995; Ruone et al. 2003). We performed ChIP using antibodies directed against Nhp6 in NHP6A Δnhp6B cells and hybridized the enriched DNA to high resolution (∼5 bp) Affymetrix tiling arrays (ChIP–chip) to identify Nhp6A-binding locations during exponential growth in rich media. Nhp6A-bound probes were mapped to genomic regions and viewed in the University of California at Santa Cruz (UCSC) Genome Browser (Fig. 1; see also Supplemental Figs. S7–S12). We qualitatively observed predominantly intergenic binding by Nhp6A. This observation was quantified by plotting mean Nhp6A-binding intensities in genomic segments surrounding the translation start site (TSS). Figure 2A shows that the highest average Nhp6A-binding intensities are located upstream of the TSS. To visualize Nhp6A binding 800 bp upstream of and downstream from TSSs for nondubious ORFs, we extracted and K-means-clustered mean Nhp6A-binding intensities. The resulting heat map (Fig. 2B) illustrates enrichment of Nhp6A at a large subset of promoters for protein-coding genes (∼1496). Additionally, Nhp6A binding is observed in the ORFs of ∼243 genes. In combination with the highly bound promoter cluster, this ORF cluster points to a distinct localization for Nhp6A within specific promoters and ORFs.
Figure 1.
Genome-wide binding of the yeast HMGB protein Nhp6A. A representative 50-kb segment of S. cerevisiae chromosome III displaying Nhp6A-binding intensity (log2). Genes (grey boxes) on the Watson strand within this region are above the black line, and genes on the Crick strand are below the black chromosome coordinate line. Nucleotide conservation across seven yeast species is denoted at the bottom.
Figure 2.
Nhp6A binds to discrete gene clusters. (A) Mean Nhp6A-binding intensity relative to TSSs, ORFs, and TTSs (transcription termination sites). Nhp6A binding was averaged over regions surrounding ORFs as depicted in the top panel (see the Materials and Methods). The bottom panel gives the Nhp6A signal intensity scale for each region. (B) K-means clusters (30) of Nhp6A binding flanking TSSs (arrow). Each column represents averaged probe fluorescence intensity over 100 bp as compared with the input control. Each row represents an ORF as defined in the S. cerevisiae Genome Database. Data for all nondubious ORFs were clustered together, but only the region representing Nhp6A clusters along with several unbound clusters is shown. The complete heat map is given in Supplemental Figure S1A. The number of genes within each cluster is given on the right. (C) Line graph of binding data for each GO cluster listed in D–I, displaying distinct peak levels and locations relative to the TSS. (D–I) GO function analysis of a selection of Nhp6A-binding clusters. The top four significantly enriched GO terms are adjacent to the bar plot representing −log10 (P-value), with the dashed line representing P = 0.01.
Each of the 10 Nhp6A-binding clusters was subjected to Gene Ontology (GO) function analysis using the Saccharomyces Genome Database GO term finder, and the six clusters with significant term annotations are shown (Fig. 2D–I). Distinct GO terms are seen within each cluster, suggesting a role for Nhp6A in regulating expression of specific classes of genes, such as those encoding ribosomal proteins, transporters, and oxidoreductases. Each Nhp6A-binding cluster also possesses a unique binding profile. As illustrated in Figure 2C, Nhp6A peak binding levels, widths, and locations relative to TSS vary uniquely between clusters, and these profiles are associated specifically with functionally related genes. For example, the ribosomal cluster (group 1) has peak binding ∼350 bp upstream of the TSS with a binding range from approximately −25 to −800 bp (Fig. 2C). On the other hand, the oxidoreductase cluster (group 5) exhibits peak Nhp6A binding at approximately −600 bp relative to the TSS and a range spanning from −200 to −900 bp (Fig. 2C). Our binding data suggest that Nhp6A binds within distinct promoter chromatin environments of functionally related gene clusters.
Genomic binding of Nhp6A is independent of DNA sequence
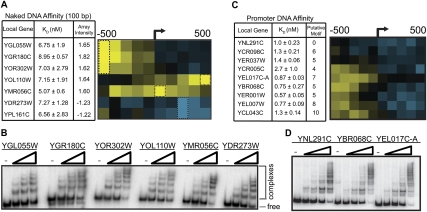
The specific Nhp6A-binding pattern led us to explore the DNA sequence at the targeted locations with the aim of identifying putative binding motifs or other DNA structural elements used for Nhp6A recruitment. To test if in vivo bound DNA sequences are directly recruiting Nhp6A, we first designed electrophoretic mobility shift assay (EMSA) probes to five 100-bp sequences representing high in vivo binding, and two 100-bp sequences that represent unbound segments. No significant binding differences were observed between these probes using purified Nhp6A (Fig. 3A,B). We also challenged prebound Nhp6A complexes with competitor DNA for increasing lengths of time and observed similar lifetimes of Nhp6A–DNA complexes for each probe (Supplemental Fig. S2C). These data provide no evidence for sequence-specific binding by Nhp6A.
Figure 3.
Nhp6A DNA binding is independent of sequence content. (A) Nhp6A affinity (KD) for 100-bp DNA sequences corresponding to regions of high and low Nhp6A binding in vivo. The heat map of ChIP–chip data for each gene is shown on the right with a black box drawn around the 100-bp region evaluated for binding by EMSA. (B) Representative EMSA gel for data summarized in A. (C) Nhp6A affinity (KD) for ∼300-bp promoter DNA sequences associated with Nhp6A in vivo, along with the corresponding ChIP–chip heat map. The number of putative Nhp6A-binding motifs for each promoter as predicted by FIRE is also shown in the third column. (D) Representative EMSA gel for data summarized in C.
In a second approach, we used a motif discovery algorithm, FIRE, to compare four broad clusters of Nhp6A-bound regions (highly bound promoters, medium-bound promoters, bound within ORFs, and unbound segments) (Supplemental Fig. S2A; Elemento et al. 2007). This in silico experiment yielded many statistically significant motifs that are enriched in Nhp6A-bound clusters and underrepresented in Nhp6A-unbound clusters (Supplemental Fig. S2B). The motifs were mapped to representative Nhp6A-binding loci, and 300-bp EMSA probes were designed to respective promoter regions containing a range of the most significantly enriched motifs. Native promoter sequences were chosen with the aim of incorporating multiple motifs and flanking sequences potentially important for targeted Nhp6A binding. We observed similar Nhp6A-binding affinity irrespective of the presence or quantity of FIRE motifs (Fig. 3C,D). While the modest decrease in KD for some probes may hint at an Nhp6A-binding element, these small differences do not correlate with motif number or in vivo levels of Nhp6A binding. The FIRE motifs may represent DNA elements recognized by other DNA-binding proteins that also inhabit Nhp6A-bound promoters and could function to recruit Nhp6A, although, with one exception, FIRE analysis of these Nhp6A-binding clusters did not return DNA motifs for known DNA-binding proteins (Supplemental Fig. S2B).
These results—in combination with previous in vitro binding, structural, and proteomic studies—suggest that the targeted Nhp6A binding observed in vivo is directed by chromatin elements and is not due to specific DNA sequence recognition (Paull and Johnson 1995; Yen et al. 1998; Formosa et al. 2001; Masse et al. 2002; Badis et al. 2008).
Nhp6A stabilizes chromatin
Since Nhp6A binds to discrete groups of genes and targeting appears independent of DNA sequence, we sought to understand the chromatin environment of Nhp6A-bound promoters. To view nucleosome positions, cross-linked chromatin from NHP6A Δnhp6B cells was micrococcal nuclease-digested, immunoprecipitated with histone H3 antibody, and hybridized to tiling arrays. Parallel experiments were performed on Δnhp6A Δnhp6B cells to determine the effects of the absence of Nhp6A on chromatin structure.
Examination of smoothed median histone H3 intensities along the length of a representative chromosome (III) reveals considerable perturbation of chromatin in the absence of Nhp6 (Fig. 4B,C; Supplemental Figs. S7–S12). Histone H3 peak intensities are often decreased and shifted, and nucleosome-depleted regions (NDRs) are often expanded. These changes can be observed at regions that are both highly enriched for Nhp6A binding as well as regions corresponding to lower Nhp6A binding (see also expanded views in Supplemental Fig. S3). For example, at the CHA1 locus, histone H3 binding is clearly altered within both the promoter region (which is highly bound by Nhp6A) and the coding region. The disruption of chromatin structure at CHA1, whose transcription is enhanced by Nhp6A, is consistent with earlier micrococcal nuclease digestion data (Moreira and Holmberg 2000).
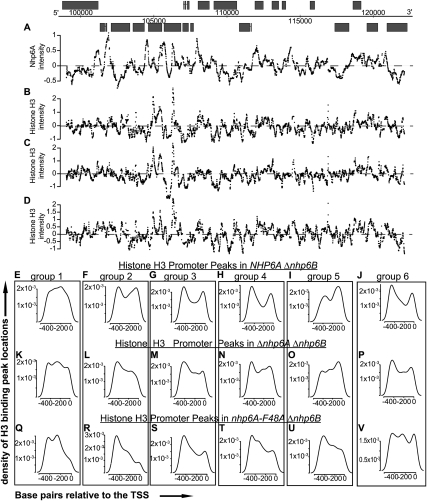
Figure 4.
Nhp6A influences promoter chromatin. (A–D) Nhp6A (A) and histone H3 binding intensity (log2) in NHP6A Δnhp6B (B), Δnhp6A Δnhp6B (C), and nhp6A-F48A Δnhp6B (D) cells along 20 kb of chromosome III. The entire chromosome III is given in Supplemental Figures S7–S12. (E–V) The density of histone H3 promoter (−500 to 0 [TSS]) peak locations are plotted for each Nhp6A-bound GO cluster in NHP6A Δnhp6B (E–J), Δnhp6A Δnhp6B (K–P), and nhp6A-F48A Δnhp6B (Q–V) cells. The interpolated lines are extrapolated to 0 at the boundaries.
To further illustrate the general changes in chromatin structure, smoothed median histone H3 intensity was plotted for each of the GO-annotated Nhp6A-binding clusters. The metagene analyses show a clear correspondence between Nhp6A binding and the NDR within the promoter regions of all six clusters (Supplemental Fig. S4). In the absence of Nhp6A, the NDR is typically expanded into the coding regions. We then plotted the histone H3 intensity peak locations in the promoter regions of all genes within the GO groups. Five of the six GO clusters exhibit two peaks flanking a valley (Fig. 4E–J), corresponding to the previously described −1 and +1 nucleosomes flanking the NDR (Lee et al. 2004, 2007; Yuan et al. 2005). Extending this analysis to histone H3 intensity peaks in Δnhp6 cells shows a disruption of the valley (five out of six) and the peak nearest the TSS (four out of six) (Fig. 4K–P).
Genome-wide transcriptional changes in Nhp6 mutants
To broadly relate Nhp6A binding with the transcriptional activity of neighboring genes, we examined Nhp6A binding at the promoters and ORFs of 5159 protein-coding genes with defined transcriptional frequencies (Holstege et al. 1998). Previous work divided these 5159 genes into five groups representing the relative abundance of mRNA. We observe mean Nhp6A binding greater than the upper bound null threshold in the two gene groups with the highest levels of transcriptional frequency (Fig. 5A). The group with the highest transcriptional activity is enriched for ribosomal proteins (P = 6.04 × 10−126), as are several Nhp6A-binding groups (Fig. 2B,D,I). Additionally, the second most highly transcribed group of genes is enriched for genes with catalytic activity (P = 8.35 × 10−6) and oxidoreductase activity (P = 0.0096), which correspond to specific Nhp6A-binding clusters (Fig. 2B,G–I). This trend between transcriptional frequency and Nhp6A binding (Fig. 5A) continues into the third most highly transcribed group of genes (four to 16 mRNAs per hour), which has an enrichment for transmembrane transporter activities (P-values ranging from 0.006 to 1 × 10−7) similar to two Nhp6A-binding clusters (Fig. 2B,E,F). To quantitatively evaluate the trend for Nhp6A to bind near transcriptionally active genes, we determined the overlap between all Nhp6A and RNA Pol II (Rpd3) (Bermejo et al. 2009) ChIP-enriched regions within 500 bp of each other (Fig. 5B). Whereas two-thirds of the RNA Pol II-binding peaks are near Nhp6A-bound regions, there are many more Nhp6A-binding sites. Altogether, these studies imply that Nhp6A often binds near transcriptionally active genes but is not exclusively associated with RNA Pol II-transcribed regions.
To more directly test the role of Nhp6A on gene expression, whole-genome transcript levels were measured. RNA from NHP6A Δnhp6B and Δnhp6 cells growing exponentially in rich media was subjected to two-color expression profiling. Differentially expressed genes were identified using limma, which allowed for multiple hypothesis testing and a rigorous false discovery rate (FDR) (Smyth and Speed 2003; Smyth 2004; Smyth et al. 2005; Ritchie et al. 2007). Using such criteria, 441 and 277 genes are up-regulated or down-regulated (P < 0.003), respectively, in the Δnhp6 strain (Fig. 5D; Supplemental Material). The 719 differentially expressed genes intersect with 31% of the genes within the Nhp6A-bound clusters identified in Figure 2B.
GO gene function analysis on all differentially expressed genes reveals an enrichment of genes annotated to oxidoreductase activity (P = 1.7 × 10−6). Among the 441 up-regulated or 277 down-regulated genes in Δnhp6 cells, we observed a significant overrepresentation of genes with oxidoreductase activity (P = 4.5 × 10−8) and substrate-specific transporter activity (P = 0.03), respectively. This correlates with GO analysis of Nhp6A-binding clusters (Fig. 2D–I), and suggests a direct role for Nhp6A promoter binding in both activating and repressing transcription. In conclusion, Nhp6A is required for proper transcriptional expression of >10% of yeast genes growing under exponential conditions in rich media, and many of these genes are annotated to specific GO groups that correlate with Nhp6A-binding clusters.
Nhp6A-bending activity is essential for coregulation of transcription
Two amino acid residues within the DNA-binding interface of Nhp6A are critical for bending DNA (Fig. 5C). Substitution of methionine 29 to alanine (M29A) reduces the formation of 98-bp microcircles by sixfold in vitro, but equilibrium binding to linear DNA is reduced only twofold. The severe phenylalanine 48 to alanine (F48A) mutation abolishes formation of 98-bp microcircles, while again lowering equilibrium binding to linear DNA by only twofold (Masse et al. 2002; Dai et al. 2005). We used these mutants to first address whether bending activity is required for transcriptional regulation, and then to address how Nhp6A bending influences target-binding specificity and chromatin structure. Specifically, genome-wide binding and transcript analysis was performed on nhp6A-M29A Δnhp6B, nhp6A-F48A Δnhp6B, and nhp6A-P18A Δnhp6B cells. Nhp6A-P18A was included to control for the modest decrease in DNA binding observed in the bending mutants because it also exhibits a twofold reduction of DNA binding but without a significant change of DNA bending (Yen et al. 1998; Allain et al. 1999). Our expression arrays revealed about five times less nhp6A-P18A mRNA as compared with the wild-type NHP6A (Supplemental Material), so the properties of the nhp6A-P18A strain also may reflect lower cellular levels of Nhp6A-P18A protein. However, of all the mutants tested, Nhp6A-P18A affects the fewest number of genes (109), and >98% of genes display wild-type expression (Fig. 5D). No differential expression of nhp6A-M29A or nhp6A-F48A as compared with wild-type NHP6A expression was observed.
The strongest bending mutant, Nhp6A-F48A, exhibits 374 differentially expressed genes, which is slightly more than half the number of genes affected in the Δnhp6-null mutant (Fig. 5D). A large fraction of these (65%) are in common with the null group (P = 2.13 × 10−149) (Fig. 5E). The weaker bending mutant, Nhp6A-M29A, generates fewer significantly affected genes (253), but again nearly 65% of these are in common with the null (P = 7.07 × 10−94). Moreover, there is a strong overlap between the genes affected by both bending mutants, as nearly 75% of Nhp6A-M29A-affected genes are in common with those of Nhp6A-F48A (P = 3.8 × 10−192). These data are consistent with the DNA-bending function of Nhp6A being an important feature in its role as a transcriptional coregulator.
The critical DNA-bending residues are not required for targeted Nhp6A binding, but affect chromatin structure
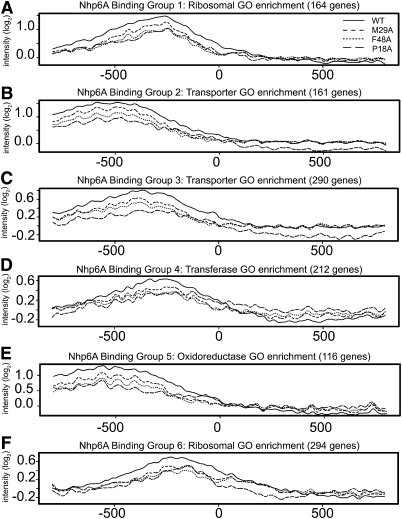
DNA-bending activity could be important for recognizing target binding sites in vivo or stabilizing chromatin structure, or function directly in transcription complex assembly and activity. To evaluate targeted binding, genome-wide binding profiles of the bending mutants Nhp6A-M29A and Nhp6A-F48A were compared with wild-type Nhp6A and Nhp6A-P18A. A similar qualitative binding pattern was obtained for all four proteins (Supplemental Fig. S5A–D), although modest reductions are present for the bending mutants when binding within the Nhp6A-bound GO clusters was quantitatively evaluated (Fig. 6A–F). Notably, Nhp6A-P18A binding is nearly equivalent to Nhp6A-F48A binding except at the transporter clusters (Fig. 6B,C), despite the decreased expression of nhp6A-P18A. Overall, the Nhp6A-bending mutants are enriched at the same promoters as wild-type Nhp6A, suggesting that DNA-bending activity is not a critical determinant in targeting Nhp6A to discrete promoter chromatin regions.
Figure 6.
In vivo targeting of Nhp6A binding is independent of DNA-bending activity. (A–F) Line plots of Nhp6A binding relative to the TSS (0) within the six GO-annotated Nhp6A-bound clusters. Wild-type Nhp6A data are plotted as a solid line, Nhp6A-M29A data are shown as a short dashed line, Nhp6A-F48A data are shown as a dotted line, and Nhp6A-P18A data are shown as a long dashed line.
Histone H3 binding in cells expressing Nhp6A-F48A was also measured. The presence of the strong bending mutant caused changes in chromatin structure that appear intermediate between the NHP6A and Δnhp6-null mutant (Fig. 4D; Supplemental Figs. S7–S12). Within promoter regions of five of the six Nhp6A-bound GO clusters, the presence of Nhp6A-F48A resulted in a decrease in histone H3 peaks corresponding to the +1 nucleosome (Fig. 4Q–V). We conclude that the bending activity of Nhp6A is important in stabilizing chromatin structure, and that this activity is particularly important for maintenance of the +1 nucleosome.
Discussion
The data presented here provide mechanistic insights into the in vivo functions of the yeast HMGB protein Nhp6A. We show that Nhp6A is targeted specifically to discrete chromosomal regions that often correlate with RNA Pol II promoters and are typically underrepresented in ORFs. Cluster analysis of Nhp6A-binding data covering promoters and 5′ ORFs reveals binding groups that are characterized by different signal intensity strengths, peak locations relative to the TSS, and widths of signal peaks. Loss of Nhp6A at these promoters typically causes delocalization of histone H3-binding peaks, especially over the +1 nucleosome position. The absence of Nhp6 results in perturbations in transcription of genes proximal to many Nhp6A-binding peak locations. Furthermore, single amino acid mutations in Nhp6A that decrease DNA bending also disrupt chromatin structure and transcription in a manner similar to deletion of nhp6A/B. This work advances our understanding of transcription and chromatin by showing that the sequence-independent binding of an HMGB protein is targeted in vivo and stabilizes nucleosomes in a DNA-bending-dependent manner to coregulate transcription.
Our analysis of Nhp6A binding was performed in the absence of its less abundant paralog, Nhp6B, which shares 89.5% amino acid identity over the HMG box but contains nine unique residues at its N terminus. ChIP–chip for Nhp6A-TAP in the presence Nhp6B gave a qualitatively similar binding profile at protein-coding genes as Nhp6A in the absence of Nhp6B (Supplemental Fig. S1B,C). A detailed study of any differences in binding selectivity between these very similar HMGB proteins may illuminate why they have been maintained in S. cerevisiae and related genomes.
Targeted Nhp6A binding directed by chromatin
The highly targeted binding of Nhp6A in vivo is remarkable given the absence of any previous evidence of sequence specificity. Thus, we used the whole-genome-binding data to further investigate the possibility of DNA sequence motifs directing Nhp6A binding. However, in vitro binding studies employing highly bound genomic segments failed to identify DNA sequences specifying selective Nhp6A binding. We conclude, therefore, that Nhp6A binding in vivo must be directed by the chromatin environment. More generally, our data demonstrate that proteins can be targeted to distinct genomic sites even though they lack any obvious specificity at the DNA sequence level. Another example is the yeast HMGB protein Hmo1, which binds DNA in an apparently sequence-neutral manner in vitro but with lower affinity than Nhp6A. Whole-genome ChIP–chip data indicate that Hmo1 also exhibits targeted binding in vivo that is distinct from Nhp6A (Hall et al. 2006; Kasahara et al. 2007; Bermejo et al. 2009; NL Dowell and RC Johnson, unpubl.).
Nhp6A stabilizes promoter chromatin
Our Nhp6A and histone H3 ChIP data show peak Nhp6A binding over the NDRs within all six GO-annotated Nhp6A-binding groups (Supplemental Fig. S4). Upon Nhp6A removal, nucleosomes do not crowd into this space, but rather histone H3 binding is decreased over an extended region. The stabilization of nucleosomes by Nhp6A in vivo was unexpected in the context of in vitro experiments showing that large amounts of Nhp6A enhance nuclease and hydroxyl radical reactivity throughout nucleosomal DNA (Ruone et al. 2003; Xin et al. 2009). These experiments have not indicated, however, that Nhp6A alters the rotational or translational positioning of the nucleosomal DNA. Mammalian HMGB1 was reported to enhance the translational movement of mononucleosomes by the Drosophila remodeling complex ACF in a manner dependent on the HMGB1 C-terminal acidic tail (Bonaldi et al. 2002). However, HMGB1 missing its acidic region, and thus more closely resembling Nhp6A, actually stabilized nucleosomes.
Our results indicate that Nhp6A functions along with sequence-specific factors (e.g., Reb1 and Abf1), DNA sequence elements (e.g., A/T tracts), histone chaperones, and ATP-dependent remodeling complexes to orchestrate chromatin structure within promoters. Like Reb1 and A/T tracts (Hartley and Madhani 2009), Nhp6A appears to be particularly important for stable positioning of the +1 nucleosome (Fig. 4E–V). The +1 nucleosome is believed to be of critical importance in phasing nucleosomes downstream within coding regions (Mavrich et al. 2008; Zhang et al. 2009), thus accounting for the NDR being extended into the 5′ coding regions of genes in the absence of Nhp6 (Supplemental Fig. S4). Previous reports have provided evidence for interactions between Nhp6A and yFACT or RSC, which enhance their activities (Brewster et al. 2001; Formosa et al. 2001; Ruone et al. 2003; Szerlong et al. 2003; Takahata et al. 2009). Our results suggest that Nhp6A also functions downstream from these chromatin remodeling complexes and potentially could be actively deposited at specific chromatin regions by these complexes to stabilize remodeled events. This mechanism is consistent with the observed decrease in nucleosome occupancy upon loss of Nhp6A.
Mapping of the chromatin environment by a number of groups using ChIP–chip has painted a comprehensive picture of nucleosome position, turnover, and modifications within gene promoters (Lee et al. 2004, 2007; Liu et al. 2005; Dion et al. 2007; Mavrich et al. 2008). In contrast, relatively few reports have demonstrated how chromatin structure is established and how nonhistone DNA-binding proteins function in the establishment and maintenance of cellular chromatin (Whitehouse et al. 2007; Parnell et al. 2008; Hartley and Madhani 2009). To our knowledge, this work represents the first report highlighting the role of an HMGB protein in maintaining chromatin structure across the genome.
Mechanisms of Nhp6A coregulation of transcription
Earlier studies have shown a stimulatory effect of Nhp6 on in vitro transcription reactions, but the mechanisms of action in these systems are not well understood (Yen et al. 1998; Kruppa et al. 2001; Kassavetis and Steiner 2006). A defining characteristic of HMGB proteins is their robust DNA-bending activity, and several reports have demonstrated the requirement for HMGB-mediated bending for enhanceosome assembly, activated transcription, and recombination (Yen et al. 1998; Mitsouras et al. 2002; Dai et al. 2005). In the present study, we evaluated the global transcription effects of two Nhp6A mutations in the DNA-binding interface that compromise DNA bending (M29A and F48A). These Nhp6A-bending mutants display broad transcription defects that closely mimic the Δnhp6-null mutant. The bending mutations have surprisingly little effect on targeted binding, but our histone H3-binding studies provide evidence demonstrating that DNA conformational changes introduced upon Nhp6A binding play a prominent role in organizing promoter chromatin structure.
Previous studies have provided clues toward understanding how Nhp6A-mediated bending of DNA may function to directly coregulate RNA Pol II transcription, in addition to its effect on nucleosome dynamics. In vitro, Nhp6A binds cooperatively with basal machinery factors like TBP to form a DNA complex with enhanced affinity for TFIIA plus TFIIB (Paull et al. 1996; Biswas et al. 2004). Additionally, model reactions have shown that DNA bending by Nhp6A facilitates interactions between proteins bound at disparate sites on DNA by stabilizing DNA loops (Paull and Johnson 1995). Mechanistic studies on individual promoters will undoubtedly reveal multiple mechanisms for how Nhp6 coregulates transcription, as has already been observed from studies at several genes (Stillman 2010).
Implications for HMGB binding and function in metazoans
Nhp6A is considered the functional yeast homolog of mammalian HMGB1, an abundant constituent of chromatin in all cell types. Although HMGB1 contains two HMG boxes, whereas Nhp6A contains only one HMG box, Nhp6A binds DNA more avidly in vitro because of its N-terminal basic tail. Like Nhp6A, HMGB1 binds DNA with no apparent sequence specificity, exhibits robust DNA-bending activity, and has been reported to function in a number of transcription, repair, and recombination reactions. Their overall similarities would suggest that the targeted binding we observe with yeast Nhp6A in vivo and its effects on promoter chromatin structure will likely be conserved with HMGB1.
Materials and methods
Nhp6A ChIP
Cells were grown in YP + 2% dextrose (YPD) to an OD595 of 0.8–1, and Nhp6A ChIP was performed as described previously (Hecht et al. 1996) with the following modifications. For each ChIP, 50 μL of immobilized Protein A beads was preincubated (overnight at 4°C) with 15 μL of Nhp6 polyclonal antibody (Paull et al. 1996). After washing the beads, equal aliquots of chromatin (500–1000 μg; Bradford assay) were added to the tubes and shuttled through the ChIP protocol.
Nucleosome DNA preparation
Cells were grown, cross-linked, harvested, and lysed in an identical manner as for Nhp6A ChIP, except that the cells were washed and resuspended into MNase digestion buffer (0.075% NP-40, 50 mM NaCl, 50 mM HEPES at pH 7.5, 5 mM MgCl2, 5 mM CaCl2). The input sample chromatin was sonicated for 30 min (1 min ON/30 sec OFF) with a Diogenode sonicator. Micrococcal nuclease (30 U; Takara) was added to the experimental samples and incubated for 20 min at 37°C. Digestion was quenched with 50 mM EDTA. The MNase-digested chromatin was then taken through the standard immunoprecipitation procedure described above. Histone H3 antibody (Abcam ab1791) was used to enrich for nucleosomes.
ChIP tiling array data analysis
Nhp6A and histone H3 raw Affymetrix data (minimum of two biological replicates) were processed, loess-normalized to sonicated input DNA, and analyzed in R using the Starr package (http://www.bioconductor.org). Nhp6A and histone H3 probe intensities were smoothed with a sliding median window of 150 and 75 bp, respectively. To identify general Nhp6A binding relative to genome elements, we isolated all genes 300–5000 bp in length that also have no genes within 500 bp upstream of or 300 bp downstream from the respective TSS or transcription termination site (TTS). For each of these 1015 genes, we plotted mean Nhp6A signal intensity in 10 regions as depicted in Figure 2A to identify bound regions. To identify each specific genomic region bound by Nhp6A, we followed several criteria: (1) A signal threshold based on the null distribution of probe intensities was exceeded, (2) a minimum of five neighboring probes intensities exceeded the threshold, and (3) at least 500 bp of spacing existed between ChIP-enriched regions. Nhp6A-bound regions were mapped to yeast chromosomes, and UCSC Genome Browser files (provided in the Supplemental Material) were created using IRanges (http://www.bioconductor.org). A custom Python script extracted and averaged probe intensities over defined intervals (100 bp) flanking TSSs of protein-coding genes. These data were clustered (K-means) using CLUSTER (Eisen et al. 1998) and were visualized with Java Treeview (Saldanha 2004).
Smoothed normalized histone H3 intensity data were plotted along yeast chromosome III using the GenomeGraphs package (http://www.bioconductor.org). Histone H3 peak locations were identified by running a narrow smoother (5% of available probes at each gene) over the probe intensities at each gene (−800 to +800) and then extracting the peak locations over defined intervals. The densities of peak locations were then plotted along the same defined interval (−500 to 0).
Rpb3 (RNA Pol II) data (Bermejo et al. 2009) were normalized using the Starr package, and binding peaks (and peak overlap) for Rpb3 and Nhp6A were identified using the ChIPpeakAnno package (http://www.bioconductor.org).
Microarray probe preparation: linear amplification, fragmentation, and labeling
ChIP DNA was linearly amplified according to the Affymetrix-provided protocol (http://www.affymetrix.com/support/downloads/manuals/chromatin_immun_ChIP.pdf). The GeneChip WT Double-Stranded DNA Terminal Labeling Kit (Affymetrix, #900812) was used to fragment the DNA to appropriate hybridization length and to biotin-label with terminal deoxynucleotidyl transferase. These samples were hybridized (Affymetrix S. cerevisiae 1.0R Tiling Arrays), washed, and scanned by the University of California at Los Angeles Clinical Microarray Core according to protocol.
RNA isolation, amplification, labeling, hybridization, and scanning of Agilent expression array
RNA was isolated from cells (OD595 = 0.8–1.0 grown in YPD) using the hot phenol method (Köhrer and Domdey 1991). The Agilent Quick Amp Two-Color Labeling Kit (#5190-0444) was used to fluorescently label the RNA. The Agilent protocol (http://www.chem.agilent.com) was followed for labeling and hybridizing the cDNA to the 4 × 44,000 (V1) S. cerevisiae gene expression arrays. The arrays were scanned on the Agilent G2565BA fluorescent scanner.
Agilent expression array data analysis
Data extraction and quality assessment was performed with Agilent Feature Extraction software. Identification of genes differentially expressed between wild-type and mutant strains was done in R using the limma package (Smyth et al. 2005). All data (minimum of two biological replicates) were loess-normalized and background-subtracted within each array and fit to a linear model with a simple design matrix comparing mutant signals to wild-type signals (Smyth and Speed 2003; Smyth 2004; Ritchie et al. 2007). An empirical Bayes method was used to calculate a moderated t-statistic and the associated P-values (Smyth 2004). The Benjamini and Hochberg method was used to control the FDR, and differentially expressed genes were analyzed using the S. cerevisiae Genome Database for GO enrichment (Hochberg and Benjamini 1990; Dwight et al. 2002). Complete lists of differentially expressed genes and associated statistics are provided (Supplemental Material). Raw Nhp6A and histone H3 CEL and expression array files have been deposited in Gene Expression Omnibus (GSE23608).
EMSAs
The final reagent concentrations of the EMSA reaction mix were as follows: 20 mM Tris-HCl (pH 8), 1 mM EDTA, 2 mM DTT, 100 μg/mL BSA, 5% glycerol, 40 mM NaCl, and PAGE-purified 32P-labeled DNA probes. Purified recombinant Nhp6A (Yen et al. 1998) was added at increasing concentrations and incubated for 10 min at room temperature. The reactions were then loaded on a 5% acrylamide (19:1), 0.5× TBE gel and electrophoresed at 35 mA.
Yeast strains
Yeast strains were derived from SEY6210 (MAT-α ura3-52 leu2-3,112 his3-Δ200 trp1-Δ201 lys2-801 suc2-Δ9 gal3) (Robinson et al. 1988). RJY6006 (nhp6B∷LEU2) and RJY6009 (nhp6A∷URA3 nhp6B∷LEU2) are described in Paull et al. (1996). RJY6809 (nhp6A-F48A∷URA3) and RJY6810 (nhp6A-M29A∷URA3) contain integrated copies of the respective mutations in RJY6009 and were kindly provided by Dr. Janet Treger. RJY6684 contains nhp6A-P18A under the control of the NHP6A promoter on the CEN6 plasmid pRJ1364 in RJY6009.
Acknowledgments
We thank Dr. Kathrin Plath for critical reading of the manuscript, Dr. Roberto Ferrari for technical help on the expression array experiments, and the UCLA Clinical Microarray Core facility for expert technical assistance. This work was supported by NIH grant GM038509. A.S.S. was supported by the National Institutes of Health Medical Scientist Training Program and a Jonsson Comprehensive Cancer Center Foundation Fellowship.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1948910.
Supplemental material is available at http://www.genesdev.org.
References
- Agresti A, Lupo R, Bianchi M, Müller S 2003. HMGB1 interacts differentially with members of the Rel family of transcription factors. Biochem Biophys Res Commun 302: 421–426 [DOI] [PubMed] [Google Scholar]
- Allain F, Yen Y, Masse J, Schultze P, Dieckmann T, Johnson R, Feigon J 1999. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J 18: 2563–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan E, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson C, Gossett A, Hasinoff M, Warren C, et al. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R, Brewer L, Corzett M 2000. DNA condensation by protamine and arginine-rich peptides: Analysis of toroid stability using single DNA molecules. Mol Reprod Dev 56: 230–234 [DOI] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Gonzalez-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, et al. 2009. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell 138: 870–884 [DOI] [PubMed] [Google Scholar]
- Bianchi M, Agresti A 2005. HMG proteins: Dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 15: 496–506 [DOI] [PubMed] [Google Scholar]
- Biswas D, Imbalzano A, Eriksson P, Yu Y, Stillman D 2004. Role for Nhp6, Gcn5, and the Swi/Snf complex in stimulating formation of the TATA-binding protein-TFIIA-DNA complex. Mol Cell Biol 24: 8312–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldi T, Längst G, Strohner R, Becker P, Bianchi M 2002. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J 21: 6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi M, Taraseviciene L, Nordeen S, Allegretto E, Edwards D 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol 18: 4471–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster N, Johnston G, Singer R 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol Cell Biol 21: 3491–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol 19: 5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan C, Kolodrubetz D, Snyder M 1994. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol Cell Biol 14: 2391–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Wong B, Yen Y, Oettinger M, Kwon J, Johnson R 2005. Determinants of HMGB proteins required to promote RAG1/2-recombination signal sequence complex assembly and catalysis during V(D)J recombination. Mol Cell Biol 25: 4413–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion M, Kaplan T, Kim M, Buratowski S, Friedman N, Rando O 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Dwight S, Harris M, Dolinski K, Ball C, Binkley G, Christie K, Fisk D, Issel-Tarver L, Schroeder M, Sherlock G, et al. 2002. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res 30: 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q, Grosschedl R 1999. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol 11: 233–240 [DOI] [PubMed] [Google Scholar]
- Eisen M, Spellman P, Brown P, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S 2007. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell 28: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Eriksson P, Wittmeyer J, Ginn J, Yu Y, Stillman D 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J 20: 3506–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fury W, Batliwalla F, Gregersen P, Li W 2006. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc 1: 5531–5534 [DOI] [PubMed] [Google Scholar]
- Guth S, Wegner M 2008. Having it both ways: Sox protein function between conservation and innovation. Cell Mol Life Sci 65: 3000–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D, Wade J, Struhl K 2006. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol Cell Biol 26: 3672–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley P, Madhani H 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96 [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y 1990. More powerful procedures for multiple significance testing. Stat Med 9: 811–818 [DOI] [PubMed] [Google Scholar]
- Holstege F, Jennings E, Wyrick J, Lee T, Hengartner C, Green M, Golub T, Lander E, Young R 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Jayaraman L, Moorthy N, Murthy K, Manley J, Bustin M, Prives C 1998. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev 12: 462–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Ohtsuki K, Ki S, Aoyama K, Takahashi H, Kobayashi T, Shirahige K, Kokubo T 2007. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol Cell Biol 27: 6686–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G, Steiner D 2006. Nhp6 is a transcriptional initiation fidelity factor for RNA polymerase III transcription in vitro and in vivo. J Biol Chem 281: 7445–7451 [DOI] [PubMed] [Google Scholar]
- Köhrer K, Domdey H 1991. Preparation of high molecular weight RNA. Methods Enzymol 194: 398–405 [DOI] [PubMed] [Google Scholar]
- Kruppa M, Moir R, Kolodrubetz D, Willis I 2001. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol Cell 7: 309–318 [DOI] [PubMed] [Google Scholar]
- Lee C, Shibata Y, Rao B, Strahl B, Lieb J 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse R, Davis R, Hughes T, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Liu C, Kaplan T, Kim M, Buratowski S, Schreiber S, Friedman N, Rando O 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328 doi: 10.1371/journal.pbio.0030328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse J, Wong B, Yen Y, Allain F, Johnson R, Feigon J 2002. The S. cerevisiae architectural HMGB protein NHP6A complexed with DNA: DNA and protein conformational changes upon binding. J Mol Biol 323: 263–284 [DOI] [PubMed] [Google Scholar]
- Mavrich T, Ioshikhes I, Venters B, Jiang C, Tomsho L, Qi J, Schuster S, Albert I, Pugh B 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K, Prives C 2002. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol Cell Biol 22: 6797–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsouras K, Wong B, Arayata C, Johnson R, Carey M 2002. The DNA architectural protein HMGB1 displays two distinct modes of action that promote enhanceosome assembly. Mol Cell Biol 22: 4390–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira JM, Holmberg S 2000. Chromatin-mediated transcriptional regulation by the yeast architectural factors NHP6A and NHP6B. EMBO J 19: 6804–6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell T, Huff J, Cairns B 2008. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J 27: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull T, Johnson R 1995. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J Biol Chem 270: 8744–8754 [DOI] [PubMed] [Google Scholar]
- Paull T, Carey M, Johnson R 1996. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev 10: 2769–2781 [DOI] [PubMed] [Google Scholar]
- Ritchie M, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth G 2007. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Robinson J, Klionsky D, Banta L, Emr S 1988. Protein sorting in Saccharomyces cerevisiae: Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol 8: 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruone S, Rhoades A, Formosa T 2003. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J Biol Chem 278: 45288–45295 [DOI] [PubMed] [Google Scholar]
- Saldanha A 2004. Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Smyth G 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Smyth G, Speed T 2003. Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Smyth G, Michaud J, Scott H 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075 [DOI] [PubMed] [Google Scholar]
- Stillman D 2010. Nhp6: A small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim Biophys Acta 1799: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerlong H, Saha A, Cairns B 2003. The nuclear actin-related proteins Arp7 and Arp9: A dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J 22: 3175–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman D 2009. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell 34: 405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Travers A 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem Sci 26: 167–174 [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando O, Delrow J, Tsukiyama T 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035 [DOI] [PubMed] [Google Scholar]
- Xin H, Takahata S, Blanksma M, McCullough L, Stillman D, Formosa T 2009. yFACT induces global accessibility of nucleosomal DNA without H2A–H2B displacement. Mol Cell 35: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Ban T, Wang Z, Choi M, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, et al. 2009. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462: 99–103 [DOI] [PubMed] [Google Scholar]
- Yen Y, Wong B, Johnson R 1998. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J Biol Chem 273: 4424–4435 [DOI] [PubMed] [Google Scholar]
- Yuan G, Liu Y, Dion M, Slack M, Wu L, Altschuler S, Rando O 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630 [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Falciola L, Helmer-Citterich M, Mavilio F, Bianchi M 1996. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J 15: 4981–4991 [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone–DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]