Abstract
The conserved TREX mRNA export complex is known to contain UAP56, Aly, Tex1, and the THO complex. Here, we carried out proteomic analysis of immunopurified human TREX complex and identified the protein CIP29 as the only new component with a clear yeast relative (known as Tho1). Tho1 is known to function in mRNA export, and we provide evidence that CIP29 likewise functions in this process. Like the known TREX components, a portion of CIP29 localizes in nuclear speckle domains, and its efficient recruitment to mRNA is both splicing- and cap-dependent. We show that UAP56 mediates an ATP-dependent interaction between the THO complex and both CIP29 and Aly, indicating that TREX assembly is ATP-dependent. Using recombinant proteins expressed in Escherichia coli, we show that UAP56, Aly, and CIP29 form an ATP-dependent trimeric complex, and UAP56 bridges the interaction between CIP29 and Aly. We conclude that the interaction of two conserved export proteins, CIP29 and Aly, with UAP56 is strictly regulated by ATP during assembly of the TREX complex.
Keywords: CIP29, Aly, UAP56, TREX complex, Tho1, helicase
The TREX complex, which is conserved from Saccharomyces cerevisiae to humans, was first identified as a complex that functions in mRNP export (Reed and Hurt 2002; Aguilera 2005; Kohler and Hurt 2007). The yeast TREX also plays roles in 3′ end formation, mRNP biogenesis, and quality control (Aguilera 2005; Kohler and Hurt 2007). Although less is known about the metazoan TREX complex, it may likewise play roles in these other processes (Johnson et al. 2009). TREX is recruited cotranscriptionally to mRNA in yeast and to the 5′ end of mRNA during splicing in humans (Reed and Hurt 2002; Aguilera 2005; Kohler and Hurt 2007). The known conserved components of TREX are the multisubunit THO complex, Tex1 (a protein of unknown function), the DEAD-box helicase UAP56, and Aly (the latter three proteins are known as Tex1, Sub2, and Yra1, respectively, in yeast) (Reed and Hurt 2002; Aguilera 2005; Kohler and Hurt 2007). In yeast, the THO complex consists of four tightly associated subunits (Tho2, Hpr1, Mft1, and Thp1) (Piruat and Aguilera 1998; Jimeno et al. 2006). Likewise, the metazoan THO complex consists of a set of tightly associated proteins, three of which (fSAP79, fSAP35, and fSAP24; known now as THOC5, THOC6, and THOC7, respectively) do not appear to be conserved in yeast and two of which are orthologs of Tho2 (THOC2) and Hpr1 (THOC1) (Rehwinkel et al. 2004; Masuda et al. 2005). In yeast, Aguilera and coworkers (Piruat and Aguilera 1998) identified a protein known as Tho1 during the same genetic screen that they used to identify the THO complex. Subsequent characterization of Tho1 revealed that it functions in mRNP biogenesis and export, but this protein was not identified as a component of the THO/TREX complex (Piruat and Aguilera 1998; Jimeno et al. 2006). However, Tho1 is a multicopy suppressor of THO complex mutants and is recruited to mRNA in a THO complex-dependent manner (Piruat and Aguilera 1998; Jimeno et al. 2006). In humans, a counterpart of yeast Tho1 was identified based on sequence alignment (Jimeno et al. 2006). This protein, CIP29, was first reported as a cytokine-induced protein and later was linked to several cancers (Choong et al. 2001; Fukuda et al. 2002; Hashii et al. 2004; Leaw et al. 2004). Like yeast Tho1, CIP29 contains a SAF motif and binds to DNA, which led to the speculation that CIP29 functions in transcription (Aravind and Koonin 2000; Hashii et al. 2004). CIP29 was also proposed to function in splicing, export, or translation, as it binds RNA and interacts with UAP56 (Leaw et al. 2004; Sugiura et al. 2007). However, at present, the function of CIP29 is not known.
In light of the connections between yeast Tho1 and mRNA export (Piruat and Aguilera 1998; Jimeno et al. 2006), we investigated the function of CIP29. Using an antibody raised against CIP29, as well as antibodies to UAP56 and THOC2, we carried out immunoprecipitations (IPs) from nuclear extract followed by mass spectrometry. Analysis of these data led to the identification of six putative new components of the human TREX complex. Of these, CIP29 was the only protein with a clear yeast ortholog. We show that both CIP29 and Aly associate with TREX in an ATP-dependent manner, and UAP56 mediates the association between these proteins and the THO complex to form TREX. Moreover, using purified recombinant proteins, we show that CIP29, Aly, and UAP56 assemble into an ATP-dependent trimeric complex. Together, our data indicate that TREX is dynamically remodeled in an ATP-dependent manner, and UAP56, Aly, and CIP29 are key players in this remodeling.
Results
CIP29 associates with the human TREX complex
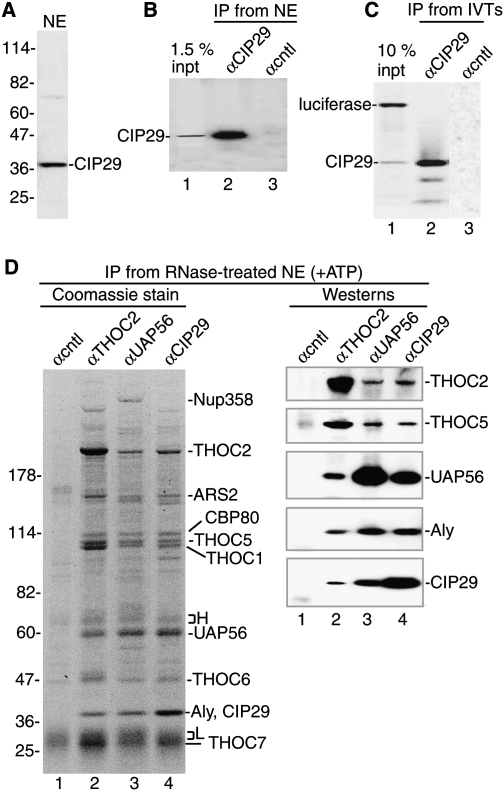
To investigate the function of CIP29, we raised an antibody against the full-length protein. The antibody detected one main band on a Western blot, immunoprecipitated CIP29 from nuclear extract, and specifically immunoprecipitated in vitro translated CIP29 (Fig. 1A–C), indicating that the antibody is specific for CIP29 and suitable for Western and IP. To characterize CIP29, we carried out side-by-side IPs using antibodies to THOC2, UAP56, or CIP29 from RNase-treated nuclear extract (extract was incubated in ATP prior to IP) (see below). Coomassie gel analysis revealed that the set of proteins and their relative levels were strikingly similar between the three IPs (Fig. 1D, left panel). All of the THO complex proteins, Aly, UAP56, and the previously identified TREX-associated proteins (CBP80 and ARS2) were detected in all three IPs by mass spectrometry of individual bands and/or comigration on the gel (Fig. 1D, left panel). Mass spectrometry also revealed that CIP29 and Aly comigrate by SDS-PAGE. The nuclear pore complex-associated protein Nup358 was detected only in the UAP56 IP (Fig. 1D, left panel), which may indicate that UAP56 has a function at the pore (Forler et al. 2004). Further work is needed to determine whether this association between Nup358 and UAP56 is related to the association between Nup358 and the mRNA export receptor NXT/TAP (Forler et al. 2004; Levesque et al. 2006). Westerns confirmed that TREX proteins specifically associate not only with THOC2 and UAP56, but also with CIP29 (Fig. 1D, right panel).
Figure 1.
CIP29 associates with the human TREX complex. (A) Western of nuclear extract (1 μL) using an antibody against CIP29. Markers in kilodaltons are indicted. (B) IPs from nuclear extract using the CIP29 antibody (lane 2) or a negative control antibody (fSAP130) (lane 3) followed by Western with the CIP29 antibody; 1.5% of nuclear extract input is shown in lane 1. (C) A mixture of in vitro translated (IVT) CIP29 and luciferase (lane 1) was used for IP with the CIP29 antibody (lane 2) or a negative control antibody (fSAP130) (lane 3). (D, left panel) Coomassie-stained gel of IPs using indicated antibodies. H and L indicate the heavy and light chains of antibodies, respectively. Molecular weight markers in kilodaltons are shown. IPs in the left panel were used for Westerns with the indicated antibodies (shown in the right panel).
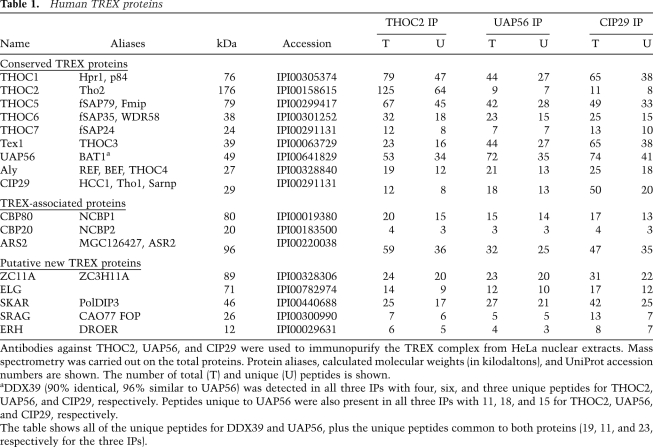
We further characterized CIP29 and the TREX complex by carrying out total mass spectrometry of the THOC2, UAP56, and CIP29 IPs (Table 1; Supplemental Table S1). All of the known TREX proteins and CIP29 were the highest hits in all three IPs, based on peptide number normalized to protein size (Table 1, conserved TREX proteins). CBC (CBP20 and CPB80) and ARS2 were also detected in all three IPs (Table 1), consistent with the cap-dependent recruitment of TREX to mRNA (Cheng et al. 2006) and the observation that ARS2 interacts with CBC (Gruber et al. 2009). Several other proteins were detected in the THOC2, UAP56, and CIP29 IPs, but were not listed in Table 1 because they were either unique to one of the three IPs (Supplemental Table S1, THOC2-, UAP56-, or CIP29-specific proteins) or present in only two of the three IPs. Proteins commonly found in IPs of RNPs and/or that were low hits based on peptide number and protein size are also listed in Supplemental Table S1. The mRNA export adaptor UIF identified recently in 293T cells (Hautbergue et al. 2009) was not detected in our mass spectrometry data, possibly because this protein is present at low levels in HeLa cells (SA Wilson, unpubl.). No peptides from the EJC (exon junction complex) (Maquat and Carmichael 2001) were detected in any of our IPs, indicating that TREX and the EJC do not associate in RNase-treated nuclear extracts. The protein SKAR (Table 1), which is an S6 kinase substrate, was shown previously to coimmunoprecipitate with the EJC component eIF4A3 in whole-cell lysate (Ma et al. 2008). We do not detect an interaction between SKAR and EJC components in nuclear extracts (data not shown), possibly indicating that SKAR interacts with the EJC during a remodeling event in the cytoplasm (Ma et al. 2008).
Table 1.
Human TREX proteins
Antibodies against THOC2, UAP56, and CIP29 were used to immunopurify the TREX complex from HeLa nuclear extracts. Mass spectrometry was carried out on the total proteins. Protein aliases, calculated molecular weights (in kilodaltons), and UniProt accession numbers are shown. The number of total (T) and unique (U) peptides is shown.
aDDX39 (90% identical, 96% similar to UAP56) was detected in all three IPs with four, six, and three unique peptides for THOC2, UAP56, and CIP29, respectively. Peptides unique to UAP56 were also present in all three IPs with 11, 18, and 15 for THOC2, UAP56, and CIP29, respectively.
The table shows all of the unique peptides for DDX39 and UAP56, plus the unique peptides common to both proteins (19, 11, and 23, respectively for the three IPs).
The observation that all known TREX proteins were the highest hits in the THOC2 and UAP56 IPs validates this approach for identifying candidate new TREX components. In addition to CIP29, five other proteins were the highest hits common to the THOC2, UAP56, and CIP29 IPs (Table 1, putative new TREX proteins). Other findings are consistent with the possibility that these five proteins are TREX components. ZC11A and SKAR are present in nuclear speckles, SKAR is highly related in sequence to Aly, and SKAR and ERH interact directly (Smyk et al. 2006; Ma et al. 2008; K Dufu and R Reed, unpubl.). ELG is a stable mRNP component that is not associated with the EJC (Merz et al. 2007), and SRAG interacts with UAP56 (Lehner et al. 2004). Further studies are required to determine whether these proteins as well as any of the proteins in Supplemental Table S1 are bona fide TREX components or associate with TREX in some manner. It is also possible that there is more than one type of TREX complex and/or that some of the proteins are involved in regulation of the TREX complex. Significantly, however, among all of the proteins identified by mass spectrometry, CIP29 was the only putative new TREX component with a clear ortholog in yeast, and this yeast ortholog is both genetically and functionally linked to mRNA export (Jimeno et al. 2006). Thus, in this study, we further characterized CIP29.
CIP29 is present in nuclear speckle domains, and efficient recruitment of CIP29 to mRNA is splicing- and cap-dependent
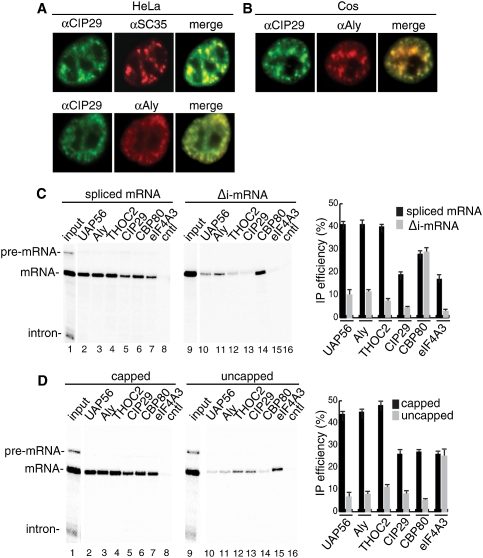
Previous characterization of the TREX complex showed that its components are distributed evenly throughout the nucleoplasm, with a portion concentrated in nuclear speckle domains (Reed and Hurt 2002; Moore and Proudfoot 2009). Extensive work shows that these domains, which are usually detected by their colocalization with the splicing factor SC35, are enriched in the pre-mRNA processing machinery polyA+ RNA as well as TREX components (Reed and Hurt 2002; Moore and Proudfoot 2009). In addition, efficient recruitment of the TREX complex to mRNA is both splicing- and cap-dependent (Reed and Hurt 2002; Kohler and Hurt 2007). Immunofluorescence (IF) studies in HeLa cells using the CIP29 antibody showed an even distribution of CIP29 in the nucleoplasm, in addition to colocalization with SC35 and Aly (Fig. 2A). Similarly, CIP29 colocalized with Aly in nuclear speckle domains in Cos cells (Fig. 2B). These data indicate that CIP29 shares the subcellular localization pattern previously identified for TREX proteins. CIP29 is also recruited more efficiently to spliced mRNA than to the corresponding mRNA lacking an intron (Δi-mRNA). Specifically, both spliced mRNA and Δi-mRNA were immunoprecipitated with similar efficiency by the CBP80 antibody (Fig. 2C, lanes 6,14). In contrast, antibodies to CIP29, TREX proteins, and the EJC component eIF4A3 immunoprecipitated spliced mRNA more efficiently than Δi-mRNA (Fig. 2C). Efficient recruitment of CIP29 is also cap-dependent, as capped mRNA was more efficiently immunoprecipitated by the CIP29 antibody, as well as by TREX and CBP80 antibodies, than was uncapped mRNA (Fig. 2D). In contrast, both capped and uncapped mRNAs were immunoprecipitated by the eIF4A3 antibody, which is expected for EJC components (Fig. 2D; Cheng et al. 2006; Merz et al. 2007). Together, the observations that CIP29 is present in nuclear speckles and that its efficient recruitment to mRNA is both cap- and splicing-dependent are consistent with the possibility that CIP29 is a newly identified conserved component of the TREX complex.
Figure 2.
CIP29 is present in nuclear speckle domains, and its efficient recruitment to mRNA is both splicing- and cap-dependent (A) IF of HeLa cells was performed using CIP29, SC35, or Aly antibodies. (B) Same as A, except using Cos cells and CIP29 and Aly antibodies. (C) IPs of spliced or Δi AdML mRNA with indicated antibodies. The inputs (25%) (shown in lanes 1,9) and IPs were run on a 6.5% denaturing gel. (D) Same as C, except that capped and uncapped AdML spliced mRNA was used. Quantitations of three independent experiments are shown. Error bars represent standard deviations (n = 3).
Evidence that CIP29 functions in mRNA export
We next asked whether CIP29 functions in mRNA export (Fig. 3). Although CIP29 was knocked down efficiently by siRNAs, no effect on polyA+ RNA export or export of a reporter was observed (data not shown), possibly due to redundancy with other proteins.
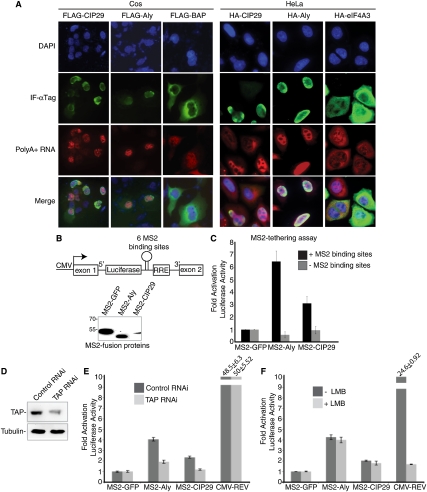
Figure 3.
Evidence that CIP29 functions in mRNA export. (A) IF and FISH for polyA+ RNA in cells transfected with CMV constructs encoding Flag- or HA-tagged CIP29, Aly, BAP, or eIF4A3 in Cos or HeLa cells as indicated. DAPI staining was used to identify the nucleus. Bar, 10 μm. The Cos cells were treated with actinomycin D for 2 h prior to FISH to reduce the nascent RNA levels. Note that the cytoplasmic signals are dampened in the control and nontransfected cells when images are captured at the same exposure as shown for all of the data except Flag-BAP. This dampening occurs because the microscope automatically adjusts the image using the strong signal detected in the nuclei in cells with an export block. For the Flag-BAP FISH data, we showed an overexposure relative to that for Flag-CIP29 and Flag-Aly in order to detect the cytoplasmic signals. (B) Schematic of MS2/RRE reporter construct. The intron contains six MS2-binding sites and one RRE, and encodes luciferase. Westerns of the MS2 fusion proteins used in the MS2-tethering assay are shown. (C) Quantitation of luciferase activity in 293 cells transfected with plasmids encoding the indicated MS2 fusion proteins. (D) Western for TAP in TAP knockdown cells. Tubulin was used as loading control. (E) MS2/RRE tethering assay in TAP knockdown cells. (F) MS2/RRE-tethering assay in Leptomycin B-treated cells. MS2 assay results represent the average values obtained from nine independent transfections, and error bars show the standard deviations.
Thus, to investigate a role for CIP29 in mRNA export, we transfected Cos or HeLa cells with a CMV construct encoding tagged full-length CIP29 (Fig. 3A). CMV-Aly was used as a positive control in both cell types. CMV-BAP (bacterial alkaline phosphatase) and CMV-eIF4A3 were used as negative controls in Cos and HeLa cells, respectively. IF was used to identify the cells expressing each tagged protein (Fig. 3A). Significantly, FISH indicated that polyA+ RNA export was inhibited in both Cos and HeLa cells expressing either the CIP29 or the Aly construct (Fig. 3A). In contrast, polyA+ RNA was exported when the eIF4A3 construct was expressed in HeLa cells or when the BAP construct was expressed in Cos cells (Fig. 3A, note that cytoplasmic signals are generally dampened in the images of the nontransfected or control cells; see the legend). Together, these data are consistent with the possibility that CIP29 functions in mRNA export.
To further investigate a potential role for CIP29 in mRNA export, we carried out an MS2-tethering assay previously used in studies of export proteins (Hargous et al. 2006; Hautbergue et al. 2009). In our study, we tested CMV constructs encoding MS2-CIP29, MS2-Aly (as a positive control), and MS2-GFP (as a negative control). We also transfected CMV-REV, as the REV protein is exported via the CRM pathway when bound to the RRE (Rev response element) (Malim and Cullen 1991). A schematic of the luciferase reporter containing six MS2-binding sites and the RRE in the intron of a pre-mRNA is shown in Figure 3B, and an isogenic reporter lacking the MS2 sites was used as a negative control. To control for variations in transfection efficiencies, a β-galactosidase construct was cotransfected, and luciferase levels were normalized to β-galactosidase levels. Western analysis of the transfected MS2 fusion proteins is shown in Figure 3B. When either the MS2-Aly or the MS2-CIP29 construct was cotransfected with the reporters, luciferase activity was observed only with the reporter containing the MS2 sites, indicating that the unspliced pre-mRNA was exported via binding to these MS2 fusion proteins (Fig. 3C; Hautbergue et al. 2009). Only background levels of luciferase activity were observed with the MS2-GFP construct, indicating that the enhanced luciferase expression was specific to Aly and CIP29 (Fig. 3C). The observation that MS2-Aly has a stronger phenotype in this assay than MS2-CIP29 may be because MS2-Aly expresses at significantly higher levels than MS2-CIP29 (Fig. 3B). We next asked whether MS2-Aly or MS2-CIP29 mediated export of the MS2 reporter via the mRNA export pathway. To do this, we carried out RNAi of the mRNA export receptor TAP. Western analysis revealed that the levels of this protein were significantly knocked down (Fig. 3D). Significantly, the levels of export of the luciferase reporter were significantly reduced for both MS2-Aly and MS2-CIP29, but were unaffected for CMV-REV (Fig. 3E). In contrast, the CRM inhibitor Leptomycin B potently decreased luciferase activity for CMV-REV, but not for MS2-Aly and MS2-CIP29 (Fig. 3F). Together, the CIP29 overexpression studies and the MS2-tethering assay support the conclusion that CIP29, like its yeast homolog (Jimeno et al. 2006), has a role in mRNA export.
CIP29 and Aly interact with the THO complex via UAP56
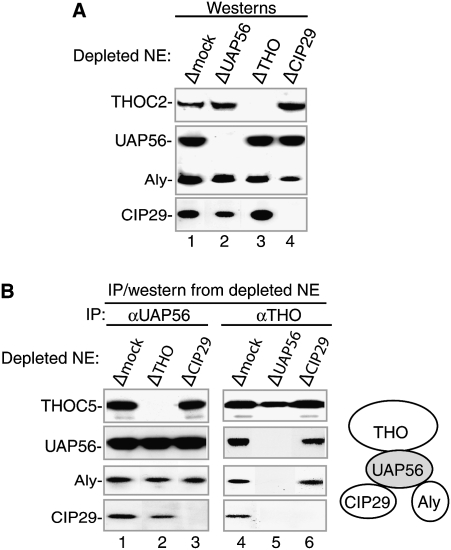
To further investigate the role of CIP29, we asked which TREX components are required for interaction with CIP29 (Fig. 4). To do this, IPs were carried out using nuclear extracts that were immunodepleted of CIP29, UAP56, or the THO complex (note that the entire THO complex is coimmunodepleted by antibodies to the different THO complex proteins) (Masuda et al. 2005; K Dufu and R Reed, unpubl.). Mock-depleted extract was used as a control. As shown in Figure 4A, Western analysis of the depleted extracts revealed that THOC2, UAP56, Aly, and CIP29 were all present in the Δmock extract (lane 1), and only the target proteins were depleted in the ΔUAP56, ΔTHO, and ΔCIP29 extracts (lanes 2–4). When UAP56 or THO antibodies were used for IPs from the depleted extracts, Westerns revealed that the THO complex, UAP56, Aly, and CIP29, were all immunoprecipitated in Δmock extract (Fig. 4B, lanes 1,4). In the ΔTHO extract, UAP56, Aly, and CIP29 were immunoprecipitated by the UAP56 antibody (Fig. 4B, lane 2), indicating that the THO complex is not required for the interaction between UAP56 and CIP29 or Aly. In the ΔCIP29 extract, the THO complex, UAP56, and Aly were all immunoprecipitated by both UAP56 and THO antibodies (Fig. 4B, lanes 3,6), indicating that CIP29 is not required for the association between the THO complex, UAP56, and Aly. In striking contrast, in the ΔUAP56 extract, neither CIP29 nor Aly was immunoprecipitated by the THO antibody, whereas the THO complex was (Fig. 4B, lane 5). We conclude that UAP56 is required for the interaction between CIP29 and the THO complex. Consistent with previous work (Masuda et al. 2005), UAP56 is also required for the interaction between Aly and the THO complex. As shown in the schematic (Fig. 4B, right), this analysis defines two conserved proteins—Aly and CIP29—whose association with the THO complex is mediated by UAP56. Whether CIP29 and Aly bind simultaneously to UAP56 is addressed below.
Figure 4.
UAP56 mediates the interaction between the THO complex and both CIP29 and Aly. (A) Immunodepleted extracts were used for Westerns with the indicated antibodies. (B, lanes 1–6) IPs from immunodepleted extracts were used for Westerns with the indicated antibodies. Schematic of the interactions among CIP29, Aly, UAP56, and the THO complex is shown on the right.
CIP29 and Aly are recruited to the TREX complex in an ATP-dependent manner
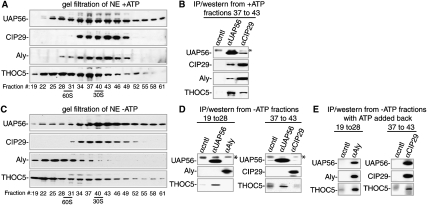
As UAP56 is an ATPase, we next investigated whether its association with the THO complex, CIP29, or Aly was affected by ATP (Fig. 5). Accordingly, RNase-treated nuclear extract was incubated in either the presence or absence of ATP followed by IP/Westerns. This analysis revealed that UAP56 associated with the THO complex in an ATP-independent manner (Fig. 5A, lanes 1–4). Strikingly, however, neither CIP29 (Fig. 5B, lanes 1–4) nor Aly (Fig. 5C, lanes 1–4) associated with UAP56 in the absence of ATP. To further characterize the ATP dependence of the association of CIP29 and Aly with UAP56, we incubated nuclear extract in the presence of ATP, size-fractionated the extract by gel filtration, and then carried out Westerns across the column. As shown in Figure 6A, UAP56, CIP29, and Aly largely coeluted in the middle of the column together with a significant level of the THO complex (which was also present in fractions containing higher-molecular-weight complexes). IP/Westerns from fractions 37–43 showed that UAP56, CIP29, Aly, and the THO complex coimmunoprecipitated in the presence of ATP (Fig. 6B). We next examined the association of TREX complex proteins after gel filtration of nuclear extract lacking ATP. These data showed that CIP29 and UAP56 peaked near the middle of the column (fractions 34–49), similar to their profile in the presence of ATP, whereas the peaks of Aly and the THO complex shifted to a region containing larger complexes (fractions 19–31) relative to their profile in the presence of ATP (Fig. 6C). Significantly, UAP56 and Aly did not coimmunoprecipitate in fractions 19–28, and UAP56 and CIP29 did not coimmunoprecipitate in fractions 37–43 (Fig. 6D), consistent with our observation that these interactions are ATP-dependent in the unfractionated nuclear extract (Fig. 5B,C). In contrast, UAP56 did coimmunoprecipitate with the THO complex in the absence of ATP (Fig. 6D), consistent with the observation that this interaction is ATP-independent in the unfractionated nuclear extract (Fig. 5A). Importantly, when ATP was added back to fractions 19–28, Aly and UAP56 now coimmunoprecipitated (Fig. 6E). Likewise, addition of ATP to fractions 37–43 resulted in coimmunoprecipitation (co-IP) of CIP29 and UAP56 (Fig. 6E). These data confirm the ATP dependence of the association between UAP56 and both CIP29 and Aly, and also reveal ATP-dependent remodeling of the TREX complex and/or of the TREX complex and other components in the nuclear extract (see below; Discussion).
Figure 5.
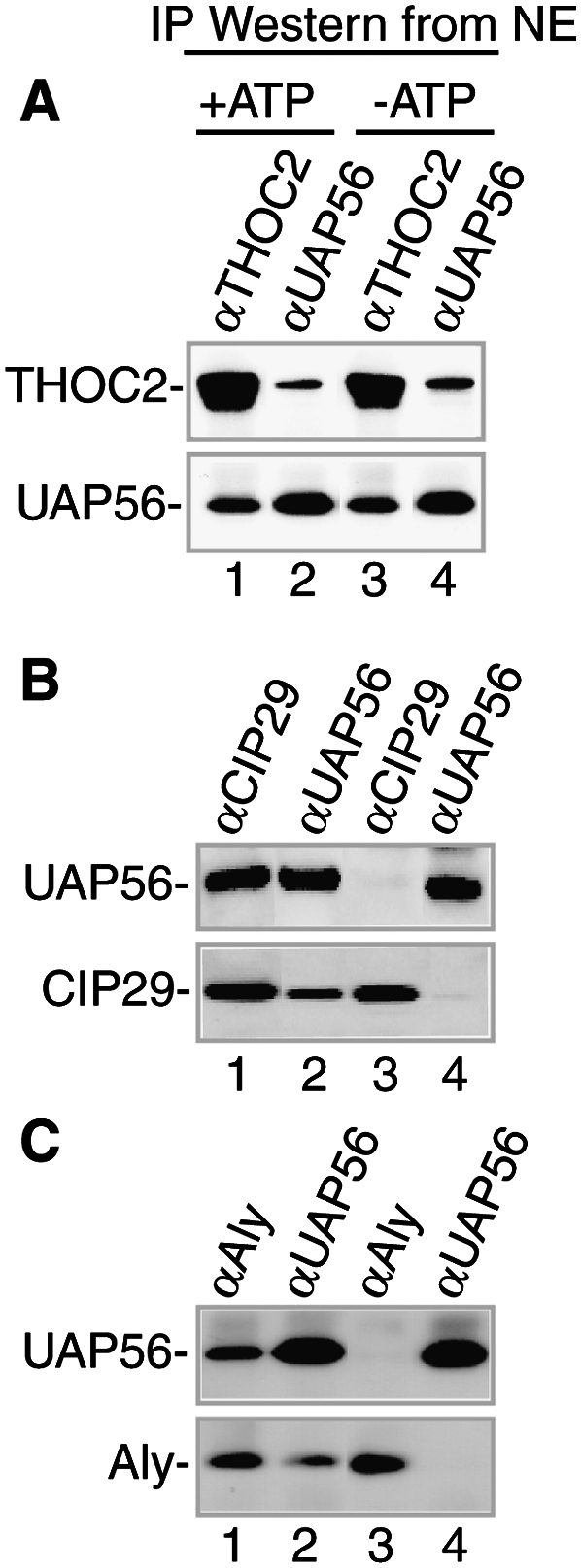
UAP56 interacts with both CIP29 and Aly in an ATP-dependent manner in the nuclear extract. IPs from nuclear extract with the indicated antibodies in the presence or absence of ATP were used for Westerns with antibodies against THOC2 and UAP56 (A), UAP56 and CIP29 (B), and UAP56 and Aly (C).
Figure 6.
ATP-dependent remodeling of the TREX complex in nuclear extract. (A) Nuclear extracts were separated on a gel filtration column in the presence of ATP followed by Westerns of fractions using the indicated antibodies. (B) IPs from +ATP fractions were used for Western analysis with the indicated antibodies. (C) Same as A, except nuclear extracts were separated in the absence of ATP. (D) IPs from −ATP fractions were used for Western analysis with the indicated antibodies. (E) IPs from −ATP fractions with ATP added back were used for Western analysis with the indicated antibodies. (*) Antibody heavy chain. The 60S and 30S size markers are from the spliceosome and hnRNP complex, respectively.
CIP29 and Aly interact directly and simultaneously with UAP56
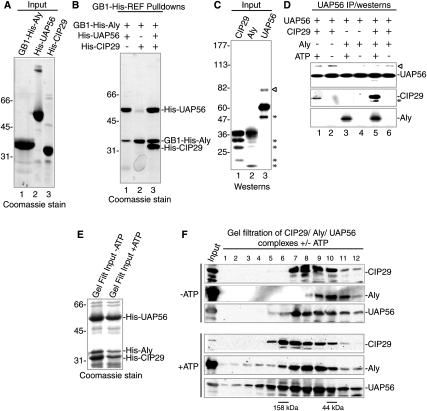
In light of our observation that UAP56 mediates the association between the THO complex and both CIP29 and Aly (Fig. 4), we next asked whether these two proteins can bind simultaneously to UAP56. To do this, we expressed GB1-His-Aly, His-tagged UAP56, and His-tagged CIP29 in Escherichia coli. The purified proteins are shown in Figure 7A (lanes 1–3). GB1-His-Aly was then bound to IgG beads and used for pull-downs. Consistent with previous work showing that UAP56 interacts directly with Aly (Luo et al. 2001), UAP56 was pulled down by GB1-His-Aly (Fig. 7B, lane 1). However, no interaction was detected between GB1-His-Aly and CIP29 (Fig. 7B, lane 2). In contrast, both UAP56 and CIP29 were pulled down by GB1-His-Aly to form a trimeric complex (Fig. 7B, lane 3). We conclude that CIP29 and Aly do not associate with each other directly, but do interact directly and simultaneously with UAP56 to form a trimeric complex.
Figure 7.
CIP29, Aly, and UAP56 assemble an ATP-dependent trimeric complex. (A,B) CIP29 and Aly interact directly and simultaneously with UAP56. (A) Coomassie-stained gel of purified GB1-His-Aly, His-UAP56, and His-CIP29 used for pull-downs in B. (B) Coomassie-stained gel of GB1-His-Aly pull-downs from a mixture of GB1-His-Aly and His-UAP56 (lane 1), GB1-His-Aly and His-CIP29 (lane 2), or GB1-His-Aly, His-UAP56, and His-CIP29 (lane 3). (C,D) CIP29, Aly, and UAP56 form an ATP-dependent trimeric complex. (C) Westerns of purified CIP29, Aly, and UAP56 used for IPs in D. (D) Westerns of UAP56 IPs from a mixture of purified UAP56 and CIP29 (lanes 1,2), UAP56 and Aly (lanes 3,4), or UAP56, CIP29, and Aly (lanes 5,6) with the indicated antibodies. IPs were carried out in the presence (lanes 1,3,5) or absence (lanes 2,4,6) of ATP. (E) Coomassie-stained gel of purified His-UAP56, His-CIP29, and His-Aly used for gel filtration studies in F. (F) Gel filtration of His-UAP56, His-CIP29, and His-Aly trimeric complex in the presence or absence of ATP.
CIP29, Aly, and UAP56 form an ATP-dependent trimeric complex
In our studies above, using unfractionated nuclear extract or nuclear extract fractionated by gel filtration, we observed that association of UAP56 with both CIP29 and Aly was ATP-dependent. Thus, we next asked whether ATP affected interactions between the three purified recombinant proteins. In the first assay, we purified GST fusions of CIP29, Aly, and UAP56 and then cleaved off the GST tag with thrombin. Western analysis of these proteins is shown in Figure 7C (lanes 1–3). When these proteins were mixed together at high concentrations (50 ng/μL) and UAP56 was used for IPs, the trimeric complex containing all three proteins was observed (data not shown). Strikingly, however, when the proteins were diluted 10-fold, we detected an ATP-dependent interaction between UAP56 and CIP29, but no interaction was detected between these proteins in the absence of ATP (Fig. 7D, lanes 1,2). Likewise, the interaction between UAP56 and Aly was ATP-dependent with the dilute proteins (Fig. 7D, lanes 3,4). When all three proteins were mixed together, we observed that the presence of Aly stimulated the interaction between UAP56 and CIP29 (Fig. 7D, lanes 1,5). Moreover, formation of the trimeric complex was also completely ATP-dependent using the dilute proteins (Fig 7D, lanes 5,6). The observation that formation of the trimeric complex is ATP-independent when the recombinant proteins are concentrated is consistent with previous studies of ATPases showing that concentrated proteins can bypass the ATP requirement for interactions (Crute et al. 1983; Snyder et al. 2004).
As another approach for assaying the role of ATP in formation of the Aly/CIP29/UAP56 complex, His-tagged versions of the proteins were purified from E. coli, mixed together in the presence or absence of ATP (Fig. 7E), and then fractionated by gel filtration on a Superose 12 column (Fig. 7F). Significantly, Western analysis revealed that all three proteins eluted in higher-molecular-weight fractions in the presence versus the absence of ATP (Fig. 7F). ATP had no effect on the elution profile of the individual purified proteins (data not shown). As shown in Figure 7F, CIP29, Aly, and UAP56 did not coelute entirely even in the presence of ATP. This may be because, after gel filtration, there is a heterogeneous mixture of the ATP-dependent UAP56-CIP29, UAP56-Aly, and trimeric complex as well as isolated proteins. However, the observation that all three proteins elute in higher-molecular-weight fractions only in the presence of ATP is consistent with UAP56, CIP29, and Aly forming an ATP-dependent trimeric complex.
Discussion
In this study, we identified CIP29 as a new TREX component conserved from yeast to humans. The conclusion that CIP29 is a bona fide TREX component is based on the observations that CIP29 is present in nuclear speckle domains, is efficiently recruited to mRNA in a splicing- and cap-dependent manner, coimmunoprecipitates with all known TREX components, and associates with them in a stoichiometry similar to that of previously characterized TREX components. Proteomic analysis of CIP29, UAP56, and THO complex IPs revealed that all of the known TREX components and CIP29 were present in all three IPs. Moreover, our data indicate that CIP29 functions in mRNA export. In addition to CIP29, our proteomic analysis identified five putative new TREX components, and further work is required to determine their relationship to the TREX complex. In contrast to CIP29, none of the five putative new TREX proteins have yeast orthologs readily detectable by sequence alignment. Thus, our work likely completes the identification of the sequence-conserved TREX components. Although it is not yet known whether the CIP29 ortholog Tho1 is a component of the yeast TREX complex, this possibility is consistent with the observation that yeast Tho1 functions in mRNP biogenesis and export and is recruited to mRNA by the THO complex (Jimeno et al. 2006).
UAP56, CIP29, and Aly form an ATP-dependent trimeric complex
The DEAD-box helicase UAP56 has been studied intensely since it was first identified as a splicing factor (Fleckner et al. 1997; Shen et al. 2008) and later as an essential mRNA export factor (Gatfield et al. 2001; Luo et al. 2001; Kapadia et al. 2006; Taniguchi and Ohno 2008). Our work indicates that UAP56 mediates the interaction with CIP29 and the THO complex to form TREX. Consistent with our previous work (Masuda et al. 2005), we also found that UAP56 mediates the interaction between Aly and the THO complex. Unexpectedly, IP/Western studies in nuclear extracts revealed that the association of both CIP29 and Aly with UAP56 is ATP-dependent, whereas the interaction between the THO complex and UAP56 does not require ATP. Thus, our study indicates that assembly of the TREX complex is ATP-dependent. When we fractionated the nuclear extract by gel filtration, we observed that the associations among the THO complex, CIP29, UAP56, and Aly differed in the presence or absence of ATP. Thus, together, our data indicate that the TREX complex undergoes ATP-dependent remodeling among TREX components and/or other components in the nuclear extract.
As our data revealed that UAP56, which is an ATPase (Shen et al. 2008), mediates an interaction between CIP29 and Aly, we asked whether the association among any of these proteins, when purified from E. coli, was affected by ATP. When the three proteins were all present in high concentrations, we observed direct ATP-independent interactions between UAP56 and CIP29, and UAP56 and Aly, and also a trimeric complex containing all three proteins. Our data also showed that UAP56 interacts simultaneously and directly with both CIP29 and Aly, but CIP29 and Aly do not interact with each other directly. These results are consistent with previous work indicating no effect of ATP on Aly–UAP56 or CIP29–UAP56 interactions (Luo et al. 2001; Leaw et al. 2004; Sugiura et al. 2007). However, as previous studies of ATPases showed that an ATP requirement for protein–protein interactions could be bypassed when proteins are mixed together in excess (Crute et al. 1983; Snyder et al. 2004), we repeated our analysis of purified CIP29, Aly, and UAP56 using dilute proteins. Strikingly, this analysis revealed that the association between CIP29 and UAP56, as well as the association between Aly and UAP56, was strictly ATP-dependent. Moreover, our data indicated that Aly, CIP29, and UAP56 form an ATP-dependent trimeric complex. Further support for the conclusion that Aly, CIP29, and UAP56 interact in an ATP-dependent manner came from analysis of the purified proteins by gel filtration. These data revealed that the three proteins eluted in a higher-molecular-weight region of the column in the presence versus the absence of ATP, consistent with the conclusion that ATP leads to interactions among the proteins. Thus, together, our data using purified proteins may be recapitulating, at least in part, the ATP requirement for association between Aly and CIP29 with UAP56 in the nuclear extract. Consistent with our observation that CIP29 and Aly associate with UAP56 in an ATP-dependent manner, previous work showed that CIP29 stimulates the helicase activity of UAP56 (Sugiura et al. 2007) and that Aly stimulates the ATPase activity of UAP56 (Taniguchi and Ohno 2008). In addition, Ohno and coworkers (Taniguchi and Ohno 2008) found that ATP stimulates the association of both UAP56 and Aly with RNA. These previous observations, together with our work, indicate that CIP29 and Aly are key regulators of UAP56, and further studies are needed to understand how these proteins participate in the functions of UAP56 during formation of the TREX complex and at other steps along the export pathway.
Precedent for formation of an ATP-dependent trimeric complex involving a helicase and two accessory factors comes from extensive studies of the prototypic DEAD-box helicase eIF4A, which functions to unwind secondary structure at the 5′ end of mRNA to promote scanning for the initiation codon during translation initiation (Marintchev et al. 2009 and references therein). In this case, eIF4A associates with both eIF4G (4G) and eIF4H (4H; homolog of eIF4B) in an ATP-dependent manner (Marintchev et al. 2009). Thus, it is possible that Aly and CIP29 are counterparts of 4G and 4H. Continuing the analogy, it is possible that, as with the eIF4A/4G/4H helicase complex, the UAP56/Aly/CIP29 complex may function as a helicase to unwind RNA secondary structure at the 5′ end of mRNA. Similar to the eIF4A/4G/4H helicase complex, the UAP56/Aly/CIP29 helicase complex is recruited to the 5′ end of the RNA via the 5′ cap. Just as the eIF4A/4G/4H complex unwinds RNA to create a landing pad for ribosome scanning, the UAP56/Aly/CIP29 helicase complex may unwind the 5′ end mRNA secondary structure to allow recruitment of the THO complex, TAP recruitment, and/or proper transit through the nuclear pore complex.
It is well established that multiple steps in spliceosome assembly are ATP-dependent (Wahl et al. 2009), and our work shows that assembly of TREX is also ATP-dependent. Numerous DEAD-box helicases function throughout the spliceosome assembly pathway, explaining in part the many ATP-dependent steps. As with the spliceosome, our work shows that TREX assembly requires ATP and the DEAD-box helicase UAP56. In light of their major role in remodeling complexes, it is likely that UAP56 functions in remodeling TREX, and, in this case, at least two proteins, CIP29 and Aly, play roles. Our gel filtration analysis of TREX components showed that they do not elute as one single complex and that they elute differently in the presence or absence of ATP. Thus, it is possible that there are multiple forms of TREX, possibly reflecting ATP-dependent dynamic changes in this complex during its recruitment to mRNA and in subsequent remodeling steps in the mRNA export pathway. Different forms of TREX may also function in exporting different mRNAs and/or regulating the export of certain mRNAs.
Materials and methods
Plasmids
The CIP29 clone was obtained from Origene (catalog no. SC309293). The GST-CIP29 plasmid was constructed by inserting a PCR fragment containing the CIP29 ORF into the EcoRI and NotI sites of pGEX4T-1. GST-Aly and GST-UAP56 plasmids were described (Bruhn et al. 1997; Fleckner et al. 1997; Luo et al. 2001). The plasmids encoding AdML pre-mRNA and Δi-mRNA (Luo et al. 2001) were digested with BamHI and transcribed with T7 RNA polymerase. To construct the plasmids encoding HA-CIP29, HA-Aly, and HA-eIF4A3, a 3′ HA tag was added to each ORF by PCR. These PCR fragments were then inserted into a vector designated pCDCI, which was constructed by inserting a BglII/NotI fragment from pCI-neo (Promega) into the same sites of pcDNA3.1/Hygro (Invitrogen). PCR fragments were inserted into pCDCI using the XhoI/XbaI sites for HA-CIP29, XhoI/EcoRV sites for HA-eIF4A3, and BamHI/XbaI sites for HA-Aly. MS2-GFP, MS2-Aly, His-UAP56, pET24b-GB1-6His-ALY, and pLUCSALRRE6MS2 (MS2 reporter) plasmids were described (Hautbergue et al. 2009). The reporter plasmid lacking MS2 sites was described (Williams et al. 2005). The MS2-CIP29 plasmid was constructed by inserting a PCR fragment containing the CIP29 ORF into XbaI/NotI of pCINeo-MS2. The His-CIP29 plasmid was constructed by inserting a PCR fragment containing the CIP29 ORF into NdeI/XhoI of pET24b.
Antibodies
A rabbit polyclonal antibody to CIP29 was raised against full-length GST-CIP29. Antibodies to Aly, UAP56, THOC2, THOC5, CBP80, and eIF4A3 were described (Zhou et al. 2000; Choong et al. 2001; Ferraiuolo et al. 2004; Masuda et al. 2005; Cheng et al. 2006). The negative control antibodies used in the protein and RNA IPs were against fSAP130 (Das et al. 2000) and Smad (generous gift from M. Whitman), respectively. The SC35 antibody was from Sigma.
Protein IPs, mass spectrometry, and immunodepletions
Antibodies were coupled to protein A Sepharose at a 1:2 ratio and covalently cross-linked using dimethylpimelimidate (Sigma). A 1-mL splicing reaction containing 300 μL of HeLa nuclear extract, 300 μL of SDB (20 mM HEPES at pH 7.9, 100 mM KCl), 500 μM ATP, 3.2 mM MgCl2, 20 mM creatine phosphate, and 50 ng/μL RNase A was incubated for 30 min at 30°C. The reaction mixture was added to 500 μL of IP buffer (1× PBS, 0.1% Triton, 0.2 mM PMSF, protease inhibitor EDTA-free [Roche]) and 80 μL of antibody-cross-linked beads. IPs were carried out overnight at 4°C, followed by six 1.5-mL washes with IP buffer. Proteins were eluted with 80 μL of SDS loading buffer followed by incubation for 20 min at room temperature. DTT was then added to a final concentration of 5 mM, and 7 μL of each IP was fractionated on a 4%–12% SDS gradient gel. Gels were stained with Coomassie. For IPs under −ATP conditions, ATP, MgCl2, and creatine phosphate were omitted. For mass spectrometry, 20 μL of the THOC2, UAP56, and CIP29 IPs were TCA-precipitated, and the total samples were used (Gygi et al. 1999). Keratins and abundant cellular proteins that were likely contaminants, including desmoplakin, actin, tubulin, myosin, and translation proteins, were omitted from Table 1. Immunodepletion of proteins were carried out according to Zhou and Reed (1998).
RNA IPs
AdML pre-mRNA was incubated for 2 h in standard 25-μL splicing reaction mixtures (Krainer et al. 1984). Rabbit polyclonal antibodies (5–10 μL) were coupled to 10 μL of protein A Sepharose beads. Splicing reaction mixtures (10 μL) were added to 200 μL of RNA-binding buffer (20 mM HEPES at pH 7.9, 150 mM KCl, 0.1% Triton, 2.5 mM EDTA, 5 mM DTT) and mixed with 10 μL of protein A Sepharose beads coupled to antibodies. IPs were performed for 2 h at 4°C. Beads were washed four times with 1 mL of binding buffer. Total RNA was analyzed on 6.5% denaturing polyacrylamide gels and visualized by PhosphorImager.
Cell culture and transfection
HeLa and Cos cells were cultured in DMEM supplemented with 10% FBS. For overexpression studies in HeLa cells, 1.2 μg of plasmids encoding HA-tagged proteins was transfected using Lipofectamine 2000 into cells that were 60% confluent on 35-mm dishes and incubated for 24 h. For overexpression studies in Cos cells, 800 ng of plasmids encoding Flag-tagged proteins was transfected using Turbofect (Fermentas) and incubated for 36 h.
FISH and IF
To detect polyA+ RNA, an HPLC-purified oligo dT(70) probe labeled at the 5′ end with Alexa Fluor 546 NHS Ester was used. HeLa cells were washed once with PBS, fixed for 10 min with 4% paraformaldehyde, washed three times with PBS, permeabilized with 0.5% Triton X-100 for 5 min, and washed twice with PBS and then once with 2× SSC for 10 min at room temperature. An oligo dT(70) probe was then added at 1 ng/μL followed by incubation for 16–24 h at 42°C. Finally, the cells were washed for 15 min each, twice with 2× SSC, once with 0.5× SSC, and once with PBS at room temperature. Images were captured with an EM-CCD camera on an inverted microscope (200M, Zeiss) using Metamorph software (Molecular Devices). Detection of polyA+ RNA in Cos cells was carried out as described previously and included a 2-h actinomycin D treatment of cells prior to fixation to reduce nascent RNA (Hautbergue et al. 2009). For IF, CIP29 (1:500), HA (1:200), and Flag M2 (1:500) were used as primary antibodies. The secondary antibodies were mouse Alexa-488 and rabbit Alexa-647 (Invitrogen) diluted to 1:1000 with IF solution (PBS, 0.1% Triton X-100, 2 mg/mL RNase-free BSA; Ambion).
MS2 assay
MS2-tethered pre-mRNA reporter assays were carried out as described with normalization for transfection efficiency using a β-galactosidase reporter (Hargous et al. 2006; Hautbergue et al. 2009). The TAP RNAi plasmid was constructed using pcDNA6.2-GW/EmGFP-miR (Invitrogen) containing the following target sequence: 5′-AACACGATGATGAACGCGTTA-3′. Leptomycin B was used at 10 nM. In all experiments, 50 ng of pLUCSALRRE6MS2 and 50 ng of β-galactosidase reporter plasmid were used per well of a 24-well plate. In Figure 3C, 700 ng of MS2 fusion expression vector was used. In Figure 3E, 350 ng of MS2 fusion expression vector was used and 350 ng of either control RNAi plasmid (pCDNA6.2-GW/EMGFP-miR-neg; Invitrogen) or TAP RNAi plasmid. In Figure 3F, 350 ng of MS2 fusion plasmid was used or 200 ng of CMV-REV expression vector (Williams et al. 2005), and the total DNA made up to 800 ng with pCINEO. All assays were carried out 48 h post-transfection.
Gel filtration of nuclear extracts and IPs from gel filtration column fractions
Nuclear extract was separated on a Sephacryl S500 (GE Healthcare, 17-0613-01) gel filtration column in the presence or absence of ATP. For +ATP conditions, a 2.5-mL splicing reaction containing 1500 μL of nuclear extract, 500 μM ATP, 3.2 mM MgCl2, and 20 mM creatine phosphate was incubated for 30 min at 30°C before loading the column. The gel filtration column buffer (20 mM HEPES, 60 mM KCL, 2.5 mM EDTA, 0.1% Triton) contained 1.5 mM ATP, 9.6 mM MgCl2, and 20 mM creatine phosphate. For −ATP conditions, ATP, MgCl2, and creatine phosphate were omitted from the incubation and column. For IPs, 1.2 mL of the indicated factions was used. For gel filtration of E. coli-purified His-UAP56, His-Aly, and His-CIP29, proteins were mixed together and dialyzed overnight in gel filtration buffer (20 mM Tris at pH 8.0, 100 mM NaCl, 1 mM DTT ± 2 mM ATP, 3 mM MgCl2) prior to separation on a Superose 12 column.
Pull-downs and IPs using purified proteins
Pull-downs using GB1-tagged proteins were carried out as described previously (Hautbergue et al. 2009). For IPs using purified proteins, GST was cleaved off of GST-Aly, GST-CIP29, and GST-UAP56 using biotinylated thrombin (Novagen). The proteins were further purified over glutathione and strepavidin beads to remove GST and biotinylated thrombin, respectively. A 500-μL reaction mixture containing 5 ng/μL each untagged protein, 1.25 mM ATP, 8 mM MgCl2, and 400 μL of protein-binding buffer (25 mM HEPES at pH 7.4, 150 mM KCL, 5% glycerol, 0.5% NP-40, 0.2 mM PMSF) was incubated for 20 min at 30°C. Ten microliters of antibody-cross-linked beads was added to each reaction mixture, and IPs were carried out overnight at 4°C followed by six 1.5-mL washes with protein-binding buffer. For IPs under −ATP conditions, ATP and MgCl2 were omitted.
Acknowledgments
We are grateful to H. Lei and A. Dias for comments on the manuscript. We thank C. Leinhard, G. Wagner, A. Johnson, and H. Cheng for useful discussions, and G. Hautbergue for help with the gel filtration analysis of purified proteins. HeLa cells were obtained from the National Cell Culture Center (Minneapolis, MN). This work was supported by an NIH grant to R.R. S.W. acknowledges support from the Wellcome Trust and BBSRC.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1898610.
Supplemental material is available at http://www.genesdev.org.
References
- Aguilera A 2005. Cotranscriptional mRNP assembly: From the DNA to the nuclear pore. Curr Opin Cell Biol 17: 242–250 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV 2000. SAP—A putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci 25: 112–114 [DOI] [PubMed] [Google Scholar]
- Bruhn L, Munnerlyn A, Grosschedl R 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev 11: 640–653 [DOI] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–1400 [DOI] [PubMed] [Google Scholar]
- Choong ML, Tan LK, Lo SL, Ren EC, Ou K, Ong SE, Liang RC, Seow TK, Chung MC 2001. An integrated approach in the discovery and characterization of a novel nuclear protein over-expressed in liver and pancreatic tumors. FEBS Lett 496: 109–116 [DOI] [PubMed] [Google Scholar]
- Crute JJ, LaDuca RJ, Johanson KO, McHenry CS, Bambara RA 1983. Excess β subunit can bypass the ATP requirement for highly processive synthesis by the Escherichia coli DNA polymerase III holoenzyme. J Biol Chem 258: 11344–11349 [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R 2000. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell 5: 779–787 [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Lee CS, Ler LW, Hsu JL, Costa-Mattioli M, Luo MJ, Reed R, Sonenberg N 2004. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci 101: 4118–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcarcel J, Green MR 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev 11: 1864–1872 [DOI] [PubMed] [Google Scholar]
- Forler D, Rabut G, Ciccarelli FD, Herold A, Kocher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E 2004. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol Cell Biol 24: 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Wu DW, Stark K, Pelus LM 2002. Cloning and characterization of a proliferation-associated cytokine-inducible protein, CIP29. Biochem Biophys Res Commun 292: 593–600 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol 11: 1716–1721 [DOI] [PubMed] [Google Scholar]
- Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, et al. 2009. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R 1999. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH 2006. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J 25: 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashii Y, Kim JY, Sawada A, Tokimasa S, Hiroyuki F, Ohta H, Makiko K, Takihara Y, Ozono K, Hara J 2004. A novel partner gene CIP29 containing a SAP domain with MLL identified in infantile myelomonocytic leukemia. Leukemia 18: 1546–1548 [DOI] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Walsh MJ, Snijders AP, Chang CT, Jones R, Ponting CP, Dickman MJ, Wilson SA 2009. UIF, a new mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol 19: 1918–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S, Luna R, Garcia-Rubio M, Aguilera A 2006. Tho1, a novel hnRNP, and Sub2 provide alternative pathways for mRNP biogenesis in yeast THO mutants. Mol Cell Biol 26: 4387–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Cubberley G, Bentley DL 2009. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol Cell 33: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia F, Pryor A, Chang TH, Johnson LF 2006. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene 384: 37–44 [DOI] [PubMed] [Google Scholar]
- Kohler A, Hurt E 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773 [DOI] [PubMed] [Google Scholar]
- Krainer AR, Maniatis T, Ruskin B, Green MR 1984. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36: 993–1005 [DOI] [PubMed] [Google Scholar]
- Leaw CL, Ren EC, Choong ML 2004. Hcc-1 is a novel component of the nuclear matrix with growth inhibitory function. Cell Mol Life Sci 61: 2264–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Semple JI, Brown SE, Counsell D, Campbell RD, Sanderson CM 2004. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics 83: 153–167 [DOI] [PubMed] [Google Scholar]
- Levesque L, Bor YC, Matzat LH, Jin L, Berberoglu S, Rekosh D, Hammarskjold ML, Paschal BM 2006. Mutations in tap uncouple RNA export activity from translocation through the nuclear pore complex. Mol Biol Cell 17: 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo MJ, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J 2008. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133: 303–313 [DOI] [PubMed] [Google Scholar]
- Malim MH, Cullen BR 1991. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: Implications for HIV-1 latency. Cell 65: 241–248 [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG 2001. Quality control of mRNA function. Cell 104: 173–176 [DOI] [PubMed] [Google Scholar]
- Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G 2009. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136: 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz C, Urlaub H, Will CL, Luhrmann R 2007. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA 13: 116–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Piruat JI, Aguilera A 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J 17: 4859–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Hurt E 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523–531 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Herold A, Gari K, Kocher T, Rode M, Ciccarelli FL, Wilm M, Izaurralde E 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11: 558–566 [DOI] [PubMed] [Google Scholar]
- Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR 2008. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev 22: 1796–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyk A, Szuminska M, Uniewicz KA, Graves LM, Kozlowski P 2006. Human enhancer of rudimentary is a molecular partner of PDIP46/SKAR, a protein interacting with DNA polymerase δ and S6K1 and regulating cell growth. FEBS J 273: 4728–4741 [DOI] [PubMed] [Google Scholar]
- Snyder AK, Williams CR, Johnson A, O'Donnell M, Bloom LB 2004. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: II. Uncoupling the β and DNA binding activities of the γ complex. J Biol Chem 279: 4386–4393 [DOI] [PubMed] [Google Scholar]
- Sugiura T, Sakurai K, Nagano Y 2007. Intracellular characterization of DDX39, a novel growth-associated RNA helicase. Exp Cell Res 313: 782–790 [DOI] [PubMed] [Google Scholar]
- Taniguchi I, Ohno M 2008. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol Cell Biol 28: 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Williams BJ, Boyne JR, Goodwin DJ, Roaden L, Hautbergue GM, Wilson SA, Whitehouse A 2005. The prototype γ-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem J 387: 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Reed R 1998. Human homologs of yeast prp16 and prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J 17: 2095–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405 [DOI] [PubMed] [Google Scholar]