Abstract
G protein-coupled receptor kinase 2 (Gprk2/GRK2) plays a conserved role in modulating Hedgehog (Hh) pathway activity, but its mechanism of action remains unknown. Here we provide evidence that Gprk2 promotes high-level Hh signaling by regulating Smoothened (Smo) conformation through both kinase-dependent and kinase-independent mechanisms. Gprk2 promotes Smo activation by phosphorylating Smo C-terminal tail (C-tail) at Ser741/Thr742, which is facilitated by PKA and CK1 phosphorylation at adjacent Ser residues. In addition, Gprk2 forms a dimer/oligomer and binds Smo C-tail in a kinase activity-independent manner to stabilize the active Smo conformation, and promotes dimerization/oligomerization of Smo C-tail. Gprk2 expression is induced by Hh signaling, and Gprk2/Smo interaction is facilitated by PKA/CK1-mediated phosphorylation of Smo C-tail. Thus, Gprk2 forms a positive feedback loop and acts downstream from PKA and CK1 to facilitate high-level Hh signaling by promoting the active state of Smo through direct phosphorylation and molecular scaffolding.
Keywords: Hedgehog, Smo, Gprk2/GRK2, GPCR, conformation, phosphorylation
The Hedgehog (Hh) family of secreted proteins plays important roles during embryonic development and adult tissue homeostasis (Ingham and McMahon 2001; Jiang and Hui 2008; Varjosalo and Taipale 2008). Aberrant Hh signaling has been implicated in numerous human disorders, including birth defects and cancers (Villavicencio et al. 2000; Taipale and Beachy 2001). In embryonic development, Hh often functions as a morphogen to specify different cell fates in a concentration-dependent manner (Jiang and Hui 2008). For example, in Drosophila wing development, posterior (P)-compartment cells express and secrete Hh proteins that move into the anterior (A) compartment to form a local concentration gradient that induces the expression of distinct Hh target genes at different concentrations. Low levels of Hh signaling suffice to activate decapentaplegic (dpp), whereas intermediate and high levels of Hh signaling activity are required to activate patched (ptc) and engrailed (en), respectively (Strigini and Cohen 1997; Ohlmeyer and Kalderon 1998; Methot and Basler 1999; Jia et al. 2004).
The reception system for the Hh signal consists of three transmembrane proteins: a 12-transmembrane protein, Patched (Ptc), as the Hh receptor; a single-span transmembrane protein, Ihog, as a coreceptor; and a seven-transmembrane protein, Smoothened (Smo), as the obligatory Hh signal transducer (Jiang and Hui 2008; Varjosalo and Taipale 2008). Ptc inhibits Smo substoichiometrically through a poorly defined mechanism in the absence of Hh (Taipale et al. 2002). Hh interacts physically with Ptc and Ihog to alleviate Ptc inhibition of Smo (Chen and Struhl 1996; Stone et al. 1996; Casali and Struhl 2004; Zheng et al. 2010), allowing Smo to activate the latent Zn finger transcription factor Cubitus interruptus (Ci)/Gli (Jiang and Hui 2008; Varjosalo and Taipale 2008).
In Drosophila wing development, Ci plays dual roles that are performed by two distinct forms (Aza-Blanc et al. 1997; Methot and Basler 1999). In the absence of Hh, the full-length Ci (CiF) undergoes extensive phosphorylation by PKA, GSK3, and CK1, which targets CiF for SCFSlimb-mediated proteolytic processing to generate a truncated repressor form (CiR) (Jia et al. 2002, 2005; Price and Kalderon 2002; Smelkinson et al. 2007). CiF forms complexes with the Ser/Thr kinase Fused (Fu), the kinesin-related protein Costal2 (Cos2), and Suppressor of fused (Sufu), which impedes CiF nuclear translocation and blocks its activity in the nucleus (Robbins et al. 1997; Methot and Basler 2000; Wang et al. 2000; Wang and Holmgren 2000; Wang and Jiang 2004; Sisson et al. 2006). In addition, Cos2 facilitates Ci phosphorylation by recruiting PKA, GSK3, and CK1 (Zhang et al. 2005). In the presence of Hh, activated Smo dissociates or changes the composition of Ci–Cos2–kinase complexes, thereby impeding Ci phosphorylation and processing (Zhang et al. 2005). Smo may also regulate Ci processing through Gαi (Ogden et al. 2008). Furthermore, high levels of Hh convert CiF into an active but labile form (CiA) by alleviating Sufu-mediated repression through Fu (Ohlmeyer and Kalderon 1998).
How Smo is activated and how Smo transduces different levels of Hh signaling activity are still poorly understood. In Drosophila, Hh induces Smo phosphorylation at multiple sites in its C-terminal tail (C-tail) by PKA and CK1, which activates Smo by promoting both its cell surface accumulation and active conformation (Jia et al. 2004; Zhang et al. 2004; Apionishev et al. 2005; Zhao et al. 2007). In addition, the levels of Smo activity appear to correlate with its levels of phosphorylation (Jia et al. 2004; Zhao et al. 2007). In vertebrates, Sonic hedgehog (Shh) induces ciliary accumulation of Smo, which correlates with Shh pathway activation (Jiang and Hui 2008). In addition, Shh promotes Smo to adopt an active conformation similar to that of Drosophila Smo (dSmo), although the canonical PKA/CK1 phosphorylation sites found in dSmo are not present in mammalian Smo (Zhao et al. 2007).
Several studies have suggested that G protein-coupled receptor (GPCR) kinase 2 (GRK2) promotes Shh signaling by regulating Smo (Chen et al. 2004; Meloni et al. 2006; Philipp et al. 2008), raising the possibility that GRK2 may substitute the role of PKA/CK1 to activate Smo in vertebrates. Interestingly, genetic studies in Drosophila suggest that Gprk2 also modulates Hh signaling (Molnar et al. 2007; Cheng et al. 2010). Nevertheless, the mechanism by which Gprk2/GRK2 regulates Hh signaling remains unknown in any system.
Although PKA and CK1 play an essential role in Smo activation in Drosophila, additional mechanisms may exist because the activity of a phosphomimetic Smo with all of the PKA/CK1 phosphorylation sites converted to Asp was still up-regulated by Hh (Jia et al. 2004). In addition, Hh induces Smo phosphorylation at many additional sites by unidentified kinases (Zhang et al. 2004). To search for additional components that regulate Smo and Hh signaling, we carried out a genetic modifier screen and identified Gprk2 as a positive regulator of Hh signaling and essential for optimal Smo activation. We provide evidence that Gprk2 regulates Smo conformation and activity by directly phosphorylating Smo at S741/T742, and this phosphorylation is regulated by PKA/CK1 phosphorylation at adjacent sites. Unexpectedly, we uncovered a kinase activity-independent mechanism for Gprk2 to regulate Hh signaling. We demonstrate that Gprk2 forms a dimer/oligomer and binds Smo C-tail in a manner regulated by PKA and CK1 phosphorylation to facilitate the dimerization/oligomerization of Smo C-tail, thereby promoting the active Smo conformation.
Results
Inactivation of Gprk2 enhances the defects caused by partial loss of Smo function
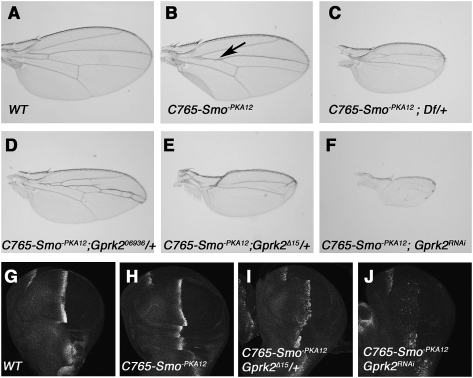
To identify additional Hh signaling components, we carried out a genetic modifier screen in which a collection of Exilexis deficiency lines were screened for modifications of a “fused wing” phenotype caused by overexpression of a dominant-negative form of Smo (Smo−PKA12) using wing-specific Gal4 drivers MS1096 or C765 (see the Materials and Methods). We identified more than a dozen deficiencies that enhanced the “fused wing” phenotype, one of which is Df(3R)Exel6219, which removes 10 genes, including the Drosophila Gprk2 (Fig. 1A–C). As mammalian GRK2 had been implicated in regulating Shh signaling (Chen et al. 2004), we proceeded to determine whether the modification was due to loss of Gprk2. We found that the heterozygote for Gprk206936, a P-element insertion allele of Gprk2 (Schneider and Spradling 1997), enhanced the phenotype caused by expressing Smo−PKA12 with C765 (C765-Smo−PKA12) (Fig. 1D). The modification by Gprk206936 is less severe compared with Gprk2 deficiency, likely due to its hypomorphic nature (Schneider and Spradling 1997). Indeed, several imprecise excision lines, including Gprk2Δ15, modified C765-Smo−PKA12 to the same extent as Gprk2 deficiency (Fig. 1E). Consistent with Gprk2Δ15 being a strong or null allele, we found that Gprk2 protein expression was diminished in Gprk2Δ15 mutant clones (Supplemental Fig. S1A). Gprk2Δ15 homozygotes were viable when grown at 25°C but were semilethal at 29°C, and escapers exhibited the “fused wing” phenotype (Supplmental Fig. S1B–D), which is indicative of partial loss of Hh signaling activity. We also generated transgenic RNAi lines targeting the Gprk2 coding sequence (Materials and Methods) and found that Gprk2 RNAi greatly enhanced the C765-Smo−PKA12 phenotype (Fig. 1F). The modification of C765-Smo−PKA12 by loss of Gprk2 is also reflected by changes in Hh target gene expression. For example, C765-Smo−PKA12 caused a reduction in ptc expression in A-compartment cells near the A/P boundary (Fig. 1H). Removal of one copy of Gprk2 further reduced, whereas Gprk2 RNAi nearly abolished, ptc expression in C765-Smo−PKA12 wing discs (Fig. 1I,J). Thus, reduction in Gprk2 activity exacerbates the Hh signaling defects caused by compromised Smo activity.
Figure 1.
Reduction of Gprk2 modifies the phenotypes caused by a dominant-negative Smo. (A,B) A wild-type wing (A) or a wing expressing UAS-Smo−PKA12 with the C765 Gal4 driver (C765-Smo−PKA12) (B). (Arrow in B) Overexpression of Smo−PKA12 causes a partial fusion between vein 3 and vein 4. (C–F) Heterozygote for a Gprk2 deficiency (C), a P-element insertion allele of Gprk2 (Gprk206936) (D), an excision allele (Gprk2Δ15) (E), or Gprk2 RNAi (F) enhanced the fused wing phenotype caused by C765-Smo−PKA12. (G–J) C765-Smo−PKA12 reduces ptc expression (H), which is enhanced by the Gprk2Δ15 heterozygote (I) or Gprk2 RNAi (J).
Gprk2 is required for high-level Hh signaling
To further investigate the role of Gprk2 in Hh signaling, we examined Hh target gene expression in wing discs in which Gprk2 is inactivated by either genetic mutations or RNAi. While levels of ptc and en expression were reduced in Gprk2Δ15 homozygous mutant discs (Supplemental Fig. S2E–E″,F–F″), dpp was expressed at levels comparable with those in wild-type discs (Supplemental Fig. S2D–D″). In Gprk2Δ15 mutant discs, CiF was accumulated in A-compartment cells near the A/P boundary similar to that in wild-type discs (indicated by arrows in Supplemental Fig. S2D–F), suggesting that loss of Gprk2 does not affect the low-level Hh signaling required for blocking Ci processing. However, CiF was elevated in A-compartment cells immediately adjacent to the A/P boundary (indicated by arrowheads in Supplemental Fig. S2D–F), implying that loss of Gprk2 prevents the conversion of CiF into the labile CiA, a process requiring high-level Hh signaling (Ohlmeyer and Kalderon 1998). Gprk2 RNAi also reduced en expression and prevented the conversion of CiF into the labile CiA but did not affect Ci processing (Supplemental Fig. S3). These data are consistent with two recent studies (Molnar et al. 2007; Cheng et al. 2010). Thus, Gprk2 is required for high levels but not low levels of Hh signaling activity.
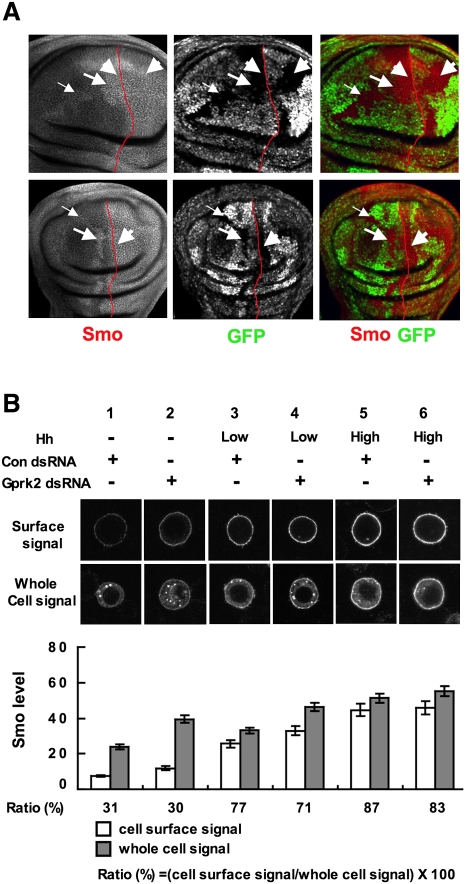
To determine whether Gprk2 is required in Hh-receiving cells to promote Hh signaling activity, we generated Gprk2Δ15 mutant clones using the FRT/FLP-mediated mitotic recombination. We found that Gprk2Δ15 mutant clones near the A/P boundary exhibited reduced levels of ptc and en expression in a cell-autonomous fashion (Fig. 2D–D″,F–F″), whereas P-compartment clones abutting the A/P boundary did not affect Hh target gene expression (Fig. 2E–E″). Taken together, these observations suggest that Gprk2 acts in the Hh-receiving cells to promote high-level Hh signaling.
Figure 2.
Genetic characterization of Gprk2. (A–C) A Gprk2-lacZ enhancer trap is expressed along the A/P border (A), and is induced ectopically by misexpression of either Hh (B) or an activated form of Ci (C) using MS1096 Gal4 driver. (D–F″) A-compartment Gprk2Δ15 mutant cells near the A/P boundary (marked by the lack of GFP) (arrows in D–D″,F–F″) exhibited reduced expression of ptc (D′) and en (F′). Insets in D′ and D″ show enlarged images of the region indicated by the arrows. (E–E″) P-compartment Gprk2Δ15 mutant cells abutting the A/P boundary did not affect ptc expression. (G–J) Wing discs expressing SmoΔSAID (G,H) or SmoSD123 (I,J) with (H,J) or without (G,I) multiple copies of a Gprk2 RNAi transgene using MS1096 were immunostained to show en expression. SmoΔSAID but not SmoSD123 induced ectopic expression of en when Gprk2 was knocked down. (K–N) Gprk2Δ15 mutant discs expressing a wild-type Gprk2 (L), a kinase-dead Gprk2 (Gprk2KM) (M), or a mammalian GRK5 (mGRK5) (N) with MS1096 were immunostained to show en expression. Expression of the wild-type Gprk2 or mGRK5 but not Gprk2KM rescued anterior en expression in Gprk2Δ15 mutant discs. The red lines demarcate the A/P border based on Ci expression (not shown).
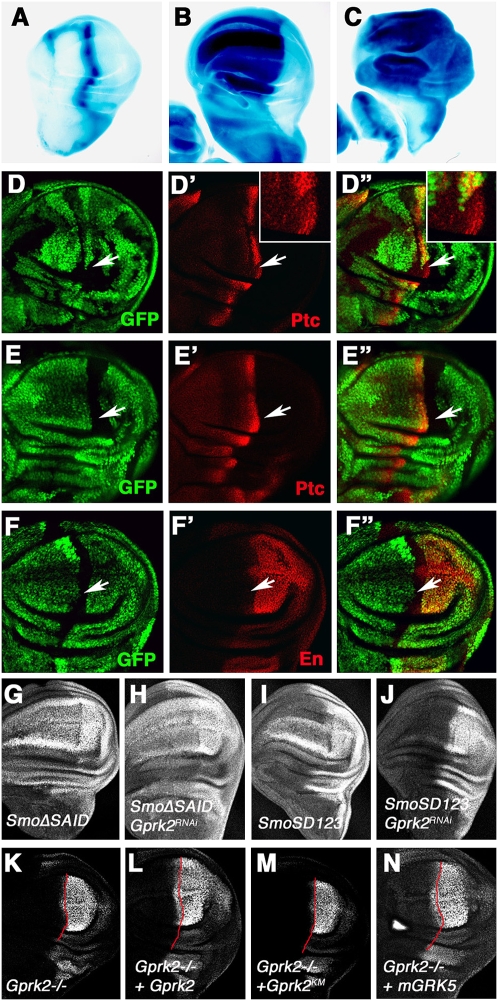
Gprk206936 harbors a lacZ reporter in the 5′ untranslated region (UTR) of the Gprk2 gene (Schneider and Spradling 1997). Both Gprk2-lacZ and Gprk2 protein are up-regulated along the A/P boundary of imaginal discs (Schneider and Spradling 1997), raising the possibility that Gprk2 transcription is regulated by Hh signaling. Indeed, Gprk2-lacZ was induced ectopically by misexpression of Hh or an activated form of Ci using the MS1096 Gal4 driver (Fig. 2A–C; Molnar et al. 2007) and its expression was suppressed by Ptc overexpression (Molnar et al. 2007), suggesting that Gprk2 transcription is up-regulated by Hh signaling.
Gprk2 regulates Smo activity
In the GPCR signaling pathway, GRKs directly phosphorylate GPCRs in response to ligand stimulation, leading to desensitization of GPCR signaling (Pitcher et al. 1998). To determine how Gprk2 participates in Hh signaling, we determined the epistatic relationship between Gprk2 and Smo, which is structurally related to GPCRs. We examined two Smo variants: a phosphomimetic form (SmoSD123) with three clusters of PKA/CK1 sites converted to Asp (Jia et al. 2004); and SmoΔ661–818 (or SmoΔSAID), in which the Smo autoinhibitory domain (SAID) was deleted (Zhao et al. 2007). Previous studies showed that both SmoSD123 and SmoΔSAID exhibit constitutive Hh signaling activity and induce the ectopic expression of high-threshold Hh-responsive genes, including en, when overexpressed in wing discs (Fig. 2G,I; Jia et al. 2004; Zhao et al. 2007). Consistent with a previous study (Molnar et al. 2007), Gprk2 RNAi diminished the ectopic en expression and reduced the ectopic ptc expression induced by SmoSD123 (Fig. 2J; data not shown), suggesting Gprk2 is indispensable for the optimal activity of SmoSD123. In contrast, the activity of SmoΔSAID was insensitive to loss of Gprk2, as SmoΔSAID induced similar levels of ectopic ptc and en expression in Gprk2 RNAi discs (Fig. 2H; data not shown). These results suggest that Gprk2 acts at a step downstream from PKA/CK1 phosphorylation to counteract Smo autoinhibition.
Gprk2 kinase activity is required for Hh signaling
To determine if Gprk2 kinase activity is required for its function in the Hh pathway, we generated UAS transgenes expressing a wild-type or a kinase-dead form of Gprk2 (Gprk2KM) (see the Materials and Methods; below). Overexpression of UAS-Gprk2 but not UAS-Gprk2KM rescued anterior en expression in Gprk2Δ15 mutant discs (Fig. 2L,M), suggesting that the kinase activity of Gprk2 is essential for high-level Hh signaling. We also generated a UAS transgene expressing a mammalian GRK5 (mGRK5), which is highly homologous to Drosophila Gprk2. We found that expression of mGRK5 also rescued anterior en expression in Gprk2Δ15 mutant discs (Fig. 2N), suggesting that mGRK5 can functionally substitute Drosophila Gprk2 to promote high-level Hh signaling activity.
Gprk2 phosphorylates Smo C-tail
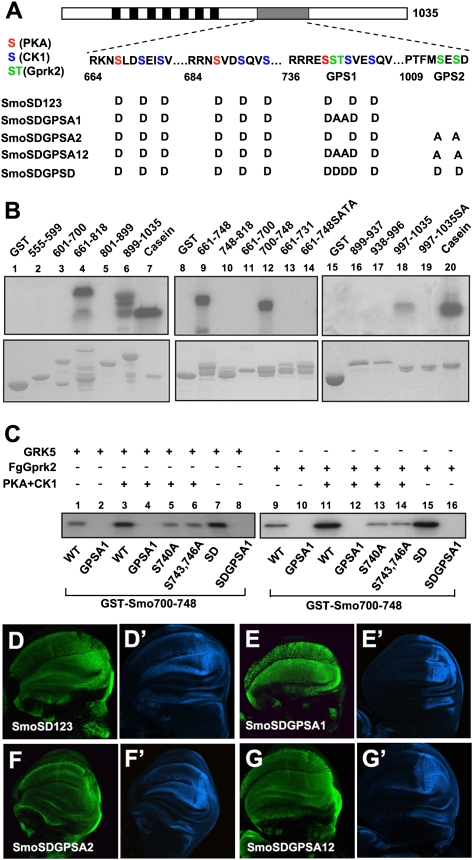
To determine whether Gprk2 directly phosphorylates Smo, we applied an in vitro kinase assay in which GST fusion proteins containing various fragments of Smo C-tail were incubated with a recombinant human GRK5 in the presence of γ-32p-ATP. Initial mapping suggested that two regions of Smo C-tail (amino acids 661–818 and amino acids 889–1035) were phosphorylated by GRK5 (Fig. 3B, lanes 4,6). Further deletion analysis mapped the GRK sites to amino acids 700–748 and amino acids 997–1035 (Fig. 3B, lanes 12,18). GST-Smo661–731 was not phosphorylated by GRK5 (Fig. 3B, lane 13), suggesting that a GRK site is located between amino acids 731 and 748 that harbors the third PKA/CK1 phosphorylation cluster (Fig. 3A; Jia et al. 2004). A previous study showed that both S741 and T742 were phosphorylated in cl8 cells exposed to Hh, but the corresponding kinase(s) remained unidentified (Zhang et al. 2004). As GRK family kinases tend to phosphorylate S/T in an acidic environment (Premont et al. 1995), we speculated that S741 and T742 could be phosphorylated by Gprk2. Indeed, mutating S741 and T742 in GST-Smo661–748 (GST-Smo661–748SATA) abolished GRK5-meidated phosphorylation in vitro (Fig. 3B, lane14). Similarly, mutating S1013 and S1015 in GST-Smo997–1035 (GST-Smo997–1035SA) abolished GRK5-mediated phosphorylation of the corresponding Smo fragment (Fig. 3B, lane 19). These results suggest that S741/T742 and S1013/S1015 are GRK phosphorylation sites. We name S741/T742 collectively as GPS1 (GRK phosphorylation sites 1) and S1013/S1015 as GPS2 (Fig. 3A).
Figure 3.
GRK phosphorylates multiple sites in Smo C-tail. (A) A schematic drawing of Smo with the sequence surrounding the PKA/CK1 sites and GRK sites (GPS1 and GPS2) shown below. PKA, CK1, and GRK sites are indicated by red, blue, and green residues, respectively. The transmembrane domains are indicated by the black boxes, and the SAID is indicated by the gray bar. Amino acid substitutions for SmoSD123 and its derivatives are listed. (B) In vitro kinase assay using a recombinant GRK5 and GST fusion proteins carrying indicated fragments from the Smo C-tail. (Lanes 12,18) Two minimal fragments, amino acids 700–748 and amino acids 997–1035, were phosphorylated by GRK5. (Lanes 14,19) Mutating the S741/T742 or S1013/S1015 abolished phosphorylation of the corresponding fragments. (C) In vitro kinase assay using recombinant GRK5 (shown in lanes 1–8) or immunoprecipitated Fg-Gprk2 with GST-Smo700–748 bearing the wild-type sequence or indicated point mutations. PKA/CK1 pretreatment was carried out in the presence of cold ATP. (D–G′) Wing discs expressing CFP-tagged SmoSD123 (D,D′), SmoSDGPSA1 (E,E′), SmoSDGPSA2 (F,F′), or SmoSDGPSA12 (G,G′) were immunostained to show the expression of CFP (green) and en (blue). SmoSD123 and SmoSDGPSA2 but not SmoSDGPSA1 or SmoSDGPSA12 induced ectopic en expression.
Because GPS1 is flanked by PKA (S740) and CK1 (S743 and S746) sites, phosphorylation of which improves the acidic environment (Fig. 3A), we tested whether phosphorylation at GPS1 could be enhanced by PKA/CK1 phosphorylation at adjacent sites. We constructed and purified GST-Smo700–748 fusion proteins with either wild-type GPS1 (WT) or GPS1 mutated to Ala (GPSA1), and carried out in vitro kinase assays using either recombinant GRK5 (Fig. 3C, lanes 1–8) or Flag-tagged Drosophila Gprk2 (FgGprk2) immunoprecipitated from S2 cells expressing FgGprk2 (Fig. 3C, lanes 9–16), with or without a prior phosphorylation by PKA and CK1 in the presence of cold ATP. We found that PKA/CK1 prior phosphorylation of GST-Smo700–748WT enhanced its phosphorylation by GRK5 or FgGprk2 (Fig. 3C, cf. lanes 3,11 and 1,9). Mutating GPS1 to Ala in GST-Smo700–748 (GST-Smo700–748SATA) abolished both basal and PKA/CK1-stimulated phosphorylation by GRK (Fig. 3C, lanes 2,4,10,12). Substitution of the PKA site (S740A) or CK1 sites (S743 and 746A) to Ala abolished PKA/CK1-stimulated GPS1 phosphorylation by GRK (Fig. 3C, lanes 5,6,13,14), whereas mutating PKA and CK1 sites to Asp (GST-Smo700–748SD) enhanced GPS1 phosphorylation in the absence of PKA and CK1 treatment (Fig. 3C, lanes 7,8,15,16). Taken together, these results demonstrate that PKA/CK1 phosphorylation at S740, S743, and S746 can stimulate GRK phosphorylation at GPS1. In contrast, we found that Gprk2 phosphorylation of GPS1 did not influence PKA/CK1 phosphorylation at adjacent sites (Supplemental Fig. S4). We also confirmed that the kinase-dead Gprk2 (Gprk2KM) did not phosphorylate GPS1 (Supplemental Fig. S5).
In vivo function of GRK phosphorylation sites
To determine the functional significance of Smo phosphorylation by Gprk2, we mutated GPS1 and GPS2 to Ala (SA) individually or in combination in the context of CFP-tagged SmoSD123 to generate CFP-SmoSDGPSA1, CFP-SmoSDGPSA2, and CFP-SmoSDGPSA12 (Fig. 3A). UAS transgenes expressing various Smo constructs were introduced into the 75B1 attP locus using the PhiC31 integration system to ensure the same level of transgene expression (Bischof et al. 2007). When expressed in wing discs using the MS1096 Gal4 driver, CFP-SmoSD123 induced ectopic en expression (Fig. 3D,D′). The ectopic en expression was diminished when GPS1 or both GPS1 and GPS2 were mutated to Ala (Fig. 3E,E′,G,G′). In contrast, CFP-SmoSDGPS2 still induced ectopic en expression (Fig. 3F,F′). The ectopic ptc expression driven by SmoSD123 was slightly reduced by GPSA1 and GPSA12 mutations, but was not affected by GPSA2 mutation (data not shown). SmoSDGPSA1 and SmoSDGPSA12 still induced anterior overgrowth of wing discs (Fig. 3E,E′,G,G′) and stabilized full-length Ci (data not shown). These results suggest that phosphorylation at GPS1 is required for maximal Smo activation.
We also mutated GPS1 and GPS2 to Asp to mimic phosphorylation in the context of CFP-SmoSD123 (CFP-SmoSDGPSD) (Fig. 3A). CFP-SmoSDGPSD induced ectopic ptc and en expression at levels similar to those induced by CFP-SmoSD123 when expressed at high levels using the MS1096 Gal4 driver (data not shown). However, when expressed at lower levels using the C765 Gal4 driver, CFP-SmoSDGPSD induced ectopic ptc and en expression more robustly than CFP-SmoSD123 (Fig. 4A–D′), suggesting that phosphorylation at GPS1 increases SmoSD123 activity.
Figure 4.
Gprk2 promotes Smo activation through phosphorylation-dependent and phosphorylation-independent mechanisms. (A–D′) Wing discs expressing UAS-SmoSD123-CFP (A,A′,C,C′) or UAS-SmoSDGPSD-CFP (B,B′,D,D′) with the C765 Gal4 driver were immunostained to show the expression of ptc (A,B), en (C,D), and CFP (A′,B′,C′,D′). (E,F). en expression (visualized by anti-En antibody) in Gprk2Δ15 heterozygous (E) or homozygous (F) wing discs expressing UAS-SmoSDGPSD-CFP with the MS1096 Gal4 driver. Loss of Gprk2 diminished the ectopic en expression induced by SmoSDGPSD. (G,G′) A Gprk2Δ15 homozygous wing disc expressing SmoSDGPSD and Flag-tagged Gprk2KM with MS1096 was immunostained with En and Flag antibodies. The kinase-dead Gprk2 rescued SmoSDGPSD-induced ectopic en expression in Gprk2Δ15 mutant discs. (H–J′) Gprk2Δ15 mutant discs expressing SmoSD123 alone (I), or together with wild-type Gprk2 (H) or Gprk2KM (J,J′) under the control of MS1096 were immunostained with anti-En and anti-Flag antibodies. Wild-type Gprk2 but not Gprk2KM rescued SmoSD123-induced ectopic en expression in Gprk2Δ15 mutant discs.
Kinase-independent function of Gprk2 in Hh signaling
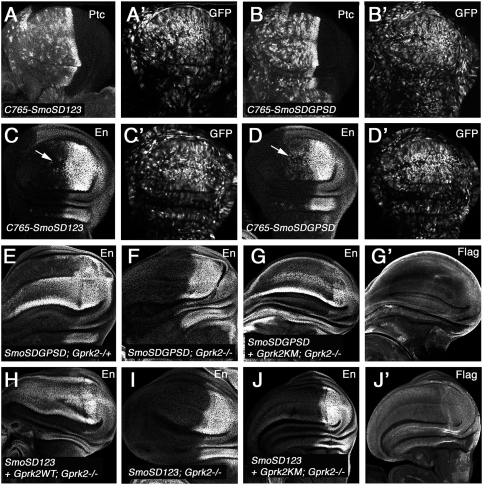
If Gprk2 promotes Smo activation solely by phosphorylating Smo C-tail, we would expect that SmoSDGPSD should bypass the requirement of Gprk2 to induce high-threshold Hh responses. To our surprise, we found that SmoSDGPSD failed to induce ectopic en expression in Gprk2 mutant discs (Fig. 4F). One possibility is that Gprk2 may phosphorylate Smo at additional sites; for example, in the intracellular loop regions that were not examined by our in vitro kinase assay. Another possibility is that Gprk2 may phosphorylate other components of the Hh pathway acting downstream from Smo. However, the observation that SmoΔSAID activity was insensitive to Gprk2 inactivation (Fig. 2G) does not favor these possibilities. A third possibility is that Gprk2 may promote Smo activity in a kinase-independent manner in addition to the phosphorylation-dependent mechanism. Indeed, kinase-independent desensitization of GPCR signaling by GRK2 was observed in cultured cells (Sallese et al. 2000). To test this latter possibility, SmoSDGPSD was coexpressed with the kinase-dead Gprk2KM in Gprk2 mutant discs. We found that coexpression of Gprk2KM restored the ectopic en expression induced by SmoSDGPSD in Gprk2 mutant discs (Fig. 4G). In contrast, when coexpressed in SmoSD123, only the wild-type Gprk2 (Gprk2WT) but not Gprk2KM rescued the ectopic en expression in Gprk2 mutant discs (Fig. 4H–J). These results demonstrate that Gprk2 uses two paralleled mechanisms to promote high levels of Hh signaling activity: a kinase-dependent mechanism mainly through phosphorylation of S741/T742, and a kinase activity-independent mechanism.
Gprk2 regulates Smo level
Hh promotes Smo cell surface accumulation, which is mediated by PKA/CK1 phosphorylation (Denef et al. 2000; Zhu et al. 2003; Jia et al. 2004; Zhao et al. 2007). To determine whether Gprk2 regulates Smo activity by promoting its cell surface expression, we examined Smo expression in wing discs carrying Gprk2 mutant clones. To our surprise, we found that Smo protein levels were elevated in anterior Gprk2 clones, which is more evident in clones near the A/P boundary where there are low levels of Hh (Fig. 5A, arrows; Molnar et al. 2007; Cheng et al. 2010). In contrast, loss of Gprk2 did not significantly affect Smo levels in A-compartment cells abutting the A/P boundary or in P-compartment cells where Smo is normally accumulated in response to high levels of Hh (Fig. 5A, arrowheads; Molnar et al. 2007; Cheng et al. 2010). Expressing wild-type Gprk2 but not Gprk2KM in Gprk2Δ15 mutant wing discs suppressed Smo up-regulation in A-compartment cells away from the A/P boundary (Supplemental Fig. S6), suggesting that the kinase activity of Gprk2 is required for destabilization of Smo in these cells.
Figure 5.
Gprk2 regulates Smo levels. (A) Wing discs carrying Gprk2Δ15 mutant clones were immunostained to show the expression of Smo (red channel) and GFP (green channel). Gprk2 mutant cells are marked by the lack of GFP expression. Smo levels were elevated in anterior Gprk2 mutant clones located near (big arrows) or away from (small arrows) the A/P boundary, but not in anterior Gprk2 mutant cells immediately abutting the A/P boundary or in P-compartment cells (arrowheads). The A/P boundary is marked by red lines based on Ci costaining (not shown). (B) S2 cells stably expressing a Myc-tagged Smo under the control of metallothionein promoter were treated with Gprk2 dsRNA or control (Luciferase) dsRNA in the absence of Hh, or in the presence of low (one-tenth of high) or high levels of Hh. Cells were immunostained with anti-SmoN antibody before membrane permeabilization to visualize cell surface Smo (top panels), or after membrane permeabilization to examine the total Smo (bottom panels). Quantification of cell surface and total Smo levels are shown (mean ± SD; n ≥ 20). The numbers below the bars indicate the percentage of Smo on the cell surface.
To determine whether loss of Gprk2 affects Smo subcellular localization, we turned to a cell-based assay using an S2 line that stably expressed a Myc-tagged Smo (Myc-Smo). Myc-Smo-expressing cells were treated with Gprk2 dsRNA or control dsRNA for 3 d, followed by treatment with conditioned medium containing different levels of Hh-N for 24 h. Cell surface or total Smo was visualized by immunostaining with anti-SmoN antibody prior to or after membrane permeabilization. As shown in Figure 5B, Gprk2 RNAi resulted in an increase in the levels of both cell surface and total Smo in the absence of Hh or in the presence of low levels of Hh (Fig. 5B, cf. lanes 2,4 and 1,3). In agreement with the in vivo results, inactivation of Gprk2 did not significantly alter the levels of either cell surface or total Smo in the presence of high levels of Hh (Fig. 5B, cf. lanes 6 and 5). Quantification analysis suggests that Gprk2 RNAi did not significantly increase the percentage of Smo on the cell surface compared with control RNAi (30% vs. 31%). In contrast, Hh treatment preferentially stabilized Smo on the cell surface as the percentage of Smo on the cell surface increased from 31% to 77% and 87% in the presence of low and high levels of Hh, respectively (Fig. 5B, lanes 3,5).
Gprk2 promotes the active Smo conformation
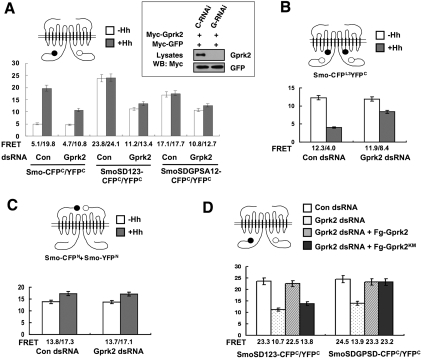
Our previous study demonstrated that Hh-induced phosphorylation of Smo triggers a conformational switch and dimerization of Smo C-tail, and that clustering of Smo C-tails promotes high-level Hh signaling activity (Zhao et al. 2007). The reduction of Smo activity in the absence of Gprk2 prompted us to investigate whether Gprk2 is required for Smo to adopt an active state. Clustering of Smo C-tails can be measured by fluorescence resonance energy transfer (FRET) analysis using a pair of C terminally CFP/YFP-tagged Smo constructs (Smo-CFPC/YFPC) (Zhao et al. 2007). Accordingly, Smo-CFPC and Smo-YFPC were transfected into S2 cells treated with Gprk2 dsRNA or control dsRNA, followed by treatment with or without Hh-conditioned medium. FRET between Smo-CFPC and Smo-YFPC (FRETC) was low in the absence of Hh but increased dramatically after Hh stimulation or when the PKA and CK1 sites were mutated to Asp to mimic phosphorylation (SmoSD123-CFPC/YFPC) (Fig. 6A; Zhao et al. 2007). Gprk2 RNAi did not change the basal FRETC but significantly reduced the Hh-induced FRETC (Fig. 6A). In agreement with the observation that Gprk2 inactivation reduces the activity of SmoSD123 in wing discs (Fig. 2I), Gprk2 RNAi significantly reduced the FRET between SmoSD-CFPC/YFPC in S2 cells (Fig. 6A). We also found that Gprk2 RNAi affected the Hh-induced decrease in the intramolecular FRET between CFP inserted in the third intracellular loop and YFP fused to the C terminus of Smo (SmoCFPL3YFPC) (Fig. 6B). In contrast, Gprk2 RNAi did not affect the FRET between the N terminally CFP/YFP-tagged Smo constructs (Smo-CFPN/YFPN) (Fig. 6C). Taken together, these results suggest that Gprk2 is not required for the constitutive dimerization of Smo through the Smo N-terminal region but is required for Smo to adopt the active conformation in response to Hh stimulation.
Figure 6.
Gprk2 regulates Smo conformation. (A–C) FRET efficiency from indicated wild-type or mutant Smo-CFPC/YFPC (A), Smo-CFPL3YFPC (B), or Smo-CFPN/YFPN (C) expressed in S2 treated with Gprk2 dsRNA or control (luciferase) dsRNA in the absence or presence of Hh treatment. Gprk2 RNAi reduced Hh-induced FRET from Smo-CFPC/YFPC (A) and attenuated Hh-induced reduction in the FRET from Smo-CFPL3YFPC (B), but did not affect the FRET from Smo-CFPN/YFPN (C). (A) Gprk2 RNAi reduced both the basal and Hh-induced FRET from SmoSD123-CFPC/YFPC or SmoSDGPSD-CFPC/YFPC. Mean ± SD; n ≥ 15. The cartoons above the graphics indicate Smo biosensors, with filled and open circles representing CFP and YFP, respectively. The inset in A shows that Gprk2 RNAi (G-RNAi) but not the control RNAi (C-RNAi) effectively knocked down transfected Myc-Gprk2 in S2 cells. Myc-GFP was cotransfected as a control for transgene expression and RNAi specificity. (D) S2 cells treated with Gprk2 or control dsRNA were transfected with SmoSD123-CFPC/YFPC or SmoSDGPSD-CFPC/YFPC with or without cotransfection of Fg-Gprk2 or Fg-Gprk2KM in the presence of Hh-conditioned medium, followed by FRET analysis. Both the wild-type and kinase-dead Gprk2 restored high FRET from SmoSDGPSD-CFPC/YFPC after endogenous Gprk2 was depleted by Gprk2 RNAi. In contrast, only the wild-type but not the kinase-dead Gprk2 rescued the FRET from SmoSD123-CFPC/YFPC after Gprk2 RNAi. Mean ± SD; n ≥ 15.
Mutating the GPS sites to Ala in the SmoSD123 background (SDGPSA12) also reduced FRETC but to a lesser extent as compared with Gprk2 RNAi (Fig. 6A). FRET between SmoSDGPSA12-CFPC/YFPC was further reduced by Gprk2 RNAi (Fig. 6A). In addition, FRET between SmoSDGPSD-CFPC/YFPC was also reduced by Gprk2 RNAi (Fig. 6D). Thus, mutating the GPS sites did not abrogate the effect of Gprk2 RNAi on Smo conformation, suggesting that Gprk2 may regulate Smo conformation and C-terminal dimerization through a mechanism paralleled to phosphorylation at the GPS sites. To determine whether Gprk2 can promote the active Smo conformation independently of its kinase activity, we asked whether overexpression of Gprk2KM could reverse the effect of Gprk2 RNAi on Smo FRET. As shown in Figure 6D, transfection of either Flag-tagged wild-type Gprk2 (Fg-Gprk2) or Gprk2KM (Fg-GprkKM) in cells treated with Gprk2 RNAi could rescue the FRET between. SmoSDGPSD12-CFPC/YFPC. In contrast, only Fg-Gprk2 but not Fg-Gprk2KM reversed the effect of Gprk2 RNAi on the FRET between SmoSD-CFPC/YFPC. Taken together, these results demonstrate that Gprk2 promotes the active Smo conformation through both phosphorylation-dependent and phosphorylation-independent mechanisms.
Gprk2 interacts with Smo
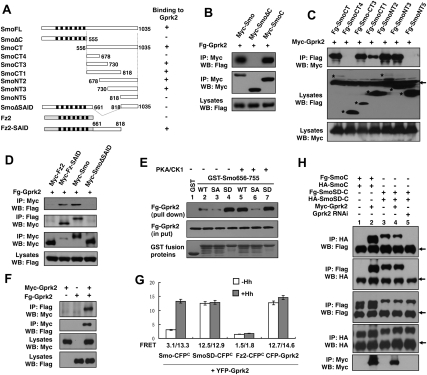
To explore the phosphorylation-independent mechanism by which Gprk2 promotes the active Smo conformation, we examined whether Gprk2 forms a complex with Smo. When expressed in S2 cells, Flag-tagged Gprk2 (Fg-Gprk2) coimmunoprecipitated with Myc-tagged full-length Smo (Myc-SmoFL) or Smo C-tail (Myc-SmoCT) but not with a truncated Smo lacking the C-tail (Myc-SmoΔC) (Fig. 7A-B), suggesting that Gprk2 interacts with Smo through its C-tail. Interaction between Smo and Gprk2 is independent of Gprk2 kinase activity, as Myc-Smo and Fg-Gprk2KM associated with each other in the coimmunoprecipitation assay (Supplemental Fig. S7). In addition, Smo interacted with mGRK5 (Supplemental Fig. S7), consistent with the observation that mGRK5 can functionally replace Gprk2 in vivo (Fig. 2M). Interaction between Smo and Gprk2 is specific because Fg-Gprk2 did not pull down Myc-tagged Frizzled2 (Fz2) (Fig. 7D), which is the closest relative of Smo. Moreover, Smo did not interact with Drosophila Gprk1 using the same assay (Supplemental Fig. S7).
Figure 7.
Gprk2 interacts with the SAID and promotes Smo C-tail dimerization. (A) Schematic drawing of full-length Smo and its deletion mutants, Fz2, and Fz2-Smo chimeric protein (Fz2-SAID), in which the SAID is fused to the C terminus of Fz2. The ability of individual constructs to interact with Gprk2 is indicated on the right. (B–D) Coimmunoprecipitation assays to determine Smo/Gprk2 interaction. S2 cells were transfected with indicated Myc-tagged or Flag (Fg)-tagged Smo or Fz2 and Gprk2 constructs, followed by immunoprecipitation and Western blot analysis with the indicated antibodies. Cell lysates were also directly immunoblotted by the indicated antibodies. Asterisks in C indicate the positions of full-length or truncated Smo. The arrow in C indicates IgG. (E) GST pull-down assay to determine Smo/Gprk2 interaction and its regulation by PKA/CK1-mediated phosphorylation. GST-Smo fusion protein containing Smo656–755 with wild-type sequence (WT); PKA sites mutated to Ala (SA) or PKA/CK1 sites mutated to Asp (SD) were purified from bacteria, treated with (+) or without (−) PKA and CK1, and incubated with cell lysates derived from S2 cells expressing a Flag-tagged Gprk2 (Fg-Gprk2). Fg-Gprk2 bound to GST-Smo fusion proteins were pulled down by glutathione beads and detected by Western blot with anti-Flag antibody. (F) Self-association of Gprk2. S2 cells were transfected with Myc-tagged or Flag-tagged Gprk2 individually or in combination. Cell lysates were subjected to immunoprecipitation and Western blot analysis with the indicated antibodies. (G) FRET analysis to determine Smo/Gprk2 interaction and Gprk2 self-association in intact cells. Numbers indicate the FRET efficiency from indicated CFP- and YFP-tagged constructs expressed in S2 cells. Mean ± SD; n ≥ 20. (H) Gprk2 promotes dimerization of Smo C-tail. Flag- and HA-tagged wild-type Smo C-tail (SmoC) or its phosphomimetic form (SmoSD-C) were transfected into S2 cells with or without Myc-Gprk2, or into S2 cells treated with Gprk2 dsRNA (Gprk2 RNAi). Cell lysates were subjected to immunoprecipitation and Western blot analysis with the indicated antibodies. Arrows indicate IgG.
Further deletion analysis suggested that two nonoverlapping regions between amino acids 661 and 818 mediate Gprk2 binding (Fig. 7A,C). Deletion of amino acids 661–818 in the context of full-length Smo (SmoΔSAID) abolished Gprk2 binding, whereas fusion of amino acids 661–818 to Fz2 (Fz2-SAID) confers Gprk2 binding to the fusion protein (Fig. 7A,D). These observations demonstrate that Smo661–818, which contains the SAID, is both necessary and sufficient for Gprk2 binding.
To further characterize the Gprk2/Smo interaction and its regulation, we carried out a GST pull-down assay in which a GST fusion protein containing the SAID from amino acids 656 to 755 (GST-Smo656–755) was incubated with extracts from S2 cells expressing Fg-Gprk2. GST-Smo656–755 but not GST pulled down Fg-Gprk2 (Fig. 7E, lanes 1,2). Interestingly, PKA and CK1 phosphorylation of GST-Smo656-755 increased its binding affinity to Fg-Gprk2 (Fig. 7E, lane 5), and mutating the three PKA sites to Ala (GST-Smo656–755SA) abolished this effect (Fig. 7E, lane 6). On the other hand, mutating the PKA/CK1 sites to Asp (GST-Smo656–755SD) increased Fg-Gprk2 binding regardless of PKA/CK1 treatment (Fig. 7E, lanes 4,7). These results demonstrate that PKA/CK1-mediated phosphorylation of the SAID enhances its binding to Gprk2.
To confirm the Smo/Gprk2 interaction in intact cells, we applied FRET analysis. S2 cells were transfected with N terminally YFP-tagged Gprk2 (YFP-Gprk2) and Smo-CFPC, SmoSD-CFPC, or Fz2-CFPC; treated with or without Hh-conditioned medium; and subjected to FRET analysis. YFP-Gprk2 was tightly associated with the plasmic membrane (Supplemental Fig. S8), likely due to its lipid modification, as observed for mammalian GRK family kinases (Pitcher et al. 1998). YFP-Gprk2 was largely colocalized with SmoSD-CFPC and Fz2-CFPC as well as Smo-CFPC after Hh treatment (Supplemental Fig. S8). FRET between Smo-CFPC and YFP-Gprk2 was low in the absence of Hh (3.1%) but increased dramatically after Hh treatment (13.3%), whereas FRET between SmoSD-CFPC and YFP-Gprk2 was high regardless of Hh treatment (Fig. 7G). In contrast, there is no significant FRET between Fz2-CFPC and YFP-Gprk2 (Fig. 7G). These observations suggest that Hh and PKA/CK1-mediated phosphorylation promotes Smo/Gprk2 association. In addition, the high FRET between Smo and Gprk2 suggests that they interact directly.
Gprk2 forms a dimer/oligomer and promotes dimerization/oligomerization of Smo C-tail
Interestingly, we also observed high FRET between CFP-Gprk2 and YFP-Gprk2 regardless of Hh treatment (Fig. 7G), suggesting that Gprk2 forms a constitutive dimer or oligomer. To further test whether Gprk2 can self-associate, we expressed two differently tagged Gprk2s in S2 cells and carried out immunoprecipitation assays. As shown in Figure 7F, Myc-tagged Gprk2 (Myc-Gprk2) pulled down Fg-Gprk2 and vice versa.
Since Gprk2 can self-associate, binding of Gprk2 to Smo C-tail may facilitate its dimerization/oligomerization, which could explain why Gprk2 is required for Hh- and phosphorylation-induced proximity of Smo C-tails. To test this possibility, we transfected S2 cells with Flag-Smo C-tail (Fg-SmoC) and HA-tagged Smo C-tail (HA-SmoC) in the absence or presence of Myc-Gprk2 cotransfection. In the absence of Myc-Gprk2, Fg-SmoC and HA-SmoC did not coimmunoprecipitate (Fig. 7H, lane 1); however, coexpression of Myc-Gprk2 allowed Fg-SmoC and HA-SmoC to associate with each other (Fig. 7H, lane 2). In agreement with our previous findings that phosphorylation-mimetic mutation promotes Smo C-tail self-association (Zhao et al. 2007), Fg-SmoSD-C and HA-SmoSD-C coimmunoprecipitated regardless of exogenously expressed Gprk2 (Fig. 7H, lanes 3,4); however, this association was diminished by Gprk2 RNAi (Fig. 7H, lane 5), suggesting that endogenous Gprk2 is required for SmoSD-C self-association. Collectively, these observations suggest that PKA/CK1-mediated phosphorylation recruits Gprk2 to promote Smo C-tail dimerization/oligomerization.
Discussion
In this study, we carried out a genetic modifier screen for novel Hh signaling components and identified Gprk2 as a positive regulator of Smo. Consistent with previous reports (Molnar et al. 2007; Cheng et al. 2010), we found that Gprk2 is required for high but not low levels of Hh signaling activity. We provided the first evidence that Gprk2 is a Smo kinase and that Gprk2 promotes maximal Smo activity by phosphorylating S741/T742 in Smo C-tail. Furthermore, we uncovered a kinase-independent function of Gprk2 in Hh signaling. We demonstrated that Gprk2 forms a dimer/oligomer and binds Smo C-tail to promote the active state of Smo. Thus, our study reveals a novel mechanism for regulating a GPCR-like protein by GRK.
Gprk2 regulates Hh signaling by phosphorylating Smo
Previous studies suggest that Drosophila Gprk2 and mammalian GRK2 are involved in Smo phosphorylation because their knockdown in cultured cells either increased Smo mobility on SDS-PAGE or decreased metabolic labeling of Smo by γ-32p-ATP (Chen et al. 2004; Cheng et al. 2010). However, these studies did not distinguish whether Gprk2/GRK2 phosphorylates Smo directly or indirectly through regulating other kinases. Neither did they reveal any biological relevance of Gprk2/GRK2-mediated phosphorylation in Hh signaling, since the relevant phosphorylation sites on Smo were not identified. In an in vitro kinase assay using purified substrates and a recombinant GRK, we found that Smo is phosphorylated by GRK at S741/T742 and S1013/S1015. Our mutagenesis study demonstrated that phosphorylation at S741/T742 is required for optimal Smo activation. Indeed, a previous study showed that Smo is phosphorylated at S741/T742 in cultured cells in the presence of Hh (Zhang et al. 2004). In further agreement with the functional significance of S741/T742 phosphorylation, conserved S/T residues are found at the corresponding location in other insect Smo proteins (FlyBase).
Interestingly, our in vitro kinase assay revealed that phosphorylation of S741/T742 by Gprk2 is regulated by PKA/CK1 phosphorylation at adjacent Ser residues, including S740, S743, and S746. Previous studies in mammalian systems suggest that GRKs tend to phosphorylate S/T residues embedded in an acidic environment (Premont et al. 1995). Phosphorylation at S740, S743, and S746 improves the acidic environment for S741/T742, which may account for the observed stimulation of S741/T742 phosphorylation by PKA/CK1. Indeed, mutating S740, S743, and S746 to Ala abolished PKA/CK1-mediated stimulation of S741/T742 phosphorylation, whereas converting these residues to acidic residues mimicked PKA/CK1-mediated stimulation. As Hh induces Smo phosphorylation by PKA and CK1, phosphorylation at S741/T742 by Gprk2 is likely to be stimulated by Hh in vivo.
A kinase-independent role of Gprk2 in Hh signaling
Although phosphomimetic mutation at S741/T742 promotes Smo activity, it does not bypass the requirement for Gprk2 for optimal Smo activation because SmoSDGPSD failed to induce ectopic en expression in Gprk2 mutant discs. This implies that Gprk2 promotes Hh signaling through a mechanism in parallel to S741/T742 phosphorylation. It is possible that Gprk2 might act at an additional step downstream from Smo activation by phosphorylating intracellular Hh signaling components, or at the level of Smo activation by phosphorylating Smo at additional sites that have been missed by our in vitro kinase assay. However, our finding that the constitutively active form of Smo lacking the SAID (SmoΔ661–818) is insensitive to Gprk2 inactivation suggests that Gprk2 acts mainly at the level of Smo, although we cannot rule out the possibility that Gprk2 may also play a minor role downstream from Smo. Interestingly, we found that the kinase-dead form of Gprk2 (Gprk2KM) can rescue the activity defect of SmoSDGPSD in Gprk2 mutants, demonstrating that Gprk2 also regulates Smo in a phosphorylation-independent manner. The observation that Gprk2KM does not rescue the activity defect of SmoSD123 in Gprk2 mutants suggests that the phosphorylation-dependent and phosphorylation-independent mechanisms act in parallel rather than redundantly to promote Smo activation. Furthermore, we obtained evidence that Gprk2 interacts with the SAID independently of its kinase activity. Therefore, we propose that Gprk2 promotes Smo activation by counteracting Smo autoinhibition through binding to and phosphorylating the SAID.
Gprk2 regulates Smo level and conformation
At least two paralleled mechanisms have been attributed to Smo activation by Hh: (1) Smo cell surface accumulation, and (2) conformation change in Smo C-tail (Jiang and Hui 2008). Intriguingly, we found that loss of Gprk2 resulted in increased rather than decreased Smo levels in cells that are not exposed to Hh or are exposed to low levels of Hh (Fig. 5A). Other groups made similar observations (Molnar et al. 2007; Cheng et al. 2010). However, unlike Hh stimulation, which preferentially stabilizes Smo on the cell surface, Gprk2 inactivation appears to stabilize Smo both inside the cell and on the cell surface (Fig. 5B). Furthermore, in the presence of high levels of Hh where Smo is accumulated at high levels on the cell surface, Gprk2 inactivation does not cause any discernible changes in either the level or subcellular distribution of Smo. Thus, the reduced Smo activity in Gprk2 mutant cells exposed to high levels of Hh is unlikely to be due to a change in Smo level or subcellular localization.
It is not clear what role Gprk2-mediated down-regulation of Smo levels might play in Hh signaling, although this may reflect an ancient mechanism by which GRK family kinases “desensitize” GPCRs. In this regard, Gprk2-mediated down-regulation could serve as a mechanism to restrict the basal level of Hh signaling activity or to terminate or tune down Hh signaling activity once the Hh signal is withdrawn. However, this negative role of Gprk2 could be masked by its positive role. The mechanism by which Gprk2 down-regulates Smo levels remains unclear, although the kinase activity of Gprk2 appears to be required. Gprk2 could phosphorylate Smo and/or other proteins to promote Smo internalization and degradation. High levels of Hh could counteract Gprk2-mediated down-regulation of Smo by preventing Gprk2-meidated Smo internalization or by promoting Smo recycling.
Our FRET analysis provided strong evidence that Gprk2 is required for Smo to adopt and/or maintain its active conformation in response to Hh stimulation. Our previous study suggested that Hh induces a conformational switch in Smo C-tail that is mediated by PKA and CK1 phosphorylation (Zhao et al. 2007). In the absence of Hh, the Smo C-tail adopts a closed conformation in which the tail folds back, resulting in a close proximity between the C terminus and the third intracellular loop. The closed conformation is maintained, at least in part, through intramolecular electrostatic interactions between the multiple Arg clusters in the SAID and multiple acidic clusters near the C terminus. Hh-induced phosphorylation at PKA and CK1 sties disrupts the intramolecular electrostatic interactions, resulting in unfolding of the C-tail, which is reflected by a decreased intramolecular FRET (FRETL3C). In addition, phosphorylation promotes dimerization of two C-tails within a Smo homodimer, leading to increased proximity of the two C termini, as reflected by an increased C-terminal FRET (FRETC). Multiple intermediate conformational states may exist, depending on the levels of Smo phosphorylation, as increasing the number of phosphomimetic mutations progressively decreased FRETL3C and gradually increased FRETC (Zhao et al. 2007). We found that both an Hh-induced decrease in FRETL3C and an Hh-induced increase in FRETC were compromised by loss of Gprk2 (Fig. 6), suggesting that Gprk2 is critical for Smo to adopt and/or maintain the fully open conformation.
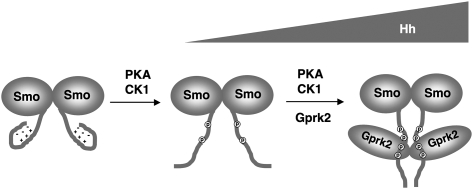
How does Gprk2 regulate Smo conformation? Our genetic and FRET analyses demonstrated that Gprk2 promotes high levels of Hh signaling activity and regulates Smo conformation through both phosphorylation-dependent and phosphorylation-independent mechanisms. Furthermore, we found that Gprk2 self-associates, binds the SAID, and promotes self-association of Smo C-tail. Interestingly, both Gprk2/SAID interaction and S741/T742 phosphorylation by Gprk2 are enhanced by PKA/CK1 phosphorylation. Taken together, we propose the following model to account for the regulation of Smo conformation by Gprk2 (Fig. 8). In response to Hh, PKA/CK1-mediated phosphorylation of Smo C-tail promotes its unfolding and dimerization; however, in the absence of Gprk2, the open conformational state of Smo is unstable and may exist in equilibrium with the closed and/or partially open conformational states. Phosphorylation of Smo by PKA/CK1 promotes the binding of Gprk2 to the SAID and phosphorylation at S741/T742, both of which may stabilize Smo in the fully open and active conformation by preventing refolding of Smo C-tail and by “cross-linking” the two C-tails within a Smo dimer via dimerization of Gprk2. In essence, Gprk2 may function as a “molecular clamp” to promote the clustering of Smo C-tails. It is also possible that Gprk2 could cross-link two or more Smo dimers to form high-order oligomers, which might be essential for high levels of Hh signaling activity. Our study thus reveals an unanticipated complexity in the regulation of Smo conformational states, and provides the first evidence that Smo conformation states are regulated by not only phosphorylation and intramolecular interactions, but also intermolecular interactions. It is possible that the closed conformation state of Smo is also regulated by intermolecular interactions in addition to intramolecular interactions. For example, it has been shown that Fu can directly bind the Smo C terminus in the absence of Hh (Malpel et al. 2007), and this interaction may help stabilize the closed conformation of Smo C-tail. Indeed, disrupting Smo/Fu interaction led to increased basal activity of Smo (Malpel et al. 2007).
Figure 8.
Model for regulating Smo activation state by Gprk2. Hh-induced phosphorylation by PKA and CK1 leads to unfolding of Smo C-tail and promotes its association with and phosphorylation by Gprk2. Both Gprk2 binding and phosphorylation stabilize Smo in the active conformation by preventing refolding. In addition, Gprk2 forms a dimer/oligomer and promotes the active state of Smo by cross-linking Smo C-tails.
Implication on mammalian Smo regulation
Recent studies have emphasized the differences between vertebrate and Drosophila Hh signaling mechanisms. The sequence divergence between Drosophila and vertebrate Smo proteins and the lack of conserved PKA/CK1 phosphorylation sites in vertebrate Smo proteins have led to the proposal that vertebrate Smo proteins are activated through fundamentally distinct mechanisms (Huangfu and Anderson 2006; Varjosalo et al. 2006). Nevertheless, our previous study revealed that Shh induces a conformational change in mSmo similar to that of dSmo, and forced clustering of mSmo also leads to pathway activation (Zhao et al. 2007). GRK2 has been implicated as a positive regulator of the Shh pathway both in cultured cells and in vivo (Chen et al. 2004; Meloni et al. 2006; Philipp et al. 2008), and mSmo phosphorylation is affected by GRK2 silencing, although direct phosphorylation of mSmo by GRK2 has not been demonstrated. It is possible that GRK2 may substitute the role of PKA and CK1 and act as a major Smo kinase in vertebrates to promote the active Smo conformation. Alternatively, GRK2 may act in conjunction with other GRKs and/or yet-to-be-identified kinases to regulate Smo conformation, subcellular localization, and activity in vertebrates. The relatively weak phenotypes exhibited by GRK2 mutants are consistent with the latter possibility (Philipp et al. 2008). Our study also raised an interesting possibility that GRK2 may regulate mSmo not only by phosphorylation, but also by a kinase-independent mechanism such as a protein–protein interaction. Further investigation of the mechanism by which GRK2 and other kinases regulate mSmo will shed an important light on how vertebrate Smo activation is achieved.
Materials and methods
Mutations and transgenes
UAS-Smo−PKA12 has been described (Jia et al. 2004). The Gal4 drivers MS1096 and C765 were described in FlyBase. The genetic modifier screen was carried out by crossing females of the genotype MS1096; UAS-Smo−PKA12/TM2 or C765, UAS-Smo−PKA12/TM6B with males of Exilexis deficiency lines obtained from the Bloomington Stock Center. The F1 progenies were scored for any modifications of the fused wing phenotype. Gprk206936 has been described (Schneider and Spradling 1997). Excision alleles of Gprk2 were generated by crossing Gprk206936 with Δ2–3 flies that carry the transposase. Progenies that lost ry+ were crossed with C765, UAS-Smo−PKA12/TM6B. Precise excision alleles no longer modified the fused wing phenotype, whereas several imprecise excision lines, including Gprk2Δ15, enhanced the wing phenotype similarly to the Gprk2 deficiency line Df(3R)Exel6219. Gprk2Δ15 mutant clones were generated in yw 122; FRT82 Gprk2Δ15/FRT82 hs-Myc-GFP.
To generate Flag- or Myc-tagged Gprk2 constructs, the Gprk2 coding sequence was amplified by PCR and inserted in-frame in the pUAST-Flag or pUAST-Myc vector (Chao and Jiang 2007). To construct the kinase-dead form of Gprk2, the conserved lysines K338 and K339 within the kinase catalytic domain of Gprk2 were mutated into Met using PCR-based site-directed mutagenesis. To construct UAS-Gprk2-CFPN/YFPN, CFP/YFP was inserted in-frame before the Gprk2 start codon. Amino acid substitutions of GPS1 and GPS2 were generated by PCR-based site-directed mutagenesis. To construct UAS-SmoGPSA-CFP/YFP and UAS-SmoSDGPSD-CFP/YFP, CFP/YFP was inserted in-frame after the C termini of the corresponding Smo variants. To construct UAS-GPRK2-RNAi, a genomic DNA fragment encoding Gprk2 amino acids 180–385 was amplified by PCR and cloned between the BglII and XhoI sites of the pUAST vector, with the corresponding cDNA fragment inserted in a reverse orientation between the XhoI and XbaI sites. The PhiC31 integration system was used to integrate Smo transgenes into the 75B1 attP locus (Bischof et al. 2007; Jia et al. 2009). To generate GST-Smo fusion constructs, DNA fragments encoding Smo C-terminal regions with either the wild-type sequence or point mutations in phosphorylation sites were amplified by PCR and inserted between BamHI and NotI sites in the pGEX-4T-2 vector.
Cell culture, transfection, immunoprecipitation, GST pull-down, Western blot, and immunostaining
Drosophila S2 cells were cultured in Drosophila SFM (Invitrogen) with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100mg/mL streptomycin at 23°C. Transfection was carried out by Calcium Phosphate Transfection Kit (Specialty Media) according to manufacturer's instructions. Hh-conditioned medium treatment was carried out as described (Lum et al. 2003). Briefly, Hh-N stably expressed S2 cells were selected in 200 μg/mL hygromycine. Hh-conditioned medium was prepared by culturing cells without hygromycine but with 0.7 mM CuSO4 for 1 d. The medium was harvested and sterilized by filtration. Unless mentioned otherwise, Hh-conditioned medium was used at a 6:4 dilution ratio by fresh medium. Immunoprecipitation and Western blot analysis were carried out using standard protocols as described previously (Zhang et al. 2005). For GST pull-down assay, GST fusion proteins bound to the glutathione beads were washed three times with ice-cold PBS containing 1% NP-40, and were incubated with cell lysates from S2 cells expressing Flag-tagged Gprk2 proteins for 1 h at 4°C with occasional mixing. The beads were washed again five times with PBS plus 1% NP-40 before separation on SDS-PAGE, followed by Western blot using anti-Flag antibody. For the Smo cell surface staining assay, S2 cells were harvested and washed with PBS, fixed with 4% formaldehyde at room temperature for 20 min, and incubated with the mouse anti-SmoN antibody in PBS at room temperature for 30 min. Cells were washed three times by PBS followed by secondary antibody staining. Immunostaining of imaginal discs was carried out as described (Jiang and Struhl 1995). Antibodies used in this study were mouse anti-En (Developmental Studies Hybridoma Bank [DSHB]), mouse anti-Ptc (DSHB), mouse anti-SmoN (DSHB), rabbit and mouse anti-Flag (Sigma), mouse anti-Myc (Santa Cruz Biotechnology), mouse anti-HA (Santa Cruz Biotechnology), mouse anti-Ci 2A1 (Motzny and Holmgren 1995), mouse anti-GFP (Millipore), rabbit anti-GFP (Santa Cruz Biotechnology), and guinea pig anti-dGprk2 (Cheng et al. 2010).
In vitro kinase assay
In vitro kinase assay using recombinant GRK5 was performed according to the manufacturer's instructions (Upstate Biotechnologies, 14-714). Gprk2 in vitro kinase reaction was performed for 1 h at 30°C in kinase buffer (20 mM Tris-HCl at pH 8.0, 2 mM EDTA, 10 mM MgCl2, 1 mM DTT), with Fg-Gprk2 or Fg-Gprk2KM immunoprecipitated from S2 cells transfected with Fg-Gprk2 or Flag-Gprk2KM expression construct, GST fusion proteins, and 0.1 mM ATP containing 10 mCi of γ-32p-ATP. PKA and CK1 were purchased from New England Biolabs. After the GST fusion proteins were incubated with PKA and CK1 in the presence of cold ATP for 30 min at 30°C, the reactions were stopped by adding 10 mM D4476 and H-89 (CK1 and PKA inhibitors, respectively). PKA/CK1-treated GST fusion proteins were further processed with GRK5 or Gprk2 phosphorylation for 1 h at 30°C in the presence of γ-32p-ATP. The reactions were finally stopped by adding 4× SDS loading buffer and were boiled for 5 min at 100°C. Phosphorylation of GST fusion proteins was analyzed by autoradiography after SDS-PAGE.
RNAi in Drosophila S2 cells
dsRNA was generated by MEGAscript High-Yield Transcription Kit (Ambion, #AM1334) according to the manufacturer's instructions. A DNA template targeting Gprk2 amino acids 124–290 was generated by PCR and used for making Gprk2 dsRNA. dsRNA targeting the Firefly Luciferase coding sequence was used as a control. For the RNAi knockdown experiments described in Figure 6, S2 cells were cultured in serum-free SFM medium containing the indicated dsRNA for 8 h at 25°C. After adding FBS to a final concentration of 10%, dsRNA-treated cells were cultured overnight before transfection with CFP/YFP-tagged Smo constructs. After additional culturing for 2∼3 d, the cells were collected for FRET assay. For Gprk2 rescue experiments in cultured S2 cells, 2 d after transfection of CFP/YFP-tagged Smo constructs, the cells were changed into fresh SFM medium containing 10% FBS, transfected with Gprk2 or Gprk2KM expression construct, and subjected to FRET assay after culturing for two additional days.
FRET analysis using confocal microscopy
FRET analysis was carried out as described previously (Zhao et al. 2007). CFP- and YFP-tagged constructs in the pUAST vector were cotransfected into S2 cells together with an ub-Gal4 expression vector. Cells were washed with PBS, fixed with 4% formaldehyde for 20 min, and mounted on slides in 80% glycerol. CFP signals were acquired with the 100× objective of a Zeiss LSM510 confocal microscope before (BP) and after (AP) photobleaching YFP. Each data set was calculated using 15–20 individual cells. In each cell, four or five regions of interest in a photobleached area were selected for analysis. The intensities of CFP signals were quantified by ImageJ software. The FRET efficiency was calculated using the formula FRET% = [(CFPAP − CFPBP)/CFPAP] × 100.
Acknowledgments
We thank Drs. Jeffrey Benovic, Robert Holmgren, and David Hipfner; the Bloomington Stock Center, and DSHB for reagents. This work was supported by grants from NIH, Welch Foundation (I-1603), and CPRIT (to J.J.). J.J. is a Eugene McDermott Endowed Scholar in Biomedical Science at UTSW.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1948710.
Supplemental material is available at http://www.genesdev.org.
References
- Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A 2005. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol 7: 86–92 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramirez-Weber F, Laget M, Schwartz C, Kornberg T 1997. Proteolysis that is inhibited by Hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci 104: 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali A, Struhl G 2004. Reading the Hedgehog morphogen gradient by measuring the ratio of bound to unbound Patched protein. Nature 431: 76–80 [DOI] [PubMed] [Google Scholar]
- Chao T, Jiang J 2007. Using immunoprecipitation to study protein–protein interaction in the Hedgehog signaling pathway. Methods Mol Biol 397: 215–229 [DOI] [PubMed] [Google Scholar]
- Chen Y, Struhl G 1996. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87: 553–563 [DOI] [PubMed] [Google Scholar]
- Chen W, Ren XR, Nelson CD, Barak LS, Chen JK, Beachy PA, de Sauvage F, Lefkowitz RJ 2004. Activity-dependent internalization of smoothened mediated by β-arrestin 2 and GRK2. Science 306: 2257–2260 [DOI] [PubMed] [Google Scholar]
- Cheng S, Maier D, Neubueser D, Hipfner DR 2010. Regulation of smoothened by DROSOPHILA G-protein-coupled receptor kinases. Dev Biol 337: 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM 2000. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102: 521–531 [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV 2006. Signaling from Smo to Ci/Gli: Conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development 133: 3–14 [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP 2001. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev 15: 3059–3087 [DOI] [PubMed] [Google Scholar]
- Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J 2002. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature 416: 548–552 [DOI] [PubMed] [Google Scholar]
- Jia J, Tong C, Wang B, Luo L, Jiang J 2004. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and Casein Kinase I. Nature 432: 1045–1050 [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, Amanai K, Jiang J 2005. Phosphorylation by double-time/CKIɛ and CKIα targets cubitus interruptus for Slimb/β-TRCP-mediated proteolytic processing. Dev Cell 9: 819–830 [DOI] [PubMed] [Google Scholar]
- Jia H, Liu Y, Yan W, Jia J 2009. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development 136: 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Hui CC 2008. Hedgehog signaling in development and cancer. Dev Cell 15: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G 1995. Protein kinase A and Hedgehog signalling in Drosophila limb development. Cell 80: 563–572 [DOI] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA 2003. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299: 2039–2045 [DOI] [PubMed] [Google Scholar]
- Malpel S, Claret S, Sanial M, Brigui A, Piolot T, Daviet L, Martin-Lanneree S, Plessis A 2007. The last 59 amino acids of Smoothened cytoplasmic tail directly bind the protein kinase Fused and negatively regulate the Hedgehog pathway. Dev Biol 303: 121–133 [DOI] [PubMed] [Google Scholar]
- Meloni AR, Fralish GB, Kelly P, Salahpour A, Chen JK, Wechsler-Reya RJ, Lefkowitz RJ, Caron MG 2006. Smoothened signal transduction is promoted by G protein-coupled receptor kinase 2. Mol Cell Biol 26: 7550–7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N, Basler K 1999. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96: 819–831 [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K 2000. Suppressor of Fused opposes Hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127: 4001–4010 [DOI] [PubMed] [Google Scholar]
- Molnar C, Holguin H, Mayor F Jr, Ruiz-Gomez A, de Celis JF 2007. The G protein-coupled receptor regulatory kinase GPRK2 participates in Hedgehog signaling in Drosophila. Proc Natl Acad Sci 104: 7963–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzny CK, Holmgren R 1995. The Drosophila cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev 52: 137–150 [DOI] [PubMed] [Google Scholar]
- Ogden SK, Fei DL, Schilling NS, Ahmed YF, Hwa J, Robbins DJ 2008. G protein Gα (i) functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 456: 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Kalderon D 1998. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396: 749–753 [DOI] [PubMed] [Google Scholar]
- Philipp M, Fralish GB, Meloni AR, Chen W, MacInnes AW, Barak LS, Caron MG 2008. Smoothened signaling in vertebrates is facilitated by a G protein-coupled receptor kinase. Mol Biol Cell 19: 5478–5489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher JA, Freedman NJ, Lefkowitz RJ 1998. G protein-coupled receptor kinases. Annu Rev Biochem 67: 653–692 [DOI] [PubMed] [Google Scholar]
- Premont RT, Inglese J, Lefkowitz RJ 1995. Protein kinases that phosphorylate activated G protein-coupled receptors. FASEB J 9: 175–182 [DOI] [PubMed] [Google Scholar]
- Price MA, Kalderon D 2002. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by glycogen synthase kinase 3 and Casein Kinase 1. Cell 108: 823–835 [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP 1997. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90: 225–234 [DOI] [PubMed] [Google Scholar]
- Sallese M, Mariggio S, D'Urbano E, Iacovelli L, De Blasi A 2000. Selective regulation of Gq signaling by G protein-coupled receptor kinase 2: Direct interaction of kinase N terminus with activated gαq. Mol Pharmacol 57: 826–831 [PubMed] [Google Scholar]
- Schneider LE, Spradling AC 1997. The Drosophila G-protein-coupled receptor kinase homologue Gprk2 is required for egg morphogenesis. Development 124: 2591–2602 [DOI] [PubMed] [Google Scholar]
- Sisson BE, Ziegenhorn SL, Holmgren RA 2006. Regulation of Ci and Su(fu) nuclear import in Drosophila. Dev Biol 294: 258–270 [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Zhou Q, Kalderon D 2007. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev Cell 13: 481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, et al. 1996. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384: 129–134 [DOI] [PubMed] [Google Scholar]
- Strigini M, Cohen SM 1997. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 124: 4697–4705 [DOI] [PubMed] [Google Scholar]
- Taipale J, Beachy PA 2001. The Hedgehog and Wnt signalling pathways in cancer. Nature 411: 349–354 [DOI] [PubMed] [Google Scholar]
- Taipale J, Cooper MK, Maiti T, Beachy PA 2002. Patched acts catalytically to suppress the activity of Smoothened. Nature 418: 892–897 [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J 2008. Hedgehog: Functions and mechanisms. Genes Dev 22: 2454–2472 [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Li SP, Taipale J 2006. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell 10: 177–186 [DOI] [PubMed] [Google Scholar]
- Villavicencio EH, Walterhouse DO, Iannaccone PM 2000. The sonic hedgehog–patched–gli pathway in human development and disease. Am J Hum Genet 67: 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Jiang J 2004. Multiple Cos2/Ci interactions regulate Ci subcellular localization through microtubule dependent and independent mechanisms. Dev Biol 268: 493–505 [DOI] [PubMed] [Google Scholar]
- Wang QT, Holmgren RA 2000. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development 127: 3131–3139 [DOI] [PubMed] [Google Scholar]
- Wang G, Amanai K, Wang B, Jiang J 2000. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes Dev 14: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Williams EH, Guo Y, Lum L, Beachy PA 2004. Extensive phosphorylation of Smoothened in Hedgehog pathway activation. Proc Natl Acad Sci 101: 17900–17907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, Jiang J 2005. Hedgehog-regulated costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev Cell 8: 267–278 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tong C, Jiang J 2007. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature 450: 252–258 [DOI] [PubMed] [Google Scholar]
- Zheng X, Mann RK, Sever N, Beachy PA 2010. Genetic and biochemical definition of the Hedgehog receptor. Genes Dev 24: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu AJ, Zheng L, Suyama K, Scott MP 2003. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev 17: 1240–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]