Abstract
Mutations in the X-linked human EPHRIN-B1 gene result in cleft palate and other craniofacial anomalies as part of craniofrontonasal syndrome (CFNS), but the molecular and developmental mechanisms by which ephrin-B1 controls the underlying developmental processes are not clear. Here we demonstrate that ephrin-B1 plays an intrinsic role in palatal shelf outgrowth in the mouse by regulating cell proliferation in the anterior palatal shelf mesenchyme. In ephrin-B1 heterozygous mutants, X inactivation generates ephrin-B1-expressing and -nonexpressing cells that sort out, resulting in mosaic ephrin-B1 expression. We now show that this process leads to mosaic disruption of cell proliferation and post-transcriptional up-regulation of EphB receptor expression through relief of endocytosis and degradation. The alteration in proliferation rates resulting from ectopic Eph–ephrin expression boundaries correlates with the more severe dysmorphogenesis of ephrin-B1+/− heterozygotes that is a hallmark of CFNS. Finally, by integrating phosphoproteomic and transcriptomic approaches, we show that ephrin-B1 controls proliferation in the palate by regulating the extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) signal transduction pathway.
Keywords: Ephrin, cell proliferation, ERK/MAPK, endocytosis, boundaries, craniofrontonasal syndrome, palate
Craniofrontonasal syndrome (CFNS) is a human congenital disorder caused by mutations in the ephrin-B1 gene that affects multiple aspects of neurological, skeletal, and craniofacial development, including palatogenesis (Twigg et al. 2004; Wieland et al. 2004). Mice harboring null mutations in ephrin-B1 display nearly identical phenotypes, including agenesis of the corpus callosum, thoracic skeletal defects, frontonasal dysplasia, and cleft palate (Compagni et al. 2003; Davy et al. 2004; Bush and Soriano 2009). Cleft palate is an extremely common congenital disorder that can occur in either syndromic or isolated contexts, and can result from the perturbation of normal palatogenesis at multiple stages. Beginning at approximately embryonic day 11.5 (E11.5) in mice, palatal shelf outgrowth is initiated from the stomodeal surface of the maxillary processes, and is populated by neural crest-derived mesenchyme surrounded by a thin layer of epithelium (Chai and Maxson 2006). Although several mechanisms may control palatal shelf outgrowth, cell proliferation of the palatal mesenchyme clearly plays a critical role in this process (Zhang et al. 2002; Rice et al. 2004; Lan and Jiang 2009). As outgrowth proceeds, the palatal shelves extend into the oral cavity and are vertically positioned lateral to the tongue by E13.5. Although the secondary palatal shelves appear continuous along their anterior–posterior (A–P) length, they are precisely patterned along this axis, and molecular control of palate formation is distinct at different A–P positions (Hilliard et al. 2005; Gritli-Linde 2007). Elevation brings the initially vertically oriented palatal shelves to a horizontal position, where continued growth results in the apposition and ultimate fusion of the palatal shelves (Chai and Maxson 2006).
Many of the anomalies exhibited by CFNS patients affect structures derived from the neural crest, raising the question of whether CFNS pathology might be secondary to ephrin-B1 influence on neural crest migration (Twigg et al. 2004; Wieacker and Wieland 2005). Indeed, deletion of ephrin-B1 in the neural crest and its derivatives resulted in similar craniofacial phenotypes as null loss of function, although this did not resolve whether ephrin-B1 acts early in neural crest migration or later in craniofacial development (Davy et al. 2004). Interestingly, although CFNS is X-linked, EPHRIN-B1+/− heterozygous females are more severely affected than hemizygous males (Wieacker and Wieland 2005). Similarly, ephrin-B1+/− heterozygous mice display additional and more severe phenotypes than ephrin-B1Y/- hemizygous or ephrin-B1−/− homozygous mutant mice, including polydactyly and frontal bone foramina (Compagni et al. 2003; Davy et al. 2004). This phenomenon is attributable to random X inactivation resulting in mosaic loss of ephrin-B1 function, and subsequent ephrin-mediated cell sorting leading to ectopic expression boundaries (Compagni et al. 2003; Davy et al. 2004). Gap junction communication is inhibited at these boundaries in the developing calvaria, decoupling ephrin-B1-expressing and -nonexpressing domains and leading to delayed ossification of the frontal bones (Davy et al. 2006). How the disruption of Eph/ephrin boundaries leads to more severe dysmorphogenesis in other contexts, however, has not been explored.
Ephrin-B1 is a member of the transmembrane B-type subfamily of Eph/ephrin signaling molecules, and is known to be capable of signaling by forward or reverse signaling mechanisms (Davy and Soriano 2005). Ephrin-B1 PDZ-dependent reverse signaling controls axon guidance of the corpus callosum, whereas forward signaling is critical for normal craniofacial development (Bush and Soriano 2009; Risley et al. 2009). Compound mutation of the high-affinity ephrin-B1 receptors EphB2 and EphB3 results in a cleft palate phenotype, indicating that these are relevant receptors for palatogenesis (Orioli et al. 1996; Risley et al. 2009).
Initial activation of forward signaling by ephrin engagement results in autophosphorylation of the Eph receptor, followed by the binding and phosphorylation of signaling adaptor molecules (Kullander and Klein 2002; Lackmann and Boyd 2008). Multiple signaling adaptors can bind to Eph receptors, although their in vivo and context-specific relevance remains mostly uncertain. Downstream transduction of the Eph/ephrin forward signal has been proposed to occur by a variety of mechanisms that differ depending on cellular context, including Rho family small GTPases, PI3K signaling pathway, integrin signaling, extracellular signal-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) pathway, and others (Kullander and Klein 2002; Lackmann and Boyd 2008; Pasquale 2008; Poliakov et al. 2008).
The ERK/MAPK signal transduction pathway is used frequently by receptor tyrosine kinase signaling, and ends with the transcriptional activation of immediate early genes (IEGs) to control cell proliferation, behavior, differentiation, and survival (Lau and Nathans 1987; Murphy and Blenis 2006). The consequence of activation of Eph/ephrin signaling on ERK/MAPK signaling is unclear, however. In neuronal cell culture, activation of EphB2 resulted in inhibition of the ERK/MAPK signaling pathway and led to neurite collapse (Elowe et al. 2001). Conversely, transfection of HEK293 cells with EphB2 activated the ERK/MAPK signaling pathway, which was required for cell repulsion (Zisch et al. 2000; Poliakov et al. 2008). The disparate and sometimes contradictory signal transduction mechanisms used by Eph/ephrin signaling may therefore require a more systems-level approach that considers context to determine the relevant mode of action (Lackmann and Boyd 2008; Jorgensen et al. 2009). Furthermore, an immediate early transcriptional response, a hallmark of activation of other receptor tyrosine kinase pathways, has not been reported downstream from Eph/ephrin signaling.
Here we elucidate mechanisms of ephrin-B1 signaling control of palatogenesis. We find that ephrin-B1 acts to intrinsically control anterior palatal shelf outgrowth and morphogenesis by regulating mesenchymal cell proliferation. We take advantage of naturally occurring mosaicism in heterozygous ephrin-B1+/− mutant mice to investigate molecular mechanisms of Eph/ephrin signaling function. Ephrin-B1+/− mutant embryos exhibit more severe dysmorphogenesis of the palatal shelves, whereby mosaic loss of function of ephrin-B1 disrupts cell proliferation rate in a nonuniform manner. Loss of ephrin-B1 expression results in up-regulation of EphB3 receptor protein as a consequence of relief of endocytosis and degradation, identifying this as a relevant in vivo mechanism for the regulation of EphB receptor expression. Integrating phosphoproteomic and transcriptomic approaches reveals that the Eph/ephrin signaling system uses the ERK/MAPK signal transduction pathway to regulate the transcription of a set of IEGs, and to control palatal shelf cell proliferation. These studies elucidate ephrin-B1 signaling control of palatal shelf outgrowth, and provide evidence that Eph/ephrins induce a growth factor signaling cascade in this developmental context.
Results
Ephrin-B1 is required in the anterior palatal shelf mesenchyme for normal outgrowth of the palatal shelves
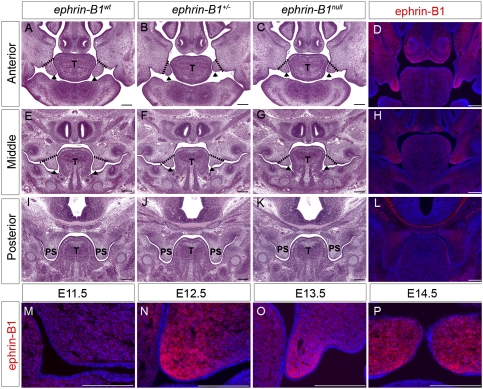
To better understand the cleft palate phenotype resulting from ephrin-B1 loss of function, we performed histological examination of frontal sections of ephrin-B1null and ephrin-B1+/− embryos at progressive stages of mouse palatogenesis. At E12.5, no differences in palatal shelf outgrowth or shape were detected in either ephrin-B1null or ephrin-B1+/− embryos, indicating that outgrowth had initiated normally (data not shown). A deficit in palatal shelf outgrowth was first apparent in ephrin-B1null mutant embryos by E13.5. This defect was limited to the anterior palate where ephrin-B1null palatal shelves were truncated and did not display the well-extended finger-like projection of anterior palatal shelves in ephrin-B1wt embryos (Fig. 1A,C). The secondary palate was more mildly affected at middle positions along the A–P axis, and the posterior palate appeared completely normal (Fig. 1E,G,I,K). Ephrin-B1+/− palates appeared more severely dysmorphic than those in ephrin-B1null mutant embryos, although the penetrance of the cleft palate phenotype is similar between the two genotypes, with ∼70% exhibiting cleft palate (Fig. 1B,C,F,G; Davy et al. 2004). By E14.5, both ephrin-B1null and ephrin-B1+/− palatal shelves were able to elevate above the tongue with normal timing, but appeared hypoplastic and did not appose at the midline (Supplemental Fig. 1). Therefore, ephrin-B1 signaling controls outgrowth of the anterior palate, but is not required for palatal shelf elevation.
Figure 1.
Ephrin-B1 is required for anterior palatal shelf morphogenesis. (A–C,E–G,I–K) Histological frontal sections of E13.5 embryonic heads reveal reduced anterior palatal shelf outgrowth in ephrin-B1+/− (arrowheads in B) and ephrin-B1null (arrowheads in C) compared with the finger-like projection in ephrin-B1wt (arrowheads in A). The palatal shelves appear slightly affected in middle positions of ephrin-B1+/− (arrowheads in F) and ephrin-B1null (arrowheads in G) mutant embryos compared with ephrin-B1wt at the same position (arrowheads in E). The posterior palate is unaffected in ephrin-B1+/− (J) and ephrin-B1null (K) mutant embryos. (D,H,L,M–P) Immunofluorescence detection of ephrin-B1 expression in ephrin-B1wt embryos. At E13.5, Ephrin-B1 expression is restricted to the anterior palatal shelf mesenchyme (D), is more weakly expressed in the middle (H), and is not detected in the posterior palate (L). Ephrin-B1 is not significantly detectable in the palatal shelf primordia at E11.5 (M), but becomes strongly detectable within the palatal mesenchyme by E12.5 (N). Ephrin-B1 expression remains highly restricted to the mesenchyme during outgrowth (E13.5) (O) and after elevation (E14.5) (P) of the palatal shelves. Bar, 200 μm. (T) Tongue; (PS) palatal shelves.
To better understand how ephrin-B1 signaling might be controlling the morphogenesis of the anterior palate, we examined its expression at progressive stages of mouse palatogenesis. Antibody staining indicated that ephrin-B1 was not significantly expressed in the primordial palatal mesenchyme at E11.5 (Fig. 1M). Robust expression of ephrin-B1 was first detected in the anterior secondary palate at E12.5, strongest at the most distal tip of the palatal shelf mesenchyme (Fig. 1N). This expression strengthens and expands to encompass most of the anterior palatal shelf mesenchyme by E13.5, is maintained after elevation at E14.5 as fusion of the palatal shelves begins, and is finally down-regulated around E15.5 (Fig. 1O,P; data not shown). At all stages, ephrin-B1 exhibited an anterior-restricted mesenchymal expression that strongly correlated spatiotemporally with the cleft palate phenotype, and suggested that ephrin-B1 controls anterior palatal shelf outgrowth intrinsically (Fig. 1D,H,L).
Since ephrin-B1 exhibited anterior-restricted expression during palatal shelf outgrowth, we asked whether A–P identity of the palatal shelves was maintained in ephrin-B1 mutants. We therefore examined the expression of several anterior- or posterior-specific markers by whole-mount and section in situ hybridization of ephrin-B1null and ephrin-B1+/− mutant embryos. The transcription factors Shox2 and Msx1 are well-studied markers of anterior palatal shelf identity, whereas the expression of the T-box transcription factor Tbx22 marks the posterior palate (Braybrook et al. 2002; Bush et al. 2002; Zhang et al. 2002; Herr et al. 2003; Yu et al. 2005). Judging by these markers, the A–P patterning of the palatal shelf was normal in both ephrin-B1null and ephrin-B1+/− mutant embryos (Supplemental Fig. 1J–O; data not shown). Since sonic hedgehog (Shh) signaling from transverse epithelial thickenings, called rugae, to the underlying mesenchyme is critical for palatal shelf outgrowth (Rice et al. 2004; Lan and Jiang 2009), we examined Shh expression by whole-mount in situ hybridization. Shh expression was not significantly affected in ephrin-B1null or ephrin-B1+/− mutant palates (Supplemental Fig. 1P–R). Anterior rugae appeared slightly disorganized in ephrin-B1+/− palatal shelves, most likely a reflection of the more severe dysmorphogenesis also observed by histology (Supplemental Fig. 1P–R).
Post-transcriptional regulation of Eph–ephrin expression borders
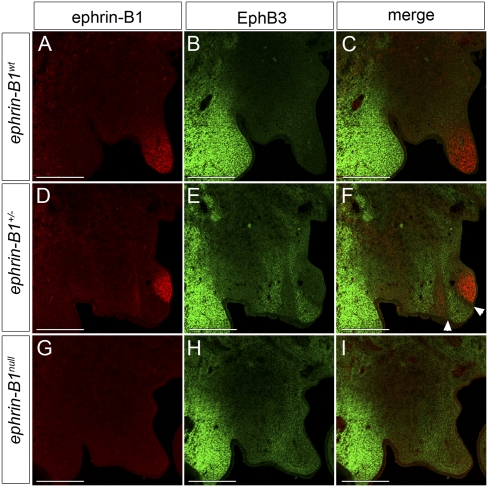
Since compound homozygous mutation of the high-affinity ephrin-B1 receptors EphB2 and EphB3 results in a cleft palate phenotype (Orioli et al. 1996), we decided to examine their expression with respect to ephrin-B1 during palatogenesis by double-antibody staining. Surprisingly, EphB3 protein was undetectable in the palatal shelf, and was instead intensely expressed in the maxillary mesenchyme lateral to the oral cavity, a region that does not express ephrin-B1 (Fig. 2A–C). Consistent with previous reports, we observed large mosaic domains of ephrin-B1 expression in ephrin-B1+/− palates (Compagni et al. 2003; Davy et al. 2006). Examination of receptor expression in ephrin-B1+/− and ephrin-B1null mutant embryos, however, revealed a striking up-regulation of EphB3 protein in ephrin-B1 mutant domains (Fig. 2D–I). In ephrin-B1+/− heterozygous mutant palates, this up-regulation was mosaic in a pattern complementary to ephrin-B1 expression, and a boundary one to two cells wide separated Ephrin-B1-positive and EphB3 up-regulated domains (Fig. 2D–F, white arrowheads in F). We could detect broad expression of EphB2 at E13.5, although this signal was complementary in intensity to ephrin-B1 expression (Supplemental Fig. 2C). EphB2 expression exhibited a similar but more subtle up-regulation in ephin-B1-negative domains in both ephrin-B1+/− and ephrin-B1null mutant palates (Supplemental Fig. 2F,I).
Figure 2.
EphB3 protein is up-regulated in ephrin-B1 mutant domains in the palate. (A–I) Immunofluorescence detection of ephrin-B1 (red) and EphB3 (green) in frontal sections of E13.5 anterior palatal shelves. (B,C) EphB3 protein expression is not detectable in the palatal shelves of ephrin-B1wt E13.5 embryos. In ephrin-B1+/− mutant embryos (D–F), the expression of ephrin-B1 is mosaic (D,F) and EphB3 protein is up-regulated in domains lacking ephrin-B1 expression within the palatal shelf mesenchyme (E,F). (F) In addition, a boundary one to two cells in width separates the ephrin-B1-expressing and EphB3-expressing domains (arrowheads). (H,I) EphB3 protein is up-regulated throughout the anterior palatal shelf mesenchyme of ephrin-B1null mutant embryos. Bar, 200 μm.
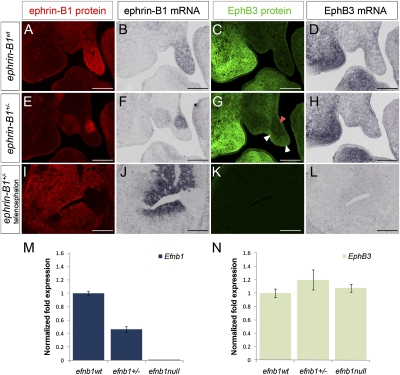
In ephrin-B1+/− embryos, sorting mediated by ephrin-B1 expression leads to enlarged mosaic patches of ephrin-B1 expression (Compagni et al. 2003; Davy et al. 2006). In the limb bud, Eph/ephrin-induced cell sorting also resulted in complementary localization of EphB receptor-expressing cells (Compagni et al. 2003). If complementary expression of EphB3 protein in the palate was due to cell sorting, EphB3 mRNA distribution should reflect the same complementary pattern of expression. We addressed this question by performing antibody staining and in situ hybridization analysis of ephrin-B1 and EphB3 in adjacent sections to compare their expression at the protein and mRNA levels. Interestingly, ephrin-B1wt embryonic palates exhibited expression of EphB3 mRNA in regions where EphB3 protein was not detectable (Supplemental Fig. 3A–D). No up-regulation of EphB3 mRNA was detected in ephrin-B1-negative regions of mutant palates, although EphB3 protein was up-regulated in both cases (Fig. 3E–H; Supplemental Fig. 3A–D). Using quantitative RT–PCR (QRT–PCR) as a more sensitive readout, we did not detect significant changes in EphB3 mRNA expression levels in either ephrin-B1+/− or ephrin-B1null palatal shelves (Fig. 3N). We asked whether this up-regulation was a consequence of loss of forward or reverse signaling by analyzing EphB3 receptor expression in ephrin-B16FΔV mouse mutants that specifically abrogate reverse signaling, leaving forward signaling intact (Bush and Soriano 2009). No up-regulation of EphB3 protein was detected in ephrin-B16FΔV/+ or ephrin-B16FΔV/Y palatal shelves, indicating that this up-regulation is not a consequence of the loss of reverse signaling (Supplemental Fig. 3E–J). These data indicate that the up-regulation of EphB3 receptor in the absence of ephrin-B1 forward signaling in the palate occurs at the post-transcriptional level. Consistent with this observation, we also found that EphB3 protein was not up-regulated in the ephrin-B1null or ephrin-B1+/− mutant telencephalon, a region that does not express EphB3 mRNA, indicating that EphB3 mRNA must be transcribed for receptor up-regulation to be possible (Fig. 3I–L).
Figure 3.
EphB3 is post-transcriptionally up-regulated in ephrin-B1 mutant palates. Adjacent frontal sections of E13.5 anterior palatal shelves analyzed by antibody staining for ephrin-B1 (red) (A,E,I), in situ hybridization for ephrin-B1 (B,F,J), antibody staining for EphB3 (green) (C,G,K), and in situ hybridization for EphB3 (D,H,L). In ephrin-B1wt embryos (A–D), ephrin-B1 protein (A) and mRNA (B) are detected throughout the palatal shelf mesenchyme. Although EphB3 protein is not detectable (C), EphB3 mRNA is expressed in the ephrin-B1wt palatal shelf (D). In ephrin-B1+/− embryos (E–H), ephrin-B1 protein (E) and mRNA (F) are expressed in an identical mosaic pattern. Whereas EphB3 protein is up-regulated specifically in ephrin-B1 mutant domains (white arrowheads in G), EphB3 mRNA does not display this complementary pattern of regulation, and instead maintains the wild-type expression pattern (H). In E13.5 ephrin-B1+/− telencephalon (I–L), mosaic domains of ephrin-B1 protein (I) and mRNA (J) but no complementary up-regulation of EphB3 protein are observed (K). (L) EphB3 mRNA is not detectable in the telencephalon at this stage. (M) QRT–PCR analysis of dissected palates indicates that ephrin-B1 is down-regulated in ephrin-B1+/− and ephrin-B1null palatal shelves relative to ephrin-B1wt (relative mean normalized to GAPDH ± SD). (N) EphB3 mRNA expression levels are not significantly different between ephrin-B1wt, ephrin-B1+/−, and ephrin-B1null mutant palatal shelves. Bar, 200 μm.
Phosphoproteomic profiling of ephrin-B1 signaling in the palate
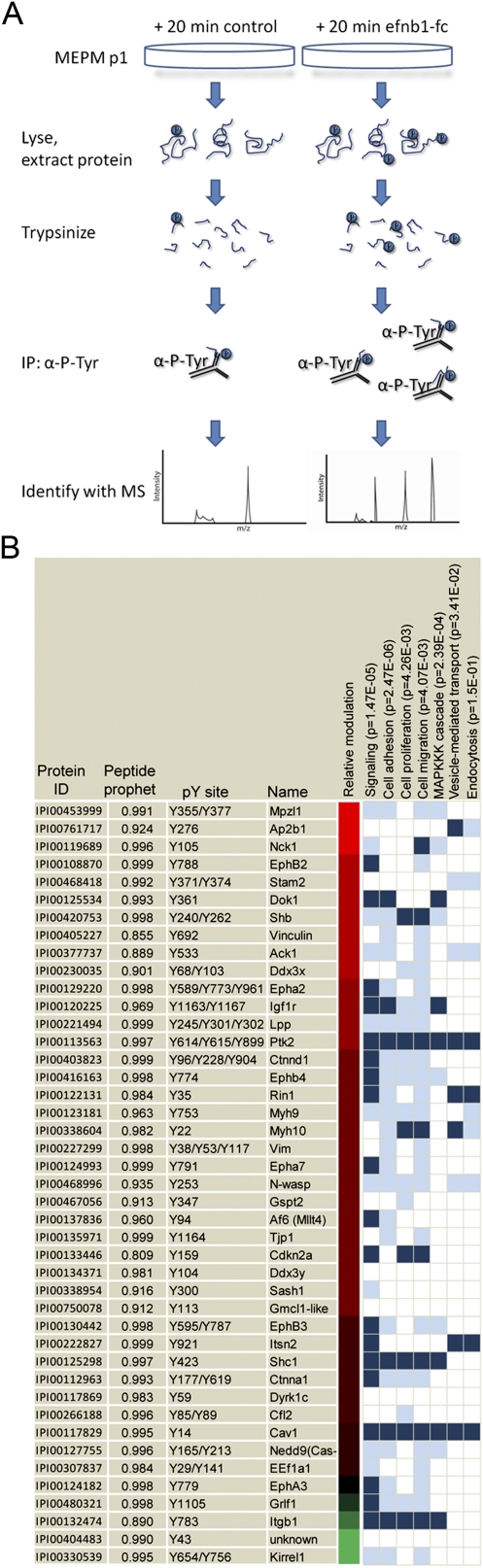
Protein phosphorylation is a central feature of receptor tyrosine kinase signaling pathways and has numerous roles in nearly all developmental and cellular processes (Hunter 2000). As such, proteomic approaches using mass spectrometry to identify tyrosine-phosphorylated proteins have become increasingly powerful for the dissection of biochemical signaling pathways (Ptacek and Snyder 2006; Huang and White 2008). We therefore undertook a proteomic profiling approach to interrogate the signal transduction pathways downstream from ephrin-B1 signaling in the palate. Toward this end, we first established primary cell cultures of mouse embryonic palate (MEP) cells as a system for identifying phosphorylation targets of ephrin-B1-activated forward signaling. In our conditions, these cells maintained expression of key markers of palatal mesenchyme cells, including Osr2, Msx1, and Pdgfrα, as well as ephrin-B1, EphB2, and EphB3 (data not shown). These cells activate forward signaling in response to preclustered ephrin-B1-Fc, and can induce the phosphorylation of unidentified target proteins (Supplemental Fig. 4A,B). To qualitatively identify phosphorylation targets of ephrin-B1 forward signaling, we adapted a proteomic approach for use in MEP cells (Fig. 4A; Zhang et al. 2005). Control induction or induction with preclustered ephrin-B1-Fc was performed for 20 min, and tyrosine-phosphorylated tryptic peptides were immunoprecipitated in parallel and identified by liquid chromatography coupled to tandem mass spectrometry with electrospray ionization (LC-ESI MS/MS). Application of this method resulted in the identification of 244 phosphorylation sites on 136 unique proteins, of which 96 could be identified with high confidence (Nesvizhskii et al. 2003). Of these, 46 were judged qualitatively differentially phosphorylated on the basis of spectral counts (Fig. 4; Supplemental Table 1). As expected, EphB2 and EphB3 were differentially phosphorylated in response to induction of forward signaling. To confirm the differential phosphorylation of other proteins identified in this experiment, we selected six candidates (Dok1, Shc1, Shb, Nck1, EphB4, and Ctnnd1) and performed immunoprecipitation followed by Western blotting with an antibody recognizing phosphorylated tyrosine. Increased tyrosine phosphorylation of candidates was observed in each of these cases, although the precise quantitative degree of phosphorylation varied due to the qualitative nature of our method (Supplemental Fig. 4B). These data therefore provide a reliable profile of the activation of Eph/ephrin forward signaling in palatal cells.
Figure 4.
Phosphoproteomic identification of phosphorylation targets of ephrin-B1 forward signaling in palate cells. (A) Schematic representation of strategy for the identification of ephrin-B1 forward signaling targets. Induction of forward signaling was performed for 20 min with 2 μg/mL preclustered ephrin-B1-Fc. Briefly, cells were lysed and protein was extracted, followed by digestion into tryptic peptides. Immunoprecipitation with an antibody against phosphorylated tyrosine was followed by elution of peptides and LC-MS/MS to identify differentially phosphorylated targets. (B) Proteins identified as phosphorylated are listed by name and protein ID. The site(s) of phosphorylation and peptide prophet score (see the Materials and Methods) are listed. Chromatic representation of the ratio of the relative number of spectral counts (induced to uninduced) was estimated based on the log10 ratio of 1 + (induced peptides to uninduced); complete spectral counts are found in Supplemental Table 1. GO annotations are listed in vertical columns, along with associated P-values. Dark blue represents official assignment to a GO category, and light blue represents manual assignments based on literature searching.
To begin to relate this signaling profile to the morphogenesis of the palate, we performed gene ontology (GO) and literature analysis of associated biological processes (Fig. 4B; Supplemental Table 2). As expected, we found that signaling proteins were strongly overrepresented in our data set (Fig. 4B). Consistent with known functions of Eph/ephrin signaling, we also found that our data set was enriched for proteins involved in cell adhesion and cell migration. Interestingly, cell proliferation and the MAPKKK cascade were also identified as overrepresented in our data set, implicating these as potentially relevant mechanisms of Eph/ephrin action in the palate. Finally, several proteins in our data set were associated with endocytosis and vesicle-mediated transport, categories that captured our interest based on the post-transcriptional regulation of EphB3 in the palate.
Endocytosis and degradation of EphB3 receptor in the palate
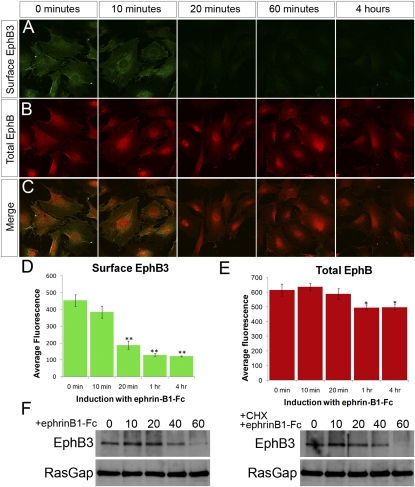
Since our proteomics data set identified components of endocytosis and protein trafficking machinery, and previous studies have shown that activation of EphB signaling by ephrin-B1 can result in receptor endocytosis, we wondered whether the post-transcriptional regulation of EphB3 receptor was related to endocytosis and degradation of the receptor (Marston et al. 2003; Zimmer et al. 2003; Deininger et al. 2008). To test whether this was the case in the palatal shelves, we induced forward signaling in MEP cells, and examined surface EphB3 by staining with an antibody recognizing the extracellular portion of EphB3 in the absence of Triton permeabilization. We found that EphB3 was detectable on the cell surface before induction, was progressively removed upon activation of signaling, and was barely detectable at 1 h, whereas total EphB remained detectable (Fig. 5 A,B).
Figure 5.
EphB3 is internalized and degraded in response to activation of forward signaling in palatal cell cultures. (A,C) Immunostaining of MEP cells with an antibody recognizing the extracellular domain of EphB3 (green), without cell permeabilization to detect cell surface EphB3. (D) Confocal quantification indicates EphB3 is removed from the cell surface over a time course of ephrin-B1 activation of forward signaling (shown in B,C,E) (mean ± SEM; [**] P < 0.0001, Student's t-test). Total EphB staining (red) diminished only slightly (mean ± SEM; [**] P < 0.02, Student's t-test). (F) Immunoprecipitation of EphB3 from MEP cell lysate followed by Western blotting indicates that total EphB3 protein is reduced over a time course of forward signaling activation. Induction of forward signaling in the presence of 10 μg/mL cycloheximide indicates that EphB3 protein reduction is not a consequence of an effect on translation rate.
To examine whether EphB3 was degraded after its internalization, we examined total EphB3 by immunoprecipitation and Western blotting. Upon activation of forward signaling, we observed a progressive decrease in the total amount of EphB3 protein (Fig. 5F). In principle, these results could be explained by constant turnover of EphB3 receptor in combination with decreased rate of translation of the receptor. Repeating inductions in the presence of cycloheximide to block protein synthesis, however, indicated that differences in translation rate were not the cause of diminished EphB3 receptor (Fig. 5F). It is possible that these results could be explained by ephrin-B1-induced cleavage and shedding of the EphB3 ectodomain, as has been shown to occur in both 293 cells and hippocampal neurons (Lin et al. 2008). Given that the kinetics of removal of EphB3 from the cell surface precede its degradation, we find this explanation unlikely. These results instead indicate that, in palatal mesenchyme cells, activation of EphB3 signaling by ephrin-B1 leads to internalization and degradation of the receptor, and are consistent with the complementary up-regulation of EphB3 protein in ephrin-B1 mutant domains resulting from relief of forward signaling-activated endocytosis and degradation of the EphB3 receptor (Fig. 7F).
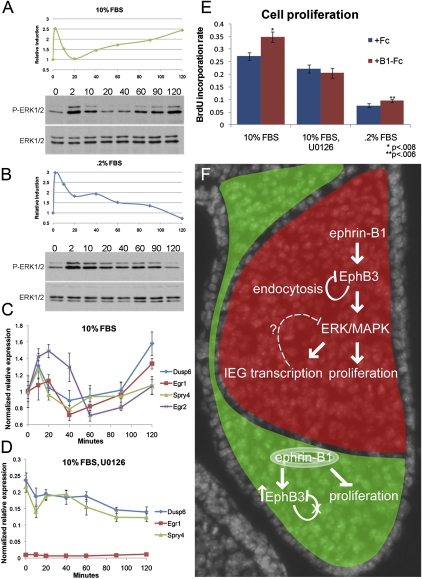
Figure 7.
Ephrin-B1 forward signaling activates ERK/MAPK signaling in palatal cells. (A,B) Phosphorylated ERK1/ERK2 were detected by Western blot and quantified using ImageJ. (A) Time course of induction with 2 μg/mL preclustered ephrin-B1-Fc in media containing 10% FBS resulted in two phases of ERK1/ERK2 activation. (B) In starvation conditions (media containing 0.2% FBS), an activation of ERK1/ERK2 was observed at 2 min, followed by decay. (C,D) Real-time QRT–PCR quantification of immediate early transcriptional targets of ephrin-B1 forward signaling in MEP cells. (C) Regulation of expression of immediate early targets shadows the activation of the ERK/MAPK signaling. QRT–PCR over a time course of forward signaling induction shows initial up-regulation, down-regulation by 1 h, and increase in expression by 2 h (relative mean normalized to GAPDH ± SD) (D) Inhibition of Ras–MAPK activation eliminates the induction of IEG transcriptional targets. Real-time QRT–PCR over a time course of forward signaling induction in the presence of 10 μM U0126 (relative mean normalized to GAPDH ± SD). (E) BrdU incorporation after 3 h of treatment with 2 μg/mL preclustered ephrin-B1-Fc indicates that cell proliferation is induced by ephrin-B1 forward signaling in media containing 10% FBS (n = 9, mean ± SEM; P < 0.008) or 0.2% FBS (n = 9, mean ± SEM; P < 0.006); this effect is eliminated when Ras–MAPK signaling is blocked by treatment with 10 μM U0126 (n = 9, mean ± SEM). (F) Model of ephrin-B1 regulation of palatal morphogenesis. Ephrin-B1+/− heterozygous females constitute mosaic expression of ephrin-B1 (red). Ephrin-B1 activates forward signaling, resulting in endocytosis and degradation of the EphB receptor. In ephrin-B1 mutant domains, relief of this feedback allows receptor up-regulation (green); several candidate mediators of this feedback were identified as phosphorylation targets of forward signaling (Rin1, Stam2, and Ap2β1). Activation of this forward signal also leads to the phosphorylation of several signaling adaptors (Shc1, Shb, and Dok1) that are capable of regulating ERK/MAPK. The transcriptional regulation of IEGs by activation of the ERK/MAPK may also play a role in feedback regulation of the pathway. Cell proliferation, which is critical for the outgrowth of the palatal shelves, is also dependent on signaling through ERK/MAPK.
Ephrin-B1 controls cell proliferation in the anterior palatal shelf
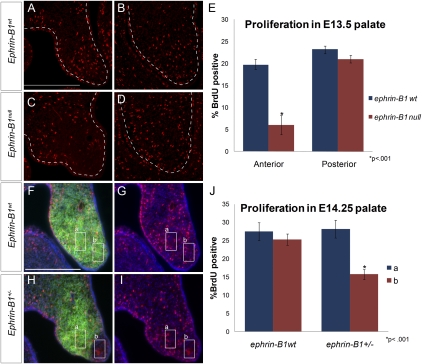
Since cell proliferation provides a critical force driving palatal shelf outgrowth, we next examined cell proliferation in ephrin-B1null mutant palatal shelves by BrdU incorporation. We found that E13.5 ephrin-B1null palatal shelves had a significantly reduced cell proliferation rate in the anterior palatal shelf mesenchyme, whereas cell proliferation in the posterior palate was unaffected (Fig. 6A–E). Our histological analysis indicated that ephrin-B1+/− palates, like other craniofacial structures, displayed a more severely dysmorphic shape than ephrin-B1null palatal shelves. Since ephrin-B1+/− mutants constitute mosaic loss of ephrin-B1 function, we wondered whether the cell proliferation rate was also disrupted in a mosaic pattern. We therefore examined double staining for ephrin-B1 and BrdU incorporation in ephrin-B1+/− heterozygous palatal shelves. Regions inside ephrin-B1-negative domains showed a significantly reduced average BrdU incorporation rate compared with neighboring ephrin-B1-positive domains of expression (Fig. 6H–J). This effect was not due to the fact that we were examining different regions of the palatal shelf, because control regions chosen from wild-type palatal shelves did not exhibit this reduction (Fig. 6F,G,J). This mosaic disruption of cell proliferation was not limited to the palate; counting regions in ephrin-B1+/− telencephalons also revealed a reduction in cell proliferation rate in ephrin-B1 mutant domains (Supplemental Fig. 5A–C).
Figure 6.
Cell proliferation rate is disrupted in ephrin-B1 mutant palates. (A–D) Frontal sections of E13.5 palatal shelves showing BrdU incorporation (red). A significant reduction in BrdU incorporation was detected in ephrin-B1null (C,E) anterior palatal shelf mesenchyme compared with ephrin-B1wt (A,E) (mean ± SEM; [*] P < 0.001, Student's t-test). No significant reduction was observed in the posterior palatal shelf mesenchyme of ephrin-B1null embryos (D,E) compared with ephrin-B1wt at this position (B,E). (F–J) Frontal sections of E14.25 palatal shelves showing double staining for BrdU incorporation (red), ephrin-B1 (green), and nuclei (DAPI, blue). Ephrin-B1-expressing (box a) and Ephrin-B1-negative (box b) regions were chosen in frontal sections of ephrin-B1+/− palates. (F,G) These regions were matched in ephrin-B1wt for control. (H,I,J) The proliferation rate was significantly reduced in ephrin-B1-negative domains compared with neighboring positive domains (n = 46 regions, 30 sections, mean ± SEM; [*] P < 0.001, Student's t-test). (F,G,J) The proliferation rate in comparable areas of ephrin-B1wt palatal shelves indicated that this was not a consequence of regional differences in the proliferation rate (n = 22 regions, 12 sections, mean ± SEM).
Since regions of palatal mesenchyme not expressing ephrin-B1 had a reduced cell proliferation rate, over time, ephrin-B1 mutant domains should be underrepresented in ephrin-B1+/− palates. Measuring ephrin-B1-expressing and -nonexpressing regions of the palate revealed that, on average, only 40% of the anterior palate was mutant for ephrin-B1, instead of the expected 50% (P < 0.001) (Supplemental Fig. 5D,E). Loss of function of ephrin-B1 in the palate therefore results in reduced cell proliferation rate, and disrupts cell proliferation rate in a nonuniform manner in ephrin-B1+/− mutant palates.
Eph/ephrin signaling activates Ras–MAPK to control palatal cell proliferation and regulate the expression of immediate early transcriptional targets
Based on GO analysis of our proteomic data, a significant number of phosphorylation targets of ephrin-B1 signaling regulate the ERK/MAPK signal transduction pathway. We therefore decided to examine activation of the pathway by Western blot analysis of phosphorylated ERK1/ERK2. We found that treatment of MEP cells with preclustered ephrin-B1-Fc resulted in rapid phosphorylation of ERK1/ERK2 in both serum-fed and serum-starved conditions, whereas treatment with an Fc control did not (Fig. 7A,B; data not shown). Interestingly, stimulation of forward signaling in serum-fed conditions resulted in an acute phase of MAPK signaling activation followed by a delayed phase, whereas the acute phase was followed by decay in serum-starved conditions (Fig. 7A,B). In contrast, induction of forward signaling revealed no change in phosphorylation of p38, JNK, or AKT (data not shown). These results indicate that, in palate cells, Eph/ephrin forward signaling activates the ERK/MAPK signal transduction pathway.
An IEG transcriptional response is a hallmark of growth factor signaling pathways, although the precise way in which this response is integrated to achieve specific cellular readout remains a question of interest (Lau and Nathans 1987; Schmahl et al. 2007). Such an immediate early transcriptional response has not been described for Eph/ephrin signaling, however, and we therefore examined the induction of forward signaling in palatal cell culture. We analyzed gene expression changes after 1-h induction of forward signaling in MEP cells by Illumina beadchip microarray. We found that, although very few genes were significantly regulated (>1.5-fold change), a set of known immediate early transcriptional targets of ERK/MAPK were significantly down-regulated (Supplemental Fig. 6A). We therefore chose to examine the expression of these genes (Egr1, Egr2, Spry4, and Dusp6) over a time course of forward signaling activation using QRT–PCR. Regulation of these targets generally shadowed the activation kinetics of ERK/MAPK signaling, such that an early induction at 10–20 min was followed by down-regulation at 40–80 min and a second phase of up-regulation by 2 h (Fig. 7C). To examine whether regulation of these immediate early targets depended on activation of ERK/MAPK signal transduction, we performed a time course in the presence of U0126, which inhibits activation of the ERK/MAPK pathway at the level of MEK. As expected, preincubation with U0126 dramatically reduced basal phosphorylation of ERK1/ERK2, and completely blocked ephrin-B1-stimulated ERK/MAPK activation (Supplemental Fig. 6B). After U0126 treatment, Egr2 mRNA was not detectable by QRT–PCR, indicating that its expression in MEP cells is absolutely dependent on Ras–MAPK, and the expression of Egr1, Spry4, and Dusp6 was also dramatically down-regulated and was not induced by activation of ephrin-B1 signaling (Fig. 7D; data not shown). Instead, Dusp6 and Spry4 were down-regulated along the time course of forward signaling activation. These results indicate that ephrin-B1 forward signaling regulates the transcription of a set of IEGs via the ERK/MAPK signaling pathway.
We next examined whether ephrin-B1 had an effect on cell proliferation in cell culture. Stimulating MEP cells in growing conditions with ephrin-B1 for 3 h resulted in an increased rate of Brdu incorporation (P < .008), indicating that activation of forward signaling was capable of inducing a proliferative response (Fig. 7E). Interestingly, induction of forward signaling in starvation conditions also induced a statistically significant proliferative response (P < .006) (Fig. 7E). In the presence of U0126, the proliferative response was alleviated, indicating that the ERK/MAPK pathway was required for transduction of the ephrin-B1 proliferative signal (Fig. 7E). Together, these results indicate that ephrin-B1 forward signaling regulates IEG transcription and cell proliferation by the ERK/MAPK signal transduction pathway.
Discussion
The Eph/ephrin family of signaling molecules has long been known as a cell positioning system that can modulate cell behavior (Lackmann and Boyd 2008). Here, we show that ephrin-B1 forward signaling also acts as a mitogen to regulate palatal shelf mesenchymal cell proliferation. Recently, evidence has been accumulating that, in some contexts, Eph/ephrin signaling may regulate cell proliferation, but the underlying mechanisms are unresolved. Anti-proliferative activities have been reported for Eph/ephrin signaling in neural progenitor cells, the epidermis, and breast cancer cells (Holmberg et al. 2005; Noren et al. 2006; Ricard et al. 2006; Genander et al. 2010). Less evidence for a positive regulation of cell proliferation exists; during hippocampal neurogenesis, loss of function of EphB1 or ephrin-B3 resulted in reduced proliferation rate, and treatment of endothelial cells with ephrin-B2 resulted in increased cell proliferation by the PI3K pathway in culture (Steinle et al. 2002; Chumley et al. 2007). In the intestinal stem cell niche, activation of EphB receptors accounts for a significant portion of ongoing mitogenic activity by regulating cyclin-D1 levels (Holmberg et al. 2006; Genander et al. 2009). Interestingly, we did not observe an effect on cyclin-D1 levels in the palate (data not shown). Instead, we now provide evidence that the mitogenic action of ephrin-B1 depends on activation of the classical ERK/MAPK signal transduction pathway.
Mass spectrometry-based proteomic approaches are beginning to provide significant insights into principles of eph/ephrin and other receptor tyrosine kinase signaling networks (Zhang et al. 2006, 2008; Jorgensen et al. 2009). These approaches generally use heterologous cell culture systems, and have revealed a strikingly complex and extensive network of targets that depends considerably on context (Ptacek and Snyder 2006; Huang and White 2008). A recent systems-level analysis of EphB2/ephrin-B1 phosphotyrosine signaling networks provided a wealth of information regarding signaling underlying cell sorting in transfected HEK293 cells (Jorgensen et al. 2009). Efforts to integrate these data with genetic studies will be critical for understanding signaling in developmental and disease processes. Here we used a phosphoproteomic approach to identify phosphorylation targets of endogenously expressed receptors in primary palatal cell culture. Not surprisingly, Eph receptors EphB2 and EphB3 were identified as phosphorylated in response to ephrin-B1 stimulation. Unexpectedly, we found that several receptors not known to be activated by ephrin-B1 were also phosphorylated, including EphB4 and two A-type receptors (EphA2 and EphA7). Whether these are autophosphorylated in response to ephrin-B1 activation or secondary targets of an ephrin-B1 signal cascade is an interesting question for further consideration. Systems level analysis of these data also revealed functional relationships to several key biological processes that are regulated by ephrin-B1 signaling in palatogenesis, including endocytosis, cell proliferation, and ERK/MAPK regulation.
Significant ongoing work indicates that endocytosis is a critical component of the regulation of Eph/ephrin signaling events (Egea and Klein 2007). In cell culture, an otherwise adhesive force provided by the interaction of receptors and ligands can be converted to a repulsive signal by bidirectional endocytosis of receptor/ligand complexes (Marston et al. 2003; Zimmer et al. 2003). Here we provide evidence that such a post-transcriptional mechanism also regulates Eph receptor expression in the palatal shelves in vivo. Our data indicate that loss of ephrin-B1 expression in the palate results in the post-transcriptional up-regulation of EphB3 by the relief of endocytosis and degradation. This model also explains the border between ephrin-B1-positive and EphB3 up-regulation domains: Signaling would be expected to induce endocytosis and degradation in cells directly neighboring an ephrin-B1-positive domain, but not beyond (Fig. 7F). Therefore, this mechanism may not only modulate adhesion versus repulsion, but may also regulate the strength or duration of an Eph/ephrin signal, as has been observed for other receptor tyrosine kinase receptors (Wiley and Burke 2001). Given the highly promiscuous nature of Eph/ephrin signaling, this regulation may be particularly critical, since Eph receptor expression is often superimposed upon the expression of multiple ligands capable of activating signaling. Regulation of receptor levels and receptor expression boundaries by this mechanism may therefore be a critical component of the Eph/ephrin signaling system in palatogenesis and elsewhere.
Efforts to elucidate the molecular mechanisms by which Eph receptor endocytosis occurs have identified several regulators of this process (Egea and Klein 2007). The Rab5-GEF Rin1 associates with and is phosphorylated by EphA4 to promote EphA4 endocytosis in HeLa cells (Deininger et al. 2008). Interestingly, Rin1 and its binding partner, Stam2, were identified in our proteomics approach as phosphorylation targets in MEP cells, and we confirmed these candidates by preliminary experiments in HEK293 cells, implicating them for further investigation as general regulators of Eph receptor internalization and trafficking. We also identified the Ap2β1 clathrin-coated adaptor as a candidate phosphorylation target. Interestingly, a recent report showed that loss of function of Ap2β1 results in an isolated cleft palate phenotype (Li et al. 2010), indicating that this adaptor's regulation of endocytosis is specifically critical for palatogenesis, perhaps by regulating EphB receptor turnover.
Ephrin-B1 signaling intrinsically regulates anterior palatal shelf outgrowth by promoting cell proliferation. This intrinsic requirement suggests that other autosomal components of this signaling pathway, including EphB2 and EphB3, might also be good candidates for either syndromic or nonsyndromic cleft palate. In fact, human EPHB2 localizes to 1p36, and monosomy at this locus results in multiple craniofacial anomalies similar to CFNS, including flat nasal bridge, midface hypoplasia, and orofacial clefting (Slavotinek et al. 1999). Furthermore, it has been demonstrated recently that loss of function of EphB2 and EphB3 also influences cell proliferation rate in the palatal shelf, although this effect was not shown to be direct (Risley et al. 2009).
Interestingly, although ephrin-B1 expression is highly restricted to the anterior palatal shelf mesenchyme, the cleft palate phenotype was complete along this axis. This is in contrast to the role of Shox2, an anterior-restricted regulator of palatal shelf outgrowth whose disruption results in a unique anterior-delimited cleft palate phenotype that can eventually close (Yu et al. 2005; Gu et al. 2008). Insufficiency of the anterior palatal shelves in ephrin-B1 mutants may therefore cross a threshold A–P position at which initiation of fusion is required. A–P marker expression was maintained in ephrin-B1 mutants, indicating that patterning of the palate along this axis is not affected by ephrin-B1 loss of function. Therefore, ephrin-B1 may act at the effector end of a genetic network controlling anterior palatal shelf outgrowth.
The mosaic disruption of ephrin-B1 signaling as a consequence of random X inactivation results in ectopic boundaries between regions with different cell proliferation rates in ephrin-B1+/− heterozygous palates, correlating with more severe dysmorphogenesis of the palatal shelves. This mosaic disruption of cell proliferation rate also corresponded with an underrepresentation of ephrin-B1 mutant cells in the palate. Such an observation is consistent with a previous study showing that human CFNS patients that were mosaic for EPHRIN-B1 mutations frequently exhibited underrepresentation of EPHRIN-B1 mutation in multiple tissues, despite displaying CFNS attributes (Twigg et al. 2006). In our study, the nonuniform disruption of proliferation rate was not limited to the palate, as the same mosaic effect was observed in the ephrin-B1+/− telencephalon. This nonuniform disruption of cell proliferation rate may therefore underlie multiple aspects of increased severity in heterozygous ephrin-B1 mutation.
We find that activation of the ERK/MAPK signaling pathway is critical for mediating an ephrin-B1 mitogenic effect in palatal cells. This is an unexpected result, given that Eph/ephrin signaling has not been reported to frequently use this signal transduction pathway. Consistent with recent findings in 293T cells, multiple signaling pathways were activated by ephrin-B1 in palatal cells (Jorgensen et al. 2009). Regulation of the ERK/MAPK signal transduction pathway may be central for mediating Eph/ephrin signaling in palatogenesis, however, since its inhibition was sufficient to attenuate the mitogenic effect. Our data indicate that ephrin-B1 forward signaling may influence the ERK/MAPK signaling pathway by multiple potential mechanisms (Fig. 7F). First, the phosphorylation of several signaling adaptors, such as Shb and/or Shc, may mediate ERK/MAPK activation, similar to their roles in Fgfr1and EphB1 signaling, respectively (Cross et al. 2002; Vindis et al. 2003). Second, the transcriptional regulation of negative feedback regulators Spry4 and Dusp6 may also modulate ERK/MAPK. Regulation of the IEGs required the ERK/MAPK signaling pathway, so it is unlikely that their regulation is the most proximal point of effect of ephrin-B1 forward signaling. Transient down-regulation of Spry4 and Dusp6 may also explain the second phase of ERK/MAPK activation. In fact, activation of Sprouty2 has been shown to be capable of inhibiting EphB2 activation and cell repulsion response in 293 cells (Poliakov et al. 2008). The second phase of MAPK activation was not necessary for mitogenesis, since its absence in starvation conditions did not eliminate the proliferative response.
Regulation of the ERK/MAPK signaling pathway provides a potential point of cross-talk with other signaling pathways critical for regulating palatal shelf outgrowth. Fgfr signaling frequently uses ERK/MAPK signal transduction, and disruption of Fgfr1 function in the neural crest-derived mesenchyme results in a cleft palate phenotype, although its role in palatogenesis has not yet been examined (Trokovic et al. 2003). Fgfr signaling has been shown to inhibit EphB2 activation by virtue of induction of Sprouty2 and Sprouty4 expression in HEK293 cells, further supporting the possibility of such cross-talk in the palate (Poliakov et al. 2008). The importance of Ras–MAPK signaling in craniofacial development has been documented recently in mice (Newbern et al. 2008). Numerous syndromes that either directly or indirectly impact regulation of ERK/MAPK signaling affect craniofacial development, and constitute neuro–cardio–facial–cutaneous syndromes (NCFS) (Bentires-Alj et al. 2006). Our data implicating ERK/MAPK signaling downstream from ephrin-B1 in palatogenesis suggest that CFNS may be related to these disorders by virtue of signaling mechanism.
Materials and methods
Mouse strains
Genotyping of ephrin-B1 null and ephrin-B16FΔV reverse-signaling mutant mice and embryos was performed as described previously (Davy et al. 2004; Bush and Soriano 2009). Since no phenotypic differences have been observed between ephrin-B1−/Y and ephrin-B1−/− mice, we refer to both genotypes as ephrin-B1null. Mice were maintained on a 129S4 coisogenic genetic background.
Histology, antibody staining, and in situ hybridization
For histology, embryos were dissected at specified stages and fixed in Bouin's fixative, dehydrated through a graded series of ethanol, and embedded in paraffin. Sections were cut at 8 μm and stained with hematoxylin and eosin. For antibody staining, embryos were removed at specified stages and fixed in 4% PFA overnight at 4°C. After dehydration in 25% sucrose, embryos were embedded in OCT and cryosections were cut at a thickness of 12 or 14 μm. Antibody staining on sections was performed according to standard protocols with antibodies recognizing ephrin-B1 (1:15; R&D Systems), EphB2 (1:20; R&D Systems), EphB3 (1:20; R&D Systems), and EphB (1:10) (Jevince et al. 2006). In situ hybridization was performed as described (Sciavolino et al. 1997).
Cell culture and immunostaining of cells
MEP cells were isolated from E13.5 embryos. Palatal shelves were dissected, rinsed in ice-cold PBS, and trypsinized in 1 mL of 0.25% trypsin/EDTA (Invitrogen 25200-072) for 10 min at 37°C with frequent trituration by pipetting. Trypsinization was stopped by adding 10 vol of media (DMEM + 10% FBS). Approximately 14 palatal shelves were plated per 10-cm dish, corresponding to a density of ∼2.5 × 104 per square centimeter, and passaged as necessary every 3–5 d by splitting at a ratio of 1:3. For induction of forward signaling, cells were stimulated with 2 μg/mL ephrin-B1-Fc (R&D Systems) that had been preclustered at a concentration of 250 μg/mL with 65 μg/mL donkey anti-human antibody (Jackson Immunoresearch) for 1 h at 4°C. For inductions in U0126, cells were pretreated for 2 h with 10 μM U0126. For immunostaining of cells, coverslips were fixed in 4% PFA and washed with PBS, blocked in 5% donkey serum in PBS (with or without 0.1% Triton), and incubated with primary antibody in PBS (with or without 0.1% Triton) for 1 h. Coverslips were washed with PBS and incubated with fluorophore-conjugated secondary antibodies. For quantification of cell surface EphB3, images were collected with a confocal laser scanning microscope (FV1000, Olympus), and average fluorescence was quantified; SEM was calculated with significance reached at P < 0.02 with a Student's t-test.
Mass spectrometry
The mass spectrometry-based phosphoproteomic approach was based on Zhang et al. (2005), with some modifications (Supplemental Material). This data set was uploaded to AmiGO to determine GO categories that were statistically overrepresented (Carbon et al. 2009). Overrepresented GO categories to which at least two proteins were annotated were examined by further literature searching whether they reached the significance level of P < 0.2. A complete list of GO terms is found in Supplemental Table 2.
Immunoprecipitation and Western blot analysis
For immunoprecipitation, cells were lysed in NP40 buffer (immunoprecipitation was performed with 1.25 μg of anti-EphB2 [R&D Systems], 1.25 μg of anti-EphB3 [R&D Systems], anti-Shc1 [1:50; Cell Signaling], 2 μg of anti-Dok-1 [Santa Cruz Biotechnology], 2 μg of anti-Ctnnd1 [BD Transduction Laboratories], anti-Nck1 [1:50; Cell Signaling]) for 4 h at 4°C, followed by incubation for 1 h with protein A/G beads; washing with NP40 buffer and elution were performed according to standard protocols. Western blotting was performed according to standard protocols with antibodies recognizing EphB2 (R&D Systems), EphB3 (R&D Systems), β-actin (Santa Cruz Biotechnology) phosphorylated tyrosine (Millipore-Upstate Biotechnology), and phosphorylated Erk1/Erk2 (Cell Signaling).
BrdU analysis of cell proliferation
For embryonic studies, 50 μg of BrdU per gram of body weight was injected intraperitoneally into pregnant females at E13.5. After 1 h, embryos were removed, fixed with 4% PFA, and processed for immunostaining by standard protocols (Supplemental Material). For cells, BrdU was added to the cell culture medium at a concentration of 10 μM at the time of signaling induction. After 3 h, cells were fixed in 2% PFA and prepared for immunostaining by standard protocols. BrdU-positive cells were counted, and the BrdU incorporation index was calculated (see Supplemental Material).
Microarray analysis and real-time QRT–PCR
Inductions of MEP cells were performed in quadruplicate with 2 μg/mL preclustered ephrin-B1-Fc for 1 h, followed by array analysis on the WG-6 version 1.1 Illumina beadchip (see Supplemental Material). Microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE23539 (http://ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE23539). Real-time QRT–PCR was performed using a Bio-Rad IQ5 cycler with PerfeCta SYBR green fastmix (Quanta Biosciences), using the standard curve method. Primers for QRT–PCR can be found in the Supplemental Material.
Acknowledgments
The G3G4 BrdU antibody developed by Stephen J. Kaufman was obtained from the DSHB. The EphB antibody was generously provided by Zaven Kaprielian. Microarray hybridization was performed by the FHCRC Genomics shared resource, and we thank Jeff Delrow and Ryan Basom for advice and assistance. Mass spectrometry was performed by the FHCRC proteomics shared resource; we thank Phil Gafken for his advice and assistance. We are grateful to Nancy Ann Oberheim, Robert Krauss, and our laboratory colleagues for helpful advice, discussions, and comments on the manuscript. Outstanding technical assistance was provided by Philip Corrin and Amelie Cornil. J.O.B. was supported by Ruth L Kirschstein NRSA Individual Fellowship F32 (DE17506) and K99 (DE020855-01) from NIH/NIDCR. This work was supported by grant R37HD25326 from the National Institute for Child Health and Human Development to P.S.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1963210.
Supplemental material is available at http://www.genesdev.org.
References
- Bentires-Alj M, Kontaridis MI, Neel BG 2006. Stops along the RAS pathway in human genetic disease. Nat Med 12: 283–285 [DOI] [PubMed] [Google Scholar]
- Braybrook C, Lisgo S, Doudney K, Henderson D, Marcano AC, Strachan T, Patton MA, Villard L, Moore GE, Stanier P, et al. 2002. Craniofacial expression of human and murine TBX22 correlates with the cleft palate and ankyloglossia phenotype observed in CPX patients. Hum Mol Genet 11: 2793–2804 [DOI] [PubMed] [Google Scholar]
- Bush JO, Soriano P 2009. Ephrin-B1 regulates axon guidance by reverse signaling through a PDZ-dependent mechanism. Genes Dev 23: 1586–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Lan Y, Maltby KM, Jiang R 2002. Isolation and developmental expression analysis of Tbx22, the mouse homolog of the human X-linked cleft palate gene. Dev Dyn 225: 322–326 [DOI] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S 2009. AmiGO: Online access to ontology and annotation data. Bioinformatics 25: 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Maxson RE Jr 2006. Recent advances in craniofacial morphogenesis. Dev Dyn 235: 2353–2375 [DOI] [PubMed] [Google Scholar]
- Chumley MJ, Catchpole T, Silvany RE, Kernie SG, Henkemeyer M 2007. EphB receptors regulate stem/progenitor cell proliferation, migration, and polarity during hippocampal neurogenesis. J Neurosci 27: 13481–13490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagni A, Logan M, Klein R, Adams RH 2003. Control of skeletal patterning by ephrinB1–EphB interactions. Dev Cell 5: 217–230 [DOI] [PubMed] [Google Scholar]
- Cross MJ, Lu L, Magnusson P, Nyqvist D, Holmqvist K, Welsh M, Claesson-Welsh L 2002. The Shb adaptor protein binds to tyrosine 766 in the FGFR-1 and regulates the Ras/MEK/MAPK pathway via FRS2 phosphorylation in endothelial cells. Mol Biol Cell 13: 2881–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A, Soriano P 2005. Ephrin signaling in vivo: Look both ways. Dev Dyn 232: 1–10 [DOI] [PubMed] [Google Scholar]
- Davy A, Aubin J, Soriano P 2004. Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev 18: 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A, Bush JO, Soriano P 2006. Inhibition of gap junction communication at ectopic Eph/ephrin boundaries underlies craniofrontonasal syndrome. PLoS Biol 4: e315 doi: 10.1371/journal.pbio.0040315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger K, Eder M, Kramer ER, Zieglgansberger W, Dodt HU, Dornmair K, Colicelli J, Klein R 2008. The Rab5 guanylate exchange factor Rin1 regulates endocytosis of the EphA4 receptor in mature excitatory neurons. Proc Natl Acad Sci 105: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea J, Klein R 2007. Bidirectional Eph-ephrin signaling during axon guidance. Trends Cell Biol 17: 230–238 [DOI] [PubMed] [Google Scholar]
- Elowe S, Holland SJ, Kulkarni S, Pawson T 2001. Downregulation of the Ras-mitogen-activated protein kinase pathway by the EphB2 receptor tyrosine kinase is required for ephrin-induced neurite retraction. Mol Cell Biol 21: 7429–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genander M, Halford MM, Xu NJ, Eriksson M, Yu Z, Qiu Z, Martling A, Greicius G, Thakar S, Catchpole T, et al. 2009. Dissociation of EphB2 signaling pathways mediating progenitor cell proliferation and tumor suppression. Cell 139: 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genander M, Holmberg J, Frisen J 2010. Ephrins negatively regulate cell proliferation in the epidermis and hair follicle. Stem Cells. 28: 1196–1205 [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A 2007. Molecular control of secondary palate development. Dev Biol 301: 309–326 [DOI] [PubMed] [Google Scholar]
- Gu S, Wei N, Yu X, Jiang Y, Fei J, Chen Y 2008. Mice with an anterior cleft of the palate survive neonatal lethality. Dev Dyn 237: 1509–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A, Meunier D, Muller I, Rump A, Fundele R, Ropers HH, Nuber UA 2003. Expression of mouse Tbx22 supports its role in palatogenesis and glossogenesis. Dev Dyn 226: 579–586 [DOI] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP 2005. Regional regulation of palatal growth and patterning along the anterior–posterior axis in mice. J Anat 207: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Armulik A, Senti KA, Edoff K, Spalding K, Momma S, Cassidy R, Flanagan JG, Frisen J 2005. Ephrin-A2 reverse signaling negatively regulates neural progenitor proliferation and neurogenesis. Genes Dev 19: 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, Genander M, Halford MM, Anneren C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisen J 2006. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 125: 1151–1163 [DOI] [PubMed] [Google Scholar]
- Huang PH, White FM 2008. Phosphoproteomics: Unraveling the signaling web. Mol Cell 31: 777–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T 2000. Signaling—2000 and beyond. Cell 100: 113–127 [DOI] [PubMed] [Google Scholar]
- Jevince AR, Kadison SR, Pittman AJ, Chien CB, Kaprielian Z 2006. Distribution of EphB receptors and ephrin-B1 in the developing vertebrate spinal cord. J Comp Neurol 497: 734–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen C, Sherman A, Chen GI, Pasculescu A, Poliakov A, Hsiung M, Larsen B, Wilkinson DG, Linding R, Pawson T 2009. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science 326: 1502–1509 [DOI] [PubMed] [Google Scholar]
- Kullander K, Klein R 2002. Mechanisms and functions of Eph and ephrin signalling. Nat Rev Mol Cell Biol 3: 475–486 [DOI] [PubMed] [Google Scholar]
- Lackmann M, Boyd AW 2008. Eph, a protein family coming of age: More confusion, insight, or complexity? Sci Signal 1: re2 doi: 10.1126/stke.115re2 [DOI] [PubMed] [Google Scholar]
- Lan Y, Jiang R 2009. Sonic hedgehog signaling regulates reciprocal epithelial–mesenchymal interactions controlling palatal outgrowth. Development 136: 1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Nathans D 1987. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: Coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci 84: 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Puertollano-Moro R, Bonifacino J, Overbeek P, Everett E 2010. Disruption of the murine Ap2b1 gene causes non-syndromic cleft palate. Cleft Palate Craniofac J doi: 10.1597/09-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KT, Sloniowski S, Ethell DW, Ethell IM 2008. Ephrin-B2-induced cleavage of EphB2 receptor is mediated by matrix metalloproteinases to trigger cell repulsion. J Biol Chem 283: 28969–28979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston DJ, Dickinson S, Nobes CD 2003. Rac-dependent trans-endocytosis of ephrinBs regulates Eph–ephrin contact repulsion. Nat Cell Biol 5: 879–888 [DOI] [PubMed] [Google Scholar]
- Murphy LO, Blenis J 2006. MAPK signal specificity: The right place at the right time. Trends Biochem Sci 31: 268–275 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Newbern J, Zhong J, Wickramasinghe RS, Li X, Wu Y, Samuels I, Cherosky N, Karlo JC, O'Loughlin B, Wikenheiser J, et al. 2008. Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. Proc Natl Acad Sci 105: 17115–17120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Foos G, Hauser CA, Pasquale EB 2006. The EphB4 receptor suppresses breast cancer cell tumorigenicity through an Abl–Crk pathway. Nat Cell Biol 8: 815–825 [DOI] [PubMed] [Google Scholar]
- Orioli D, Henkemeyer M, Lemke G, Klein R, Pawson T 1996. Sek4 and Nuk receptors cooperate in guidance of commissural axons and in palate formation. EMBO J 15: 6035–6049 [PMC free article] [PubMed] [Google Scholar]
- Pasquale EB 2008. Eph–ephrin bidirectional signaling in physiology and disease. Cell 133: 38–52 [DOI] [PubMed] [Google Scholar]
- Poliakov A, Cotrina ML, Pasini A, Wilkinson DG 2008. Regulation of EphB2 activation and cell repulsion by feedback control of the MAPK pathway. J Cell Biol 183: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J, Snyder M 2006. Charging it up: Global analysis of protein phosphorylation. Trends Genet 22: 545–554 [DOI] [PubMed] [Google Scholar]
- Ricard J, Salinas J, Garcia L, Liebl DJ 2006. EphrinB3 regulates cell proliferation and survival in adult neurogenesis. Mol Cell Neurosci 31: 713–722 [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP 2004. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest 113: 1692–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risley M, Garrod D, Henkemeyer M, McLean W 2009. EphB2 and EphB3 forward signalling are required for palate development. Mech Dev 126: 230–239 [DOI] [PubMed] [Google Scholar]
- Schmahl J, Raymond CS, Soriano P 2007. PDGF signaling specificity is mediated through multiple immediate early genes. Nat Genet 39: 52–60 [DOI] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C 1997. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn 209: 127–138 [DOI] [PubMed] [Google Scholar]
- Slavotinek A, Shaffer LG, Shapira SK 1999. Monosomy 1p36. J Med Genet 36: 657–663 [PMC free article] [PubMed] [Google Scholar]
- Steinle JJ, Meininger CJ, Forough R, Wu G, Wu MH, Granger HJ 2002. Eph B4 receptor signaling mediates endothelial cell migration and proliferation via the phosphatidylinositol 3-kinase pathway. J Biol Chem 277: 43830–43835 [DOI] [PubMed] [Google Scholar]
- Trokovic N, Trokovic R, Mai P, Partanen J 2003. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev 17: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SR, Kan R, Babbs C, Bochukova EG, Robertson SP, Wall SA, Morriss-Kay GM, Wilkie AO 2004. Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc Natl Acad Sci 101: 8652–8657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg SR, Matsumoto K, Kidd AM, Goriely A, Taylor IB, Fisher RB, Hoogeboom AJ, Mathijssen IM, Lourenco MT, Morton JE, et al. 2006. The origin of EFNB1 mutations in craniofrontonasal syndrome: Frequent somatic mosaicism and explanation of the paucity of carrier males. Am J Hum Genet 78: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindis C, Cerretti DP, Daniel TO, Huynh-Do U 2003. EphB1 recruits c-Src and p52Shc to activate MAPK/ERK and promote chemotaxis. J Cell Biol 162: 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieacker P, Wieland I 2005. Clinical and genetic aspects of craniofrontonasal syndrome: Towards resolving a genetic paradox. Mol Genet Metab 86: 110–116 [DOI] [PubMed] [Google Scholar]
- Wieland I, Jakubiczka S, Muschke P, Cohen M, Thiele H, Gerlach KL, Adams RH, Wieacker P 2004. Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am J Hum Genet 74: 1209–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley HS, Burke PM 2001. Regulation of receptor tyrosine kinase signaling by endocytic trafficking. Traffic 2: 12–18 [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, et al. 2005. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development 132: 4397–4406 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y 2002. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development 129: 4135–4146 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM 2005. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics 4: 1240–1250 [DOI] [PubMed] [Google Scholar]
- Zhang G, Spellman DS, Skolnik EY, Neubert TA 2006. Quantitative phosphotyrosine proteomics of EphB2 signaling by stable isotope labeling with amino acids in cell culture (SILAC). J Proteome Res 5: 581–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fenyo D, Neubert TA 2008. Screening for EphB signaling effectors using SILAC with a linear ion trap-orbitrap mass spectrometer. J Proteome Res 7: 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Palmer A, Kohler J, Klein R 2003. EphB-ephrinB bi-directional endocytosis terminates adhesion allowing contact mediated repulsion. Nat Cell Biol 5: 869–878 [DOI] [PubMed] [Google Scholar]
- Zisch AH, Pazzagli C, Freeman AL, Schneller M, Hadman M, Smith JW, Ruoslahti E, Pasquale EB 2000. Replacing two conserved tyrosines of the EphB2 receptor with glutamic acid prevents binding of SH2 domains without abrogating kinase activity and biological responses. Oncogene 19: 177–187 [DOI] [PubMed] [Google Scholar]