Abstract
Germ cells serve as intriguing examples of differentiated cells that retain the capacity to generate all cell types of an organism. Here we used functional genomic approaches in planarians to identify genes required for proper germ cell development. We conducted microarray analyses and in situ hybridization to discover and validate germ cell-enriched transcripts, and then used RNAi to screen for genes required for discrete stages of germ cell development. The majority of genes we identified encode conserved RNA-binding proteins, several of which have not been implicated previously in germ cell development. We also show that a germ cell-specific subunit of the conserved transcription factor CCAAT-binding protein/nuclear factor-Y is required for maintaining spermatogonial stem cells. Our results demonstrate that conserved transcriptional and post-transcriptional mechanisms regulate germ cell development in planarians. These findings suggest that studies of planarians will inform our understanding of germ cell biology in higher organisms.
Keywords: Spermatogenesis, post-transcriptional regulation, nuclear factor Y, nanos, RNAi screen, Schmidtea mediterranea
The specification and proper differentiation of germ cells are essential for the continuity of sexually reproducing species. Germ cells can maintain totipotency throughout their differentiation, and, although gametes are highly specialized cell types, this potential is released upon fertilization to generate a new organism (Seydoux and Braun 2006). Dissecting the molecular mechanisms underlying germ cell development is critical for understanding the nature of totipotency, the factors regulating meiotic progression, and the sex-specific differentiation of gametes.
Studies conducted in classic genetic model organisms have identified both evolutionarily conserved and species-specific regulators of germ cell development (Ewen-Campen et al. 2010). This work has revealed the importance of transcriptional repression of somatic fates and post-transcriptional control of gene expression in regulating germ cell differentiation (Seydoux and Braun 2006; Kimble and Crittenden 2007; Cinalli et al. 2008). Aside from a few model organisms, however, the mechanisms of germ cell development remain largely unexplored in the vast majority of metazoan phyla. Moreover, current genetic models represent only two of the major bilaterian superphyla (Ecdysozoa and Deuterostomia), but not the Lophotrochozoa (Dunn et al. 2008). Therefore, a comprehensive understanding of germ cell development in metazoans will benefit from mechanistic studies of animals representing additional evolutionary lineages.
The freshwater planarian Schmidtea mediterranea represents one promising model for studying germ cell development (Newmark et al. 2008). This member of the Lophotrochozoan phylum Platyhelminthes can regenerate new germ cells from fragments of adult tissue that lack reproductive organs (Morgan 1902; Sato et al. 2006; Wang et al. 2007). The source of the regenerated germ cells appears to be the somatic stem cells—the neoblasts—that are responsible for the animal's well-known regenerative abilities (Baguñà et al. 1989). Neoblasts express several conserved regulators of germ cell development (Shibata et al. 1999; Reddien et al. 2005a; Salvetti et al. 2005; Guo et al. 2006; Palakodeti et al. 2008; Solana et al. 2009). Thus, these pluripotent cells share many features with germ cells, and studies of planarians should help reveal the mechanisms by which somatic stem cells can produce germ cells.
Planarian germ cells express orthologs of nanos (Sato et al. 2006; Handberg-Thorsager and Saló 2007; Wang et al. 2007), a gene required for germ cell differentiation and maintenance in a wide range of animals (Kobayashi et al. 1996; Deshpande et al. 1999; Subramaniam and Seydoux 1999; Koprunner et al. 2001; Tsuda et al. 2003; Hayashi et al. 2004; Wang and Lin 2004; Sada et al. 2009). We showed that nanos is required for the development, maintenance, and regeneration of the germ cell lineage in sexually reproducing planarians (Wang et al. 2007). Surprisingly, nanos-positive cells were also observed in asexually reproducing planarians that propagate by fission and never develop gonads (Sato et al. 2006; Handberg-Thorsager and Saló 2007; Wang et al. 2007), suggesting that they possess early germ cells that fail to differentiate; nanos function is also required for maintaining these presumptive germ cells (Wang et al. 2007).
Here we used functional genomic tools to investigate systematically the molecular mechanisms underlying planarian germ cell development. We identified transcripts down-regulated after nanos RNAi-mediated germ cell loss, and validated their germ cell-enriched expression by in situ hybridization. We then performed a targeted RNAi screen to investigate the functions of these genes, revealing previously unreported roles in germ cell development for several molecules conserved between planarians and vertebrates.
Results
Identification of germ cell-specific genes in S. mediterranea
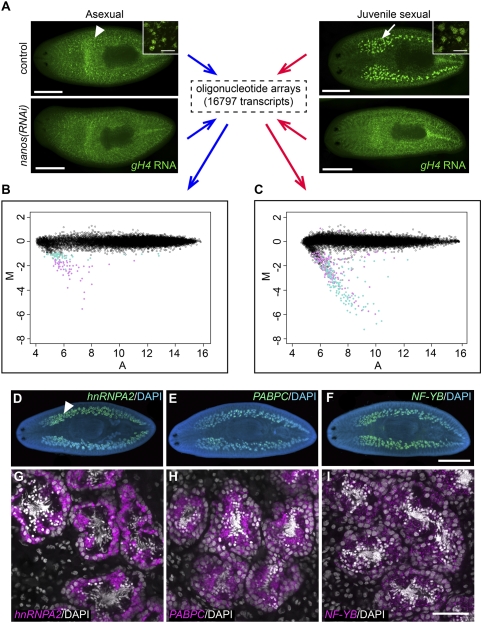
To identify genes required for germ cell development in planarians, we used microarray analyses to compare gene expression profiles of planarians with and without germ cells [control(RNAi) vs. nanos(RNAi) animals, respectively]. We generated custom oligonucleotide arrays representing 16,797 unique S. mediterranea transcripts from two EST collections (Sánchez Alvarado et al. 2002; Zayas et al. 2005) and carried out two sets of comparisons: asexual nanos(RNAi) versus control(RNAi) animals, to identify genes expressed in presumptive germ cells; and juvenile sexual nanos(RNAi) versus control(RNAi) animals, to identify genes expressed in testes primordia (Fig. 1A).
Figure 1.
Transcriptional profiling of nanos(RNAi) versus control(RNAi) animals, and validation of germ cell-enriched expression of identified genes. (A) Experimental design of microarray analyses. (Arrowhead) Presumptive germ cells; (arrow) testes primordia. (Insets) Magnified views of germ cells in control(RNAi) animals. (B,C) M-A plots showing differential expression between asexual nanos(RNAi) versus control(RNAi) animals (B), and juvenile sexual nanos(RNAi) versus control(RNAi) animals (C). M represents the log2 intensity ratios, and A represents the log2 intensity averages. (Open black circles) Transcripts that did not show significantly differential expression; (closed turquoise circles) transcripts with adjusted P-value = 0.01–0.05; (closed magenta circles) transcripts with adjusted P-value < 0.01. (D–I) Genes identified from microarray analyses were expressed in different germ cell populations in testes, as revealed by FISH. (D–F) Whole-mount images. (Arrowhead) Testes. (G–I) Magnified views of testes lobules. Bars: A, 0.5 mm; insets in A, 50 μm; D–F, 1 mm; G–I, 50 μm. See also Supplemental Figure S1.
We first determined the transcriptional profiles of asexual nanos(RNAi) and control(RNAi) planarians. The vast majority (∼99.4%) of the transcripts examined did not show differential expression between nanos(RNAi) and control animals, consistent with the observation that nanos knockdown does not detectably affect somatic cells in the animal (Wang et al. 2007). One-hundred-three genes showed significant differential expression (adjusted P < 0.05) between nanos(RNAi) and control animals (Fig. 1B; Supplemental Table S1); notably, all of these genes were down-regulated in nanos(RNAi) animals. Out of 103 top hits, 72 genes have homologs in other organisms. These genes encode proteins with a variety of diverse functions, as predicted by associated Cluster of Orthologous Groups (COG) terms (Tatusov et al. 2003). They are largely enriched for cytoskeletal components, genes involved in carbohydrate transport and metabolism, post-translational modification/protein turnover/chaperones, energy production and conversion, and RNA processing and modification (Supplemental Table S2). The nanos transcript itself was the second highest hit by fold change (M = −4.71), confirming the RNAi knockdown efficiency. The top hit was a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) homolog, Smed-GAPDHs (for spermatogenic GAPDH), which showed the largest fold change (M = −5.54) and the lowest P-value (0.00011) among all transcripts.
Using the same approach, we identified 278 genes that showed significant differential expression (adjusted P < 0.05) between juvenile sexual nanos(RNAi) and control planarians (Fig. 1C; Supplemental Table S3). Of these, 275 genes were down-regulated in nanos(RNAi) animals; 195 out of 275 have likely homologs in other organisms. Based on COG terms, the most enriched functional categories include cytoskeletal components, post-translational modification/protein turnover/chaperones, signal transduction mechanisms, carbohydrate transport, and metabolism, as well as RNA processing and modification (Supplemental Table S4). As in asexuals, Smed-GAPDHs was the top hit by both P-value and fold change. When we compared the 103 top hits from asexuals to these 278 genes, we found that 80 genes overlapped between the two data sets.
Validation of germ cell-specific expression
To validate our microarray results, we analyzed the expression patterns of the top hits by whole-mount in situ hybridization. Sexual S. mediterranea have numerous testes lobules distributed dorsolaterally and a pair of ovaries located more ventrally behind the cephalic ganglia. Out of 98 ESTs examined, 93 showed testes-specific or testes-enriched expression (Supplemental Table S1); of these, three genes were also expressed in ovaries (Supplemental Table S1). Transcripts of two other genes were detected only in ovaries (Supplemental Table S1). Thus, our microarray analyses were effective at identifying transcripts whose expression was enriched in gonads; the enrichment for testes-specific transcripts is a consequence of the large number of testes in planarians.
In order to define more precisely the cell types in which the top hits from both sets of arrays are expressed, we performed fluorescent in situ hybridization (FISH), and visualized transcript distributions by confocal microscopy (Fig. 1D–I). Mature planarian testes lobules have a peripheral layer of spermatogonial cells; as spermatogonia differentiate, spermatocytes and spermatids accumulate toward the luminal side of the testes, where mature sperm will ultimately be released into the sperm ducts (Fig. 1G; Supplemental Fig. S1A). Transcripts of several genes were detected in distinct spermatogenic populations: For example, Smed-hnRNPA2 mRNAs were enriched in spermatogonial cells and early spermatocytes (Fig. 1D,G), Smed-PABPC-1 [poly(A)-binding protein, cytoplasmic] transcripts were detected in spermatocytes and spermatids (Fig. 1E,H), and Smed-NF-YB (nuclear factor Y, subunit B) mRNAs were detected in all germ cells within the testes, except mature sperm (Fig. 1F,I).
Distinct subpopulations of spermatogonia were revealed by the expression patterns of some germ cell-enriched transcripts. For example, FISH detected both Smed-GAPDHs and Smed-rap55 (RNA-associated protein 55) mRNAs specifically in spermatogonial cells (Supplemental Fig. S1B–G). Double FISH detected nanos transcripts in a small subset of spermatogonial cells that also express GAPDHs; spermatogonia with the highest GAPDHs levels, however, were often found adjacent to the nanos-positive cells (Supplemental Fig. S1B–D). On the other hand, rap55 was detected only weakly in the nanos-positive spermatogonia (Supplemental Fig. S1E–G). These expression patterns reveal heterogeneity within the spermatogonial population: Assuming that nanos-positive spermatogonia are the spermatogonial stem cells (SSCs) of planarians (Wang and Lin 2004; Sato et al. 2006; Wang et al. 2007; Sada et al. 2009), rap55-positive cells are likely to represent differentiating spermatogonia, and GAPDHs-positive cells seem to include both SSCs and their direct division progeny.
Functional characterization of genes required for distinct stages of germ cell development
To investigate the functions of these germ cell-enriched transcripts in germ cell development, we performed RNAi experiments. We selected 109 ESTs from both asexual and juvenile sexual array top hits to generate dsRNA feeding vectors (Supplemental Table S5). Priority was given to genes with the largest fold change in expression, and genes predicted to encode proteins involved in transcription, RNA processing, translational control, and signal transduction. We also included several genes with homologs of unknown function in other organisms, and a few novel genes. Because the majority of candidate genes are expressed in planarian testes, we focused on male germ cell differentiation; furthermore, planarians have numerous testes lobules, and their position beneath the dorsal epidermis facilitates subsequent analyses. We performed RNAi by feeding dsRNA-containing food to small, juvenile sexual animals (lacking testes) for ∼1 mo. When controls reached sexual maturity, we assessed development of the testes by fixing animals and staining them with DAPI to label germ cell nuclei within the testes; we then examined them microscopically for defects in spermatogenesis. Using this method, we identified 13 genes for which RNAi knockdown yielded phenotypes at various stages of male germ cell development and spermatogenesis (Table 1).
Table 1.
Genes required for various stages of planarian germ cell development
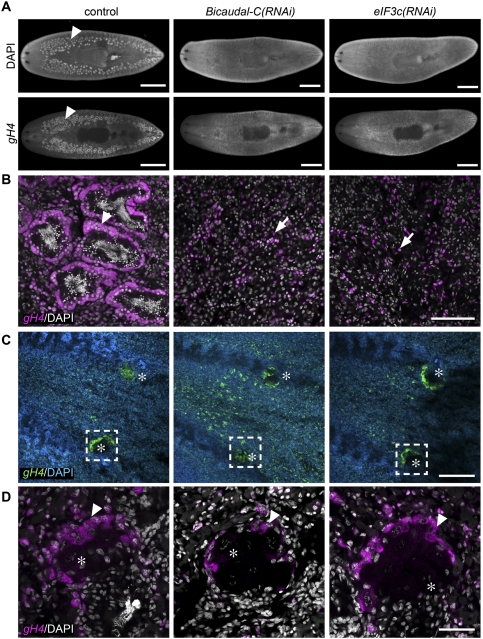
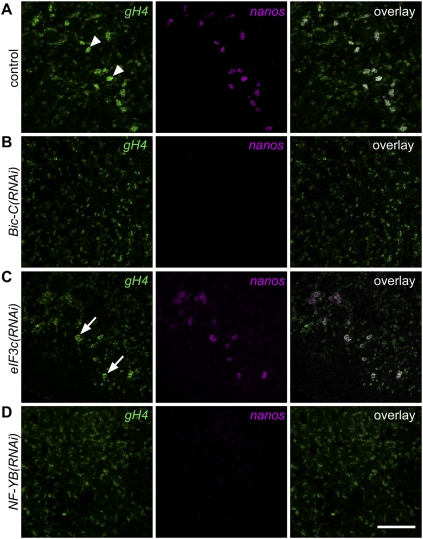
Among them, RNAi knockdown of Smed-Bicaudal-C (Bic-C) and Smed-eIF3c (eukaryotic initiation factor 3, subunit C) resulted in a “no testes” phenotype (Fig. 2A). Failure of testes development in Bic-C(RNAi) and eIF3c(RNAi) animals was confirmed by FISH to detect expression of germinal Histone H4 (gH4), a marker for spermatogonia, oogonia, and neoblasts. Whereas control animals showed normal testes morphology with an outer layer of gH4-positive spermatogonia, only individual neoblasts were detected dorsolaterally in RNAi animals (Fig. 2B). Length measurements of eIF3c(RNAi) (5.09 ± 0.75 mm, n = 15; mean ± SD), Bic-C(RNAi) (5.27 ± 0.55 mm, n = 15), and control (3.97 ± 0.26 mm, n = 16) animals revealed that eIF3c(RNAi) and Bic-C(RNAi) animals were significantly larger than controls (P < 0.0005, Student's t-test); therefore, the absence of testes was not a consequence of inhibited growth or developmental arrest. Although both Bic-C and eIF3c are also expressed in the ovaries (data not shown), defects in oogenesis were not apparent in knockdown animals: Both gH4 expression and oocyte formation appeared to be unaffected (Fig. 2C,D). Sexually mature animals with fully developed gonads treated with Bic-C or eIF3c dsRNA over a period of 1 mo also lost their testes, as shown by gH4 and DAPI staining, yet no defects in oogenesis were observed (data not shown). Therefore, we conclude that Bic-C and eIF3c are required for proper development and maintenance of planarian male germ cells.
Figure 2.
Bic-C and eIF3c are required for testes formation. (A) DAPI staining and gH4 FISH to label testes (arrowheads) in control and RNAi animals. (B) Magnified views of testes in control(RNAi) (n = 14 out of 14) and neoblasts in Bic-C(RNAi) (n = 15 out of 15) and eIF3c(RNAi) (n = 15 out of 15) animals. (Arrowhead) spermatogonia; (arrows) neoblasts. (C,D) DAPI staining and FISH to detect gH4 transcripts in ovaries of control and RNAi animals. (C) Asterisks indicate ovaries. (D) Magnified views of boxed regions in C. (Arrowheads) Oogonia. (*) Oocyte nuclei stained with DAPI. Bars: A, 1 mm; B, 100 μm; C, 200 μm; D, 50 μm.
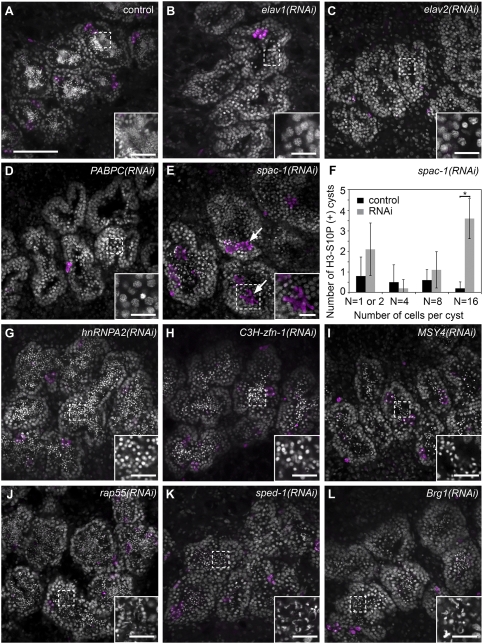
RNAi knockdowns of four genes resulted in meiotic defects in developing sexual animals. Planarian spermatogonia undergo three rounds of mitotic division with incomplete cytokinesis; the resulting eight primary spermatocytes then undergo meiosis to generate 32 spermatids (Franquinet and Lender 1973; Newmark et al. 2008). Three knockdowns (Smed-elav-1 [embryonic-lethal abnormal vision], Smed-elav-2, and Smed-PABPC) appear to block meiotic progression: No spermatids were detected in the testes of these RNAi animals (Fig. 3A–D), yet a few spermatocytes with pachytene-like nuclear morphology were observed (Fig. 3B–D, insets), suggesting that meiotic entry may occur. When compared with control animals that possess a single layer of spermatogonia around the periphery of the testis lobules (Fig. 3A), these RNAi animals exhibit expanded peripheral layers in the testes (Fig. 3B–D), suggestive of an accumulation of spermatogonia and primary spermatocytes. The expansion of spermatogonia in PABPC(RNAi) animals was confirmed by labeling with anti-Sm monoclonal Y12 (Lerner et al. 1981); this antibody recognizes symmetrical dimethylarginines (Brahms et al. 2000) and labels neoblasts and spermatogonia (Supplemental Fig. S2A,B). Mitotic spermatogonia and meiotic spermatocytes can be labeled with anti-phospho-Histone H3-S10 (H3-S10P) antibodies (Hendzel et al. 1997; Wei et al. 1998; Cobb et al. 1999; Manzanero et al. 2000); the number of cells within H3-S10P-positive clusters enables us to distinguish dividing spermatogonia from dividing primary and secondary spermatocytes (e.g., clusters of eight anti-H3-S10P-positive cells correspond to dividing primary spermatocytes). Anti-H3-S10P immunofluorescence did not reveal increased mitotic/meiotic activity between RNAi and control animals, indicating that the accumulation is likely not a result of overproliferation (Fig. 3B–D).
Figure 3.
Genes required for meiotic progression and spermatid elongation. (A–E,G–L) DAPI (gray) and H3-S10P (magenta) staining of testes in control and RNAi animals (n > 12 for each gene). (B–D) RNAi knockdowns showing meiotic failure. (E) spac-1(RNAi) animals accumulated H3-S10P-positive secondary spermatocytes (arrows). (F) Quantification of H3-S10P-positive cysts in control and spac-1(RNAi) animals; the latter showed increased numbers of H3-S10P-positive secondary spermatogenic cysts (P < 0.0005, Student's t-test). Error bars represent SEM. Twelve testis lobules from three animals were counted for each group. (G–L) RNAi knockdowns resulting in spermatid elongation defects. Bars: A, 100 μm; insets, 20 μm. See also Supplemental Figure S2.
The fourth gene, spac-1 (for secondary spermatocyte accumulation), encodes a protein containing an RRM (RNA recognition motif) domain, and is homologous to a hypothetical protein from the parasitic flatworm Schistosoma japonicum. Round spermatids could be detected in testes lobules of spac-1(RNAi) animals (Fig. 3E), suggesting that spermatocytes were able to progress through meiosis. However, a dramatic accumulation of secondary spermatocytes (clusters of 16 H3-S10P-positive cells) was observed in these testes (Fig. 3E,F; Supplemental Movies S1, S2), suggesting that meiotic progression was aberrant.
We identified six genes for which RNAi knockdown resulted in defects in spermatid elongation: Smed-hnRNPA2, Smed-C3H-zinc finger-1 (zfn-1), Smed-MSY4, Smed-rap55, Smed-Brg1, and sped-1 (spermatid elongation-defective), a novel gene encoding a predicted protein that shares no similarity with any known or predicted proteins. These RNAi animals showed varying degrees of spermatid elongation, but none of them was able to produce mature, fully elongated sperm (Fig. 3G–L): Testes of hnRNPA2(RNAi), C3H-zfn-1(RNAi), and MSY4(RNAi) animals contained only round spermatids in the testes (Fig. 3G–I); rap55(RNAi), sped-1(RNAi), and Brg1(RNAi) animals had partially elongated spermatids (Fig. 3J–L).
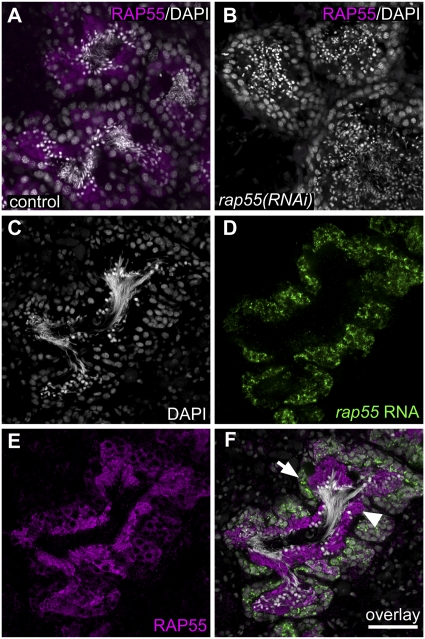
All of the aforementioned genes were expressed specifically in germ cells in testes (Fig. 1G,H; Supplemental Fig. S2C–L); PABPC-1 is also expressed in the oogonia as well as differentiating and mature oocytes (Supplemental Fig. S2M–P). Thus, the RNAi phenotypes described above appear to be the result of germ cell-intrinsic defects. Surprisingly, genes for which RNAi generated spermatid elongation defects (hnRNPA2, MSY4, C3H-zfn-1, rap55, sped-1, and Brg1) were expressed as early as the spermatogonial stage (Fig. 1G; Supplemental Fig. S2E–I, respectively). To ask whether this phenotypic delay could be due to a delay in protein translation, we examined localization of the Smed-RAP55 protein in the testes using α-Trailer-hitch (Drosophila RAP55 ortholog) (Wilhelm et al. 2005) antibodies that cross-react with Smed-RAP55 (Fig. 4A). The absence of detectable staining following rap55 RNAi (Fig. 4A,B) demonstrates the antibody specificity and confirms the effectiveness of the RNAi knockdown. Although rap55 transcripts were detected mainly in spermatogonia (Fig. 4D,F), RAP55 protein was most abundant in spermatocytes as well as round and elongating spermatids (Fig. 4E,F). Therefore, RAP55 expression appears to be regulated post-transcriptionally, and the accumulation of RAP55 protein in spermatids is consistent with the spermatid elongation phenotype seen after rap55 RNAi.
Figure 4.
Post-transcriptional regulation of rap55. (A,B) Anti-Tral antibodies (Wilhelm et al. 2005) cross-react with planarian RAP55 protein, as shown by lack of detectable anti-Tral staining in rap55(RNAi) animals [control, n = 6 out of 6; rap55(RNAi), n = 5 out of 5]. Animals were fed dsRNA-containing food three times over 12 d and were fixed 7 d after the last feeding. (C–F) Localization of rap55 transcripts and protein in testes. (C) DAPI-stained testis lobule. (D) FISH to detect rap55 transcript. (E) Anti-Tral staining to detect RAP55 protein. (F) Overlay. (Arrow) Spermatogonia; (arrowhead) spermatids. Bar, 50 μm.
NF-YB is required for the maintenance of SSCs
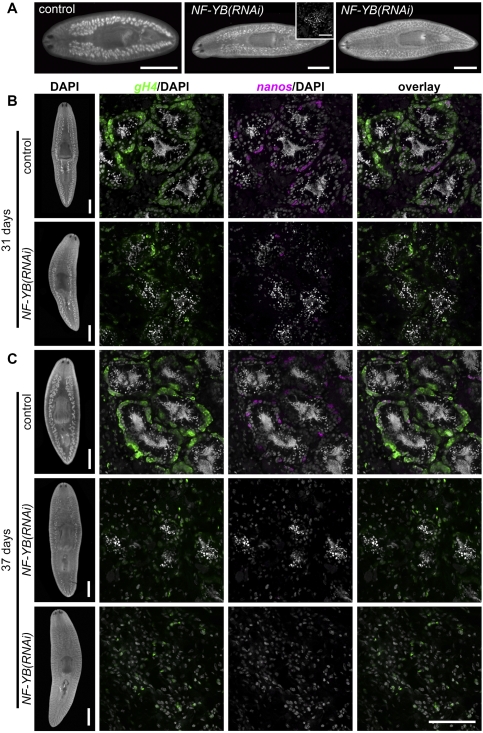
Finally, our screen identified an NF-YB (or CCAAT-binding transcription factor) homolog for which RNAi knockdown produced a unique testes degeneration phenotype. Smed-NF-YB(RNAi) animals examined after six RNAi feedings over a period of ∼1 mo had either very tiny testes (n = 6 out of 12) or no testes (n = 6 out of 12) (Fig. 5A). Animals with tiny testes appeared to contain mainly round spermatids and occasionally mature sperm in the testis lobules, but lacked spermatocytes and spermatogonial cells at the periphery (Fig. 5A, inset). Nevertheless, in some NF-YB(RNAi) animals, including those in which no testes were observed by DAPI staining, we observed well-developed copulatory apparatus and mature sperm in seminal vesicles or sperm ducts (data not shown), suggesting that these animals could undergo the initial rounds of spermatogenesis, but failed to maintain sperm production when NF-YB was knocked down. The first wave of sperm production in NF-YB(RNAi) animals could be explained by incomplete knockdown during the initial period of RNAi treatment. This scenario seems unlikely, however, because spermatids or mature sperm were not detected in other RNAi knockdowns that resulted in failure of testes formation or meiotic progression.
Figure 5.
Loss of spermatogonia after NF-YB(RNAi). (A) DAPI staining showing that NF-YB(RNAi) juveniles developed small testes or no testes. (Inset) Small testis lobule in NF-YB(RNAi) animal showing accumulation of round spermatids. Bars: 1 mm; inset, 50 μm. (B,C) Double FISH to detect nanos and gH4 transcripts in sexually mature control or NF-YB(RNAi) animals. Over time, NF-YB(RNAi) animals showed testes degeneration, loss of spermatogonia, and reduced gH4 and nanos expression. Bars: Whole-animal images, 1 mm; magnified views, 100 μm. See also Supplemental Figure S3.
To examine the role of NF-YB in spermatogonial maintenance, we performed NF-YB RNAi in mature sexual animals and examined nanos and gH4 expression (Fig. 5B,C). After four dsRNA feedings over a period of 31 d, control animals showed robust spermatogenesis in their testes, with a layer of gH4-positive spermatogonial cells at the periphery (n = 17 out of 17) (Fig. 5B, top panel). A subset of spermatogonia was nanos-positive, likely representing SSCs. NF-YB(RNAi) animals, however, had predominantly round spermatids in the lumen of their testes, surrounded by a discontinuous spermatogonial layer containing only a few gH4-positive cells (n = 19 out of 19); the number of nanos-positive putative SSCs per testis lobule was also greatly reduced (Fig. 5B, bottom panel). After five dsRNA feedings over a period of 37 d, 15 out of 19 RNAi animals had tiny residual testes, in which very little spermatogonial gH4 and nanos expression could be detected (Fig. 5C, middle panel). In the remaining RNAi animals (n = 4 out of 19), testes had completely degenerated (Fig. 5C, bottom panel). Taken together, these results suggest that NF-YB RNAi caused testes degeneration in sexually mature planarians, possibly due to depletion of SSCs. In contrast, oogenesis appeared to be normal in NF-YB(RNAi) animals, since mature oocytes could be found in the ovaries, and ovarian expression of both gH4 and nanos was similar to that observed in controls (data not shown). Thus, NF-YB appears to be required for continuous spermatogenesis and for maintenance of SSCs.
In sexual planarians, NF-YB is expressed in all germ cells in the testes (including nanos-positive cells) except sperm, and no somatic expression could be detected (Fig. 1F,I; Supplemental Fig. S3A–C). Therefore, the NF-YB(RNAi) phenotype appears to result from a germ cell-intrinsic defect. Like nanos NF-YB expression is detected in presumptive germ cells in asexuals and developing testes primordia in juvenile sexual animals (Supplemental Fig. S3D). Furthermore, it is also expressed in animals undergoing testes regression and regeneration following post-ovary amputation (Supplemental Fig. S3E). nanos-positive cells are retained throughout the course of testes regression and regeneration, whereas later differentiation markers (such as T-plastin) are lost during testes regression and are only re-expressed when spermatogonia produce differentiated progeny (Wang et al. 2007). These expression dynamics are consistent with a role for NF-YB in the SSCs.
Genes required for maintaining germ cells in asexual S. mediterranea
Since Bic-C, eIF3c, and NF-YB are required for testes formation and maintenance in sexual planarians, we asked whether they are also required for maintaining presumptive germ cells in asexual animals. After 1 mo of RNAi treatment, Bic-C(RNAi) and NF-YB(RNAi) animals lacked nanos- and gH4-positive presumptive germ cells (Fig. 6B,D); eIF3c(RNAi) animals, however, appeared to be unaffected (Fig. 6C). These observations suggest that Bic-C and NF-YB, but not eIF3c, are required for maintaining presumptive germ cells in asexual planarians; thus, both NF-YB and Bic-C appear to be critical for the maintenance of undifferentiated germ cells (SSCs in the sexual strain, and presumptive germ cells in asexual animals).
Figure 6.
NF-YB and Bic-C are required to maintain nanos and gH4 expression in presumptive germ cells in asexual planarians. (A) Presumptive germ cells in control(RNAi) animals express gH4 and nanos (arrowheads, n = 18 out of 18). (B,D) No gH4 and nanos-positive germ cells were detected in Bic-C(RNAi) (n = 18 out of 18) and NF-YB(RNAi) (n = 16 out of 16) animals. (C) eIF3c(RNAi) animals showed normal nanos and gH4 expression in presumptive germ cells (arrows, n = 17 out of 17). Bar, 100 μm.
Discussion
To dissect the mechanisms regulating germ cell development in planarians, we identified genes whose expression was germ cell-dependent. We identified a total of 297 transcripts that showed significant down-regulation in planarians lacking germ cells; we validated germ cell-enriched expression for >95% of the top hits by in situ hybridization. Our subsequent targeted RNAi screen identified 13 genes required for planarian testis formation, maintenance, meiotic progression, or spermatid elongation. Nearly all of these genes (12 out of 13) encode conserved proteins with homologs in other organisms; notably, nine of 13 genes are predicted to encode RNA-binding proteins.
These RNA-binding proteins likely play a variety of roles, including translational activation, translational repression, alternative splicing, and regulation of mRNA stability (Table 1). For example, PABPC and hnRNPA2 are involved in multiple aspects of translational regulation and mRNA metabolism (Dreyfuss et al. 1993; Kwon et al. 1999; Mangus et al. 2003). ELAV family proteins have been implicated in regulation of mRNA stability, pre-mRNA splicing, and, more recently, translational activation (Koushika et al. 1996, 2000; Fan and Steitz 1998; Soller and White 2003; Fukao et al. 2009). Bic-C was shown recently to act as a translational repressor by recruiting the CCR4 deadenylase complex, which shortens poly(A) tails of target mRNAs during Drosophila oogenesis (Chicoine et al. 2007).
Among these RNA-binding proteins, ELAV family members and MSY4 have been shown previously to function in spermatogenesis in other organisms. Recently, an elav-like gene, Melav2, was shown to be required for spermatid elongation in the flatworm Macrostomum lignano (Sekii et al. 2009). However, here we found that knockdown of Smed-elav1 and Smed-elav2, the closest homologs of Melav2 in S. mediterranea, resulted in an earlier defect—failure of meiosis. Whether this discrepancy reflects species-specific differences in the function(s) of these genes and if they are true orthologs, remain to be determined. The Y box-containing protein MSY4 is highly expressed in mouse spermatogenesis (Davies et al. 2000). An MSY4 homolog, DeY1, was identified from the planarian Dugesia etrusca and shown to be expressed specifically in differentiating germ cells in the testes (Salvetti et al. 2002). Ectopic expression of MSY4 in late-stage mouse spermatids disrupted spermatid differentiation and resulted in sterility (Giorgini et al. 2002). Here we demonstrate that RNAi knockdown of a planarian MSY4 homolog also caused failure of spermatid differentiation. Since forced expression of MSY4 in mouse spermatids resulted in translational repression of transcripts critical for spermatid maturation (Giorgini et al. 2002), the MSY4(RNAi) phenotype may result from precocious translation of spermatid mRNAs.
Orthologs of several of the genes we identified (e.g., rap55 and Bic-C) are required for oogenesis in other species (Table1; Boag et al. 2005; Chicoine et al. 2007; Marnef et al. 2009); here we report novel spermatogenic functions for these genes. In vertebrate cells, RAP55 is a component of Processing (P) bodies, sites of mRNA degradation and translational silencing (Tanaka et al. 2006; Yang et al. 2006). The Drosophila rap55 ortholog trailer hitch (tral) is required for proper dorsal–ventral egg patterning: tral mutations disrupt secretion of Gurken protein (Wilhelm et al. 2005), and have defects in gurken mRNA localization and cytoskeletal organization (Snee and Macdonald 2009). The Caenorhabditis elegans rap55 ortholog car-1 is required for proper oogenesis; RNAi knockdown results in elevated cell death in oocytes and early cytokinesis defects in progeny (Boag et al. 2005). Given these roles of Tral and CAR-1, defects in membrane trafficking and/or cytoskeletal function could account for the spermatid elongation defects observed in Smed-rap55(RNAi) animals.
In Drosophila, mutations in Bic-C disrupt oogenesis and anteroposterior patterning of the developing egg (Mahone et al. 1995). Recent work has shown that Bic-C mutants have defects in Gurken secretion resembling those observed in tral mutants; furthermore, Bic-C and tral interact genetically (Snee and Macdonald 2009), and their proteins interact biochemically (Kugler et al. 2009). Adult Drosophila males express two alternatively spliced Bic-C transcripts at very high levels; however, the functions of these male-specific isoforms remain unclear, since all Bic-C mutant alleles are male-fertile (Mahone et al. 1995). A C. elegans Bic-C homolog, gld-3, plays multiple roles in the germline: It is required maternally for germline survival and proper embryogenesis, and also promotes sperm fate and meiotic progression (Eckmann et al. 2002, 2004). GLD-3 is a divergent member of the Bic-C family, lacking the SAM (sterile α motif) domain found at the C terminus of other family members (Eckmann et al. 2002). GLD-3 interacts biochemically with FBF RNA-binding proteins, thereby inhibiting their binding to target RNAs (Eckmann et al. 2002), and with GLD-2, the catalytic subunit of a cytoplasmic poly(A) polymerase, to promote meiotic entry (Eckmann et al. 2004) and spermatogenesis (Kim et al. 2009).
Intriguingly, the human rap55 ortholog LSM14B shows dramatic up-regulation during spermatogenesis (Chalmel et al. 2007), and, in mouse testes, RAP55 protein is detected in spermatocytes and spermatids (Pepling et al. 2007)—the same cells in which the planarian RAP55 protein is detected most abundantly. Furthermore, mouse Bic-C appears to be highly expressed in spermatogonia (Chalmel et al. 2007), as it is in planarians. Thus, it will be of interest to examine whether these genes play similar roles in spermatogenesis and early germ cell development in vertebrates.
There has been speculation that, during germ cell development, the requirements to maintain genomic totipotency force germ cells to rely mainly on “RNA-centric” translational control to regulate gene expression (Seydoux and Braun 2006). Our data provide additional evidence for the critical role of post-transcriptional regulation throughout germ cell development. The majority of genes for which RNAi knockdown results in a germ cell-defective phenotype encode RNA-binding proteins. Intriguingly, most identified neoblast-specific markers also encode RNA-binding proteins (Shibata et al. 1999; Reddien et al. 2005a; Salvetti et al. 2005; Guo et al. 2006; Palakodeti et al. 2008; Solana et al. 2009; Rouhana et al. 2010), providing further support for the idea that post-transcriptional control is critical for regulating pluripotency. Here we identified candidate transcripts based on their expression in relatively early germ cells, yet many of the RNA knockdowns revealed functions in later stages of spermatogenesis. This observation suggests that, in planarians, early male germ cells store RNAs required for later stages of differentiation, and that sequential waves of post-transcriptional control drive meiotic progression and spermiogenesis, as observed in meiotic progression in Xenopus oocytes (Vasudevan et al. 2006) and throughout germ cell development in C. elegans (Kimble and Crittenden 2007). The abundance of rap55 mRNA in spermatogonia (in which the protein is barely detectable) and the paucity of rap55 mRNA in spermatids (which contain high levels of the protein) are consistent with this idea.
Our screen also identified two genes (Smed-C3H-zfn-1 and spac-1) with highest similarity to sequences from parasitic flatworms of the genus Schistosoma. These organisms are responsible for schistosomiasis, a neglected tropical disease affecting >200 million people worldwide (King 2009). Although genome sequences are available for two species of Schistosomes (Berriman et al. 2009; The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium 2009), the requirements for invertebrate intermediate hosts and vertebrate definitive hosts to propagate the life cycle complicate large-scale analysis of gene function in these animals. The ability to characterize flatworm-specific genes by examining their roles in planarians represents a promising strategy for understanding the less-accessible parasitic flatworms. Because the pathology of schistosomiasis results from inflammatory responses to the parasite's eggs (King 2009), any flatworm-specific targets that eliminate this tremendous reproductive output could be useful in controlling the disease.
Finally, we found that a planarian NF-YB homolog is required for maintaining spermatogenesis. Following NF-YB RNAi in sexual planarians, round spermatids accumulate and the gH4+ spermatogonial layer of the testis is lost. Thus, spermatogonial progeny can differentiate properly, forming spermatocytes that progress through meiosis and initiate spermatid nuclear condensation; however, the spermatogonial layer disappears, consistent with a failure to maintain SSCs. Furthermore, NF-YB RNAi in asexual planarians resulted in a loss of nanos+ gH4+ early germ cells. Together, these results suggest that this gene is required for maintaining undifferentiated male germ cells in planarians.
NF-YB encodes one subunit of the trimeric transcription factor NF-Y (or CCAAT-binding factor), which binds specifically to the CCAAT box, a motif commonly found in eukaryotic promoters (Ceribelli et al. 2008). The NF-YB(RNAi) phenotype in sexual planarians is strikingly similar to that of TAF4b and Plzf mutant mice, which initially produce sperm, but then become infertile due to progressive loss of spermatogonia (Buaas et al. 2004; Costoya et al. 2004; Falender et al. 2005). TAF4b is a component of the TFIID complex of the RNA polymerase II basal transcription machinery, and PLZF (promyelocytic leukemia zinc finger protein) is a transcriptional repressor. Both genes are required for maintaining SSCs (Buaas et al. 2004; Costoya et al. 2004; Falender et al. 2005); NF-YB may play a similar role in planarians. NF-YB transcripts are enriched in mouse spermatogonia relative to other testes cell types (Chalmel et al. 2007), so it will be of interest to determine whether it is also involved in regulating SSCs in mammalian testes. Interestingly, CCAAT boxes were overrepresented in promoters of genes up-regulated in primordial germ cells and embryonic stem (ES) cells (human and mouse) relative to differentiated cell types (Grskovic et al. 2007). Expression of NF-YA and NF-YB mRNAs decreased during the course of ES cell differentiation, and RNAi knockdowns of these genes reduced the proliferative potential of ES cells (Grskovic et al. 2007). In murine hematopoietic stem cells (HSCs), NF-Y overexpression activates HSC regulatory genes and promotes HSC self-renewal in vitro and in vivo (Zhu et al. 2005). Together, these results suggest a widely conserved role for NF-Y in regulating multiple stem cell populations.
In summary, we identified genes required for distinct stages of male germ cell development in planarians; notably, most of these genes share extensive similarity with vertebrate genes. Several microarray analyses have surveyed germ cell differentiation in various mammalian species (e.g., Schultz et al. 2003; Oatley et al. 2006; Chalmel et al. 2007); however, large-scale functional analyses remain difficult in these animals. Our work demonstrates that planarians can serve as valuable models for screening large numbers of such differentially expressed genes to dissect conserved mechanisms of germ cell development.
Materials and methods
Planarian culture
Clonal lines of hermaphroditic (Zayas et al. 2005) and asexual (Sánchez Alvarado et al. 2002) S. mediterranea were used for all experiments and maintained as per Wang et al. (2007); they were fed weekly with organic calf liver and starved at least 1 wk before use.
RNAi
RNAi feedings were performed as described (Gurley et al. 2008). Control animals were fed bacteria containing empty pPR242 vector (courtesy of Peter Reddien, Whitehead Institute/Massachusetts Institute of Technology). For experimental RNAi, corresponding cDNAs were subcloned into pPR244 using Gateway cloning (Invitrogen) as described (Reddien et al. 2005b) and confirmed by sequencing. For the RNAi screen, all experiments were performed at least twice.
Microarray analyses
nanos(RNAi) was performed as described (Wang et al. 2007) on asexuals or 1-d-old sexual hatchlings over ∼1 mo (total of six dsRNA feedings). Absence of germ cells after nanos(RNAi) treatment was confirmed by in situ hybridization to detect gH4 expression in half of the treated animals. Total RNA was then extracted from the remaining asexual nanos(RNAi) (n = 10) and control(RNAi) (n = 10) animals, or juvenile nanos(RNAi) (n = 6) and control(RNAi) (n = 6) animals using Trizol Reagent (Invitrogen); purified with RNeasy minispin columns (Qiagen) with on-column DNase treatment; and quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA samples were labeled and hybridized to custom oligonucleotide arrays (Roche Nimblegen). All array experiments were performed in duplicate using independently prepared RNA samples from independent RNAi experiments. To analyze microarray data, expression values normalized using the Robust Multichip Average (RMA) algorithm were fit to a linear model using the Bioconductor limma package (Gentleman et al. 2004; Smyth 2004) to calculate moderated t-statistics and adjusted P-values for each transcript. Microarray data have been deposited in the Gene Expression Omnibus (accession no. GSE22797).
Whole-mount in situ hybridization
Whole-mount in situ hybridization of planarians was performed as described (Pearson et al. 2009) with the following modifications for large sexual planarians: They were killed in 10% N-acetyl-cysteine, fixed in 4% formaldehyde for 25–30 min at room temperature, permeabilized with 1% SDS for 10 min at room temperature, and reduced for 10 min at room temperature. Prior to hybridization, animals were treated with proteinase K for 20 min at room temperature. Samples were imaged with a Leica M205A microscope and DFC420 camera. For FISH, after antibody incubation (α-digoxigenin-peroxidase, 1:500 [Roche]; or α-dinitro-phenol-peroxidase, 1:100 [Perkin-Elmer]) overnight at 4°C, samples were washed in MABT for 2 h, followed by two 10-min washes in PBS, and were developed with FITC-tyramide or Cy3-tyramide (Perkin-Elmer) per the manufacturer's protocol. For double FISH, after developing the first color, peroxidase was inactivated with 2% H2O2 in PBST for 1 h before blocking and second antibody incubation. FISH samples were imaged using a Zeiss 710 confocal microscope.
Whole-mount immunofluorescence
Planarians were killed with 2% HCl for 5 min on ice and fixed in 4% formaldehyde in PBS. Staining with α-H3-S10P and DAPI was performed as described (Guo et al. 2006). Staining with mAb-1 Y12 (1:500; Neomarkers) was performed similarly. Rabbit antisera were raised against both full-length and the first 130 amino acids of Drosophila Tral and affinity-purified using the latter. Staining with α-Tral (1:1000, crude antisera; 1:100, affinity-purified) was performed after FISH procedures. Images were captured with Zeiss SteREO Lumar.V12 and 710 confocal microscopes.
Gene sequences
cDNA sequences were obtained from ESTs from hermaphroditic S. mediterranea. Accession numbers are as follows: Smed-elav2, HM639987; spac-1, HM639988; Smed-hnRNPA2, HM639989; Smed-PABPC, HM100651; sped-1, HM100652; Smed-NF-YB, HM100653; Smed-MSY4, BK007100; Smed-C3H-zfn-1, BK007101; Smed-Brg1, BK007102; Smed-elav1, BK007103. Additional cDNA sequences for Bic-C, Smed-eIF3c, and Smed-rap55 were obtained by PCR from hermaphroditic S. mediterranea cDNA, cloned into the pCR2.1-TOPO vector (Invitrogen), and entered into GenBank (accession nos. HM055594, HM055595, and HM055596).
Acknowledgments
We thank David Forsthoefel, James Collins, Ryan King, and Labib Rouhana for critical reading of the manuscript and helpful discussions, and members of Alejandro Sánchez Alvarado's laboratory for sharing in situ protocols before publication. This work was supported by NIH (R01 HD043403) and NSF (IOS-0744689) to P.A.N. P.A.N. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1951010.
Supplemental material is available at http://www.genesdev.org.
References
- Baguñà J, Saló E, Auladell C 1989. Regeneration and pattern formation in planarians. III. Evidence that neoblasts are totipotent stem cells and the source of blastema cells. Development 107: 77–86 [Google Scholar]
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, et al. 2009. The genome of the blood fluke Schistosoma mansoni. Nature 460: 352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boag PR, Nakamura A, Blackwell TK 2005. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development 132: 4975–4986 [DOI] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem 275: 17122–17129 [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE 2004. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36: 647–652 [DOI] [PubMed] [Google Scholar]
- Ceribelli M, Dolfini D, Merico D, Gatta R, Vigano AM, Pavesi G, Mantovani R 2008. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol 28: 2047–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmel F, Rolland AD, Niederhauser-Wiederkehr C, Chung SS, Demougin P, Gattiker A, Moore J, Patard JJ, Wolgemuth DJ, Jegou B, et al. 2007. The conserved transcriptome in human and rodent male gametogenesis. Proc Natl Acad Sci 104: 8346–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P 2007. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell 13: 691–704 [DOI] [PubMed] [Google Scholar]
- Cinalli RM, Rangan P, Lehmann R 2008. Germ cells are forever. Cell 132: 559–562 [DOI] [PubMed] [Google Scholar]
- Cobb J, Miyaike M, Kikuchi A, Handel MA 1999. Meiotic events at the centromeric heterochromatin: Histone H3 phosphorylation, topoisomerase II α localization and chromosome condensation. Chromosoma 108: 412–425 [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP 2004. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 36: 653–659 [DOI] [PubMed] [Google Scholar]
- Davies HG, Giorgini F, Fajardo MA, Braun RE 2000. A sequence-specific RNA binding complex expressed in murine germ cells contains MSY2 and MSY4. Dev Biol 221: 87–100 [DOI] [PubMed] [Google Scholar]
- Deshpande G, Calhoun G, Yanowitz JL, Schedl PD 1999. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell 99: 271–281 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG 1993. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem 62: 289–321 [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452: 745–749 [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Kraemer B, Wickens M, Kimble J 2002. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell 3: 697–710 [DOI] [PubMed] [Google Scholar]
- Eckmann CR, Crittenden SL, Suh N, Kimble J 2004. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 168: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen-Campen B, Schwager EE, Extavour CG 2010. The molecular machinery of germ line specification. Mol Reprod Dev 77: 3–18 [DOI] [PubMed] [Google Scholar]
- Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS 2005. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev 19: 794–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 17: 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franquinet R, Lender T 1973. Étude ultrastructurale des testicules de Polycelis tenuis et Polycelis nigra (Planaires). Evolution des cellules germinales mâles avant la spermiogenèse. Z Mikrosk Anat Forsch 87: 4–22 [PubMed] [Google Scholar]
- Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T 2009. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol Cell 36: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80 doi: 10.1186/gb-200405010-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgini F, Davies HG, Braun RE 2002. Translational repression by MSY4 inhibits spermatid differentiation in mice. Development 129: 3669–3679 [DOI] [PubMed] [Google Scholar]
- Grskovic M, Chaivorapol C, Gaspar-Maia A, Li H, Ramalho-Santos M 2007. Systematic identification of cis-regulatory sequences active in mouse and human embryonic stem cells. PLoS Genet 3: e145 doi: 10.1371/journal.pgen.0030145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA 2006. A bruno-like gene is required for stem cell maintenance in planarians. Dev Cell 11: 159–169 [DOI] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sanchez Alvarado A 2008. β-Catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319: 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handberg-Thorsager M, Saló E 2007. The planarian nanos-like gene Smednos is expressed in germline and eye precursor cells during development and regeneration. Dev Genes Evol 217: 403–411 [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Kobayashi S 2004. Nanos suppresses somatic cell fate in Drosophila germ line. Proc Natl Acad Sci 101: 10338–10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106: 348–360 [DOI] [PubMed] [Google Scholar]
- Kim KW, Nykamp K, Suh N, Bachorik JL, Wang L, Kimble J 2009. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell 16: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol 23: 405–433 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kim J, Kim YJ 1999. Requirement of RBP9, a Drosophila Hu homolog, for regulation of cystocyte differentiation and oocyte determination during oogenesis. Mol Cell Biol 19: 2505–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ishida K, Kashiwabara S, Baba T 2009. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol Reprod 80: 545–554 [DOI] [PubMed] [Google Scholar]
- King CH 2009. Toward the elimination of schistosomiasis. N Engl J Med 360: 106–109 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T 1996. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380: 708–711 [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E 2001. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev 15: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Lisbin MJ, White K 1996. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol 6: 1634–1641 [DOI] [PubMed] [Google Scholar]
- Koushika SP, Soller M, White K 2000. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol Cell Biol 20: 1836–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler JM, Chicoine J, Lasko P 2009. Bicaudal-C associates with a Trailer Hitch/Me31B complex and is required for efficient Gurken secretion. Dev Biol 328: 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Barbarese E, Carson JH 1999. The cis-acting RNA trafficking signal from myelin basic protein mRNA and its cognate trans-acting ligand hnRNP A2 enhance cap-dependent translation. J Cell Biol 147: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA Jr, Steitz JA 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci 78: 2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahone M, Saffman EE, Lasko PF 1995. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J 14: 2043–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223 doi: 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanero S, Arana P, Puertas MJ, Houben A 2000. The chromosomal distribution of phosphorylated histone H3 differs between plants and animals at meiosis. Chromosoma 109: 308–317 [DOI] [PubMed] [Google Scholar]
- Marnef A, Sommerville J, Ladomery MR 2009. RAP55: Insights into an evolutionarily conserved protein family. Int J Biochem Cell Biol 41: 977–981 [DOI] [PubMed] [Google Scholar]
- Morgan TH 1902. Growth and regeneration in Planaria lugubris. Arch Entwicklungsmech Org 13: 179–212 [Google Scholar]
- Newmark PA, Wang Y, Chong T 2008. Germ cell specification and regeneration in planarians. Cold Spring Harb Symp Quant Biol 73: 573–581 [DOI] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL 2006. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci 103: 9524–9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakodeti D, Smielewska M, Lu YC, Yeo GW, Graveley BR 2008. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14: 1174–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A 2009. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn 238: 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling ME, Wilhelm JE, O'Hara AL, Gephardt GW, Spradling AC 2007. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc Natl Acad Sci 104: 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A 2005a. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1327–1330 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A 2005b. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8: 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Shibata N, Nishimura O, Agata K 2010. Different requirements for conserved post-transcriptional regulators in planarian regeneration and stem cell maintenance. Dev Biol 341: 429–443 [DOI] [PubMed] [Google Scholar]
- Sada A, Suzuki A, Suzuki H, Saga Y 2009. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science 325: 1394–1398 [DOI] [PubMed] [Google Scholar]
- Salvetti A, Lena A, Rossi L, Deri P, Cecchettini A, Batistoni R, Gremigni V 2002. Characterization of DeY1, a novel Y-box gene specifically expressed in differentiating male germ cells of planarians. Gene Expr Patterns 2: 195–200 [DOI] [PubMed] [Google Scholar]
- Salvetti A, Rossi L, Lena A, Batistoni R, Deri P, Rainaldi G, Locci MT, Evangelista M, Gremigni V 2005. DjPum, a homologue of Drosophila Pumilio, is essential to planarian stem cell maintenance. Development 132: 1863–1874 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Newmark PA, Robb SM, Juste R 2002. The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129: 5659–5665 [DOI] [PubMed] [Google Scholar]
- Sato K, Shibata N, Orii H, Amikura R, Sakurai T, Agata K, Kobayashi S, Watanabe K 2006. Identification and origin of the germline stem cells as revealed by the expression of nanos-related gene in planarians. Dev Growth Differ 48: 615–628 [DOI] [PubMed] [Google Scholar]
- The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium 2009. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature 460: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz N, Hamra FK, Garbers DL 2003. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci 100: 12201–12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekii K, Salvenmoser W, De Mulder K, Scharer L, Ladurner P 2009. Melav2, an elav-like gene, is essential for spermatid differentiation in the flatworm Macrostomum lignano. BMC Dev Biol 9: 62 doi: 10.1186/1471-213X-9-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE 2006. Pathway to totipotency: Lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Shibata N, Umesono Y, Orii H, Sakurai T, Watanabe K, Agata K 1999. Expression of vasa(vas)-related genes in germline cells and totipotent somatic stem cells of planarians. Dev Biol 206: 73–87 [DOI] [PubMed] [Google Scholar]
- Smyth GK 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article 3. doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM 2009. Bicaudal C and trailer hitch have similar roles in gurken mRNA localization and cytoskeletal organization. Dev Biol 328: 434–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J, Lasko P, Romero R 2009. Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev Biol 328: 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, White K 2003. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev 17: 2526–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871 [DOI] [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M 2006. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem 281: 40096–40106 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. 2003. The COG database: An updated version includes eukaryotes. BMC Bioinformatics 4: 41 doi: 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y 2003. Conserved role of nanos proteins in germ cell development. Science 301: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Seli E, Steitz JA 2006. Metazoan oocyte and early embryo development program: A progression through translation regulatory cascades. Genes Dev 20: 138–146 [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin H 2004. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 303: 2016–2019 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zayas RM, Guo T, Newmark PA 2007. nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci 104: 5901–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD 1998. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci 95: 7480–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell 9: 675–685 [DOI] [PubMed] [Google Scholar]
- Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB 2006. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA 12: 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas RM, Hernandez A, Habermann B, Wang Y, Stary JM, Newmark PA 2005. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: Analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci 102: 18491–18496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG 2005. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci 102: 11728–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]