Abstract
Escherichia coli DNA photolyase is an enzyme that repairs the major kind of UV-induced lesions, cyclobutane pyrimidine dimer (CPD) in DNA utilizing 350–450 nm light as energy source. The enzyme has very high photo-repair efficiency (the quantum yield of the reaction is ~0.85), which is significantly greater than many model compounds that mimic photolyase. This suggests that some residues of the protein play important roles in the photo-repair of CPD. In this paper, we have focused on several residues discussed their roles in catalysis by reviewing the existing literature and some hypotheses.
1. Introduction
The sun gives warmth and light to the living beings on the earth. However, the ultra-violet (UV) radiation in the sunlight stimulates lesions forming in DNA. The UV-induced lesions in DNA block the replication and transcription events in the living cells, cause growth delay, mutagenesis, or lethal effects to organisms [1]. In order to survive under the sunlight, the organisms have evolved several repair mechanisms to resist the harmfulness of UV. Direct reversal by DNA photolyases is one of the mechanisms. There are two types of DNA photolyases, CPD photolyases and (6-4) photolyases, which, respectively, reverse the two major UV-induced lesions in DNA, cyclobutane pyrimidine dimers (CPD), and (6-4) photoproducts, utilizing blue or near-UV light (350–450 nm) as energy source [1–4]. CPD photolyases can be further categorized into two subclasses, class I (microbial) and class II (animal and plant), based on their amino acid sequence similarity [3, 5, 6]. Flavin adenine dinucleotide (FAD) is catalytic cofactor of all photolyases [3]. And a second cofactor, usually a derivative of folate, deazaflavin or flavin, acts as a photoantenna to increase the repair efficiency of the enzymes under limiting light conditions [3, 7–9]. The repair reactions are proposed through a photon-induced electron transfer mechanism which is supported by many model compounds studies. However, the quantum yields (Φ = 0.7–0.98) in the repair of pyrimidine dimers by DNA photolyase is significantly higher [3, 10–13] than those model compounds (Φ = 0.016–0.4) [14–16]. These results indicate that some amino acid residues of photolyases play important roles in the repair reactions.
Escherichia coli DNA photolyase is a representative of them. By reviewing the existing literature and some hypotheses, we discussed the roles of some residues of E. coli photolyase in the highly efficient catalysis. This paper would provide the further insights into the catalytic mechanism of the enzyme.
2. Escherichia coli DNA Photolyase
Escherichia coli DNA photolyase is a class I CPD photolyase [17], containing 471 amino acids [18, 19] and two cofactors, FAD [20] and a folate derivative, 5,10-methenyltetrahydropterolypolyglutamate (MTHF) [7]. The enzyme was found in 1950s by Rupert et al. [21]. Its gene was first cloned by Sancar et al. [18, 19], which settled the problem that the little expression of the gene in the cells prevents the high yield of pure enzyme for researches. During the following years, the enzyme has been extensively studied. The physiological form of the enzyme contains a fully reduced FAD (FADH−) that is required for its activity both in vivo and in vitro [22]. It binds a CPD in DNA independent of light [17] and flips the dimer out of the double helix into the active site cavity to make a stable enzyme-substrate complex [23–26]. The light-dependent catalytic reaction was proposed through these steps: FADH− is excited directly by a photon or by the photoexcited MTHF cofactor and then transfers an electron to CPD to generate a charge-separated radical pair (FADH• + CPD•-); then the CPD radical anion cleaves, and the excess electron returns to FADH• to restore the reduced form and close the catalytic photocycle [3, 11, 22, 27–29]. By the techniques such as time-resolved spectroscopy, laser flash photolysis [30–35], and transient electron paramagnetic resonance [36, 37], this photon-induced electron transfer mechanism has been substantiated. However, the roles of the amino acid residues in the steps of the high efficient enzymatic reaction, such as substrate docking and splitting, electron transfer, and intermediate stabilization, need further investigation.
3. Trp277: A Residue for CPD Docking and Splitting
E. coli DNA photolyase contains 15 tryptophan residues. Trp277 lies in a highly conserved region Trp277-Tyr281, which is considered to be important for DNA binding [38]. By the site-directed mutagenesis studies, it was found when Trp277 was replaced with arginine or glutamate, the binding affinity for CPD substrate was lower for 300- or 1000-fold, respectively, although the photochemical properties and the quantum yields for catalyses (under the irradiation wavelengths at 366 nm and 384 nm) of the mutants were indistinguishable from the wild-type enzyme [38]. Later on, it was discovered that Trp277 can also directly and efficiently repair CPD under 280 nm light [39]. These results revealed that Trp277 is crucial for substrate binding, and under certain conditions it also acts as a catalytic residue.
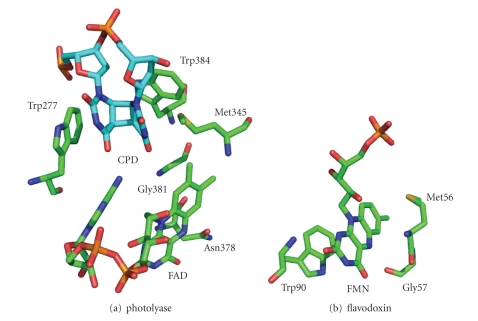
The crystal structure of E. coli photolyase (Protein Data Bank entry 1DNP) shows that a positively charged groove on the surface of the protein which might interact with the DNA backbone and a hydrophobic cavity locates at the center. The cavity has the right dimension to hold a cis,syn CPD, and Trp277 forms one side wall of it [40] (Figure 1(a)). It is proposed that photolyase binds DNA chain containing a CPD, flips it out into the cavity, and Trp277 stacks with the 5′ side of the CPD by π-π interaction [23–26, 38]. This is confirmed by the crystal structure of the complex of CPD-like lesion in DNA and photolyase from Anacystis Nidulans (Synechococcus sp.) (Protein Data Bank entry 1TEZ) [41].
Figure 1.
Conformations of (a) active sites of photolyase and (b) FMN binding sites of Clostridium beijerinckii flavodoxin. The CPD structure was extracted from the PDB entry 1SNH and coupled with E. coli DNA photolyase structure 1DNP according to the CPD like lesion-photolyase complex structure 1TEZ. The FMN binding sites of flavodoxin were extracted form the PDB entry 5ULL. This figure was prepared with PyMOL (http://www.pymol.org/).
There is another tryptophan, Trp384, in the cavity forming a wedge with Trp277 (Figure 1(a)) [40]. By the examination of the cocrystal structure of Thermus thermophilus photolyase with a thymine, it is proposed that CPD might be sandwiched by these two tryptophans [42], and its 3′ side might stack with Trp384 in a similar manner. However, from the structure 1TEZ, it is concluded that the 3′ side should stack with a methionine residue, Met345, but not Trp384. Met345 is to be discussed in the next section.
4. Met345: A Discriminating Residue of CPD Photolyase from Photolyase-Cryptochrome Super Family
A methionine residue in the active cavity of Saccharomyces cerevisiae photolyase, which corresponds to Met345 of E. coli photolyase, was predicted to interact with CPD [24]. By the structure of the complex of Anacystis Nidulans photolyase and CPD-like lesion, it was confirmed that Met345 should stack with the 3′ side of CPD [41] (Figure 1(a)). Methionine is a sulfur-containing amino acid. From the studies of the crystal structures of many proteins, Morgan and coworkers proposed that the sulfur atoms might interact with aromatic rings by the so-called sulfur-π interaction [43–51]. Although the mechanism of this interaction is still controversial, it does exist. For example, in the structure of the flavodoxin of Clostridium beijerinckii (Protein Data Bank entry 5ULL), a methionine residue is located near the xylene ring side of the flavin cofactor [52] the conformations are just like those of Met345 and the 3′ side of CPD (Figure 1(b)). The sulfur-π stack might also contribute to substrate-binding affinity. In addition, this interaction together with that between Trp277 and the 5′ side of CPD might have some effects on substrate splitting.
The electron transfer from excited FADH− to CPD is now considered to be through a direct pathway [33, 53, 54]. However, a theoretical calculation with the CPD-photolyase complex structure shows that the indirect electron transfer via protein mediators is as important as the direct electron transfer [55]. The electron-tunneling pathway analysis suggested that there were two typical electron-tunneling routes for the electron transfer of photolyase, one was an adenine route, and the other was through a methionine corresponding to Met345 of E. coli photolyase [55]. It is widely accepted that the electron transfers from FADH− to the 5′ side of CPD first, then to the 3′ side [3, 32]. And the pathway for excess electron transfer back to FADH• remains unclear till now. Considering Met345 adjacent to the 3′ side of CPD, we speculate that it might be an electron back transfer pathway.
It is intriguing that Met345 is proposed to be a residue for the discrimination of CPD photolyase from the photolyase-cryptochrome super family [55]. The super family contains CPD photolyases, (6-4) photolyases, and cryptochromes, all of which are flavoproteins. (6-4) photolyases repair (6-4) photoproduct but not CPD [4]. Cryptochromes play roles in photomorphogenesis in plants and entrain the circadian biological clocks in animal [2, 3, 56]. Recently, it is found that some cryptochromes in insects and birds might function as light-activated magnetoreceptors [57–61]. Although these proteins are functionally diverse, they have relatively high degree of homology. Met345 is conserved in all CPD photolyase whereas in (6-4) photolyases, it is replaced by a histidine which is also important for catalysis [62]. In cryptochromes, it is replaced by histidine, valine, glutamine, and so forth. It might be one of the residues responsible for the function diversity of the proteins in this super family [55].
5. Asn378: A Stabilizer of the Neutral FAD Radical
Although E. coli photolyase contain reduced FAD in vivo, it is usually purified with FAD in the blue neutral radical form (FADH•). It is known that E. coli photolyase is one of the unusual flavoproteins in which the radical is extremely stable [63]. The purified enzyme can hold its radical flavin cofactor unoxidized in aerobic conditions for several days whereas it hardly exists free in aqueous solutions because the dismutation of the radical is favored. These results indicate that the protein environment gives a strong stabilization to the radical. Clostridium beijerinckii flavodoxin is another example that has the ability to hold stable radical flavin cofactor. It was proposed that a hydrogen bond between the flavin N(5)H group and the backbone carbonyl oxygen of Gly57 in the flavodoxin is important for the modulation of the redox potentials of the cofactor and the stabilization of the radical form (Figure 1(b)) [64, 65]. Interestingly, there is a similar hydrogen bond between the flavin N(5)H group and the side carbonyl of the Asn378 residue (Figure 1(a)). We had replaced the asparagine residue with serine and found that the mutant has no stable radical state [66]. Moreover, the catalytic activity of the mutant was lost [66]. These experiments show that Asn378 is crucial both for the stabilizing the neutral flavin radical cofactor and for catalysis. It is convincible because the catalytic reaction of CPD splitting is through a radical mechanism: FADH− gives an electron to CPD and becomes FADH•. If the transient radical intermediate is well stabilized, it will give enough time for cleavage of the cyclobutane ring to achieve high repair efficiency [33, 67]. When the stabilizing effect is disrupted, the unwanted back electron transfer might be accelerated, leading to low repair efficiency.
Asn378 is a highly conserved residue in photolyases. In most class I CPD photolyases and (6-4) photolyases, the residues are unchanged or replaced with asparate [6, 68]. The residue is also conserved in mammalian cryptochromes [69]. In a plant cryptochrome Arabidopsis thaliana CRY1, it is replaced with asparate that is proposed to be responsible for the down shift of the flavin redox potentials which make the difference between the cryptochrome and photolyases [70]. Meanwhile, some insect cryptochromes have replaced the residue with cysteine, which may be responsible for a red anionic radical (FAD•-) state but not the blue neutral radical state in photolyases [69, 71]. However, in many class II CPD photolyases, the asparagine residues are not conserved [6, 72]. There is evidence that class II CPD photolyases have the similar photochemical properties and the FAD binding environments as compare to class I CPD and (6-4) photolyases [72]. Thus, a stabilizer near the flavin N(5)H group is also required in a class II enzyme. By examining a model structure of a class II enzyme (Oryza sativa CPD photolyase) calculated by comparative modeling (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi) [72, 73], we find another asparagine residue, Asn421, might be an alternative candidate for the stabilization function, which is also highly conserved in class II CPD photolyases.
From the model structure of Oryza sativa CPD photolyase, it is of interest to find that the residue corresponding to Trp277 in E. coli photolyase is not conserved, which is considered to be crucial for substrate binding (vide ante). It indicates that the class II CPD photolyases might use a different substrate binding mechanism as compared to class I group. However, it seems that the residues corresponding to Met345 and Trp384 in E. coli photolyase are conserved in the substrate binding cavity, emphasizing their important roles for substrate binding and/or catalysis. Therefore, to fully uncover details of substrate binding and FAD usage, a real crystal structure of a class II photolyase is highly awaited [72].
A mutagenesis study on Anacystis nidulans photolyase shows that two residues (Trp384 and Gly381 of E. coli photolyase) are crucial for the kinetics stability of the neutral flavin radical in the enzyme (Figure 1(a)) [74]. In Synechocystis sp. PCC6803 CRY-DASH, these residues are replaced with tyrosine and asparagine, respectively [74]. The difference might also be a reason for the diverse functions of photolyases and cryptochromes.
6. Perspectives
Although the first photolyase has been discovered more than 50 years [21], photolyases still occupies a unique position in biochemistry [1, 2]. Photolyases bind the substrate in a light-independent manner, but the catalysis is absolutely dependent on light that makes the possible to analyze the binding and catalysis steps of the enzymes independently [2, 3]. Furthermore, the level of substrate in the cell can be controlled easily by simply changing the UV dose, and the repairing of the bound substrate is ultrafast by a single light flash. All characteristics make the photolyases to be useful tools for biochemical research, especially the in vivo enzymology [2]. Cryptochromes are homologues of photolyases, which are widely concerned now for their functions in the circadian clocks of animal, the photomorphogenesis of plants and the migration of birds and insects by acting as light-activated magnetoreceptors [2, 3, 56–61]. The action mechanisms of cryptochromes are still unclear at present. However, as photolyases, flavin radical is also proposed to be crucial for their functions [57–61, 69–71, 74–76]. Thus, further exploring the photolyase system might give new insights into the researches of both photolyases and cryptochromes.
7. Summary
In this paper, we have discussed some of important residues of E. coli DNA photolyase. Evidence suggests that they play significant roles in substrate docking and splitting, electron transfer, and intermediate stabilization. Of course, due to the limits of our knowledge, there must be a lot of amino acid residues that might be much more important in the catalysis of enzyme, which are not discussed here. Together with all these amino acid residues and the cofactors of the enzyme, they built a very high efficient system for the photo-repair of cyclobutane pyrimidine dimers in DNA. Further research on this system will not only contribute to understanding its efficient catalytic mechanism, but also give new insights to the other biochemical research in the future.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (30900243; 30870062) and Bilingual Teaching Project of the Education Ministry of China.
References
- 1.Beukers R, Eker APM, Lohman PHM. 50 years thymine dimer. DNA Repair. 2008;7(3):530–543. doi: 10.1016/j.dnarep.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A. Structure and function of photolyase and in vivo enzymology: 50th anniversary. Journal of Biological Chemistry. 2008;283(47):32153–32157. doi: 10.1074/jbc.R800052200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chemical Reviews. 2003;103(6):2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 4.Kim S-T, Malhotra K, Smith CA, Taylor J-S, Sancar A. Characterization of (6-4) photoproduct DNA photolyase. Journal of Biological Chemistry. 1994;269(11):8535–8540. [PubMed] [Google Scholar]
- 5.Yasui A, Eker APM, Yasuhira S, et al. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO Journal. 1994;13(24):6143–6151. doi: 10.1002/j.1460-2075.1994.tb06961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanai S, Kikuno R, Toh H, Ryo H, Todo T. Molecular evolution of the photolyase-blue-light photoreceptor family. Journal of Molecular Evolution. 1997;45(5):535–548. doi: 10.1007/pl00006258. [DOI] [PubMed] [Google Scholar]
- 7.Johnson JL, Hamm-Alvarez S, Payne G, Sancar GB, Rajagopalan KV, Sancar A. Identification of the second chromophore of Escherichia coli and yeast DNA photolyases as 5,10-methenyltetrahydrofolate. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(7):2046–2050. doi: 10.1073/pnas.85.7.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eker APM, Kooiman P, Hessels JKC, Yasui A. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. Journal of Biological Chemistry. 1990;265(14):8009–8015. [PubMed] [Google Scholar]
- 9.Ueda T, Kato A, Kuramitsu S, Terasawa H, Shimada I. Identification and characterization of a second chromophore of DNA photolyase from Thermus thermophilus HB27. Journal of Biological Chemistry. 2005;280(43):36237–36243. doi: 10.1074/jbc.M507972200. [DOI] [PubMed] [Google Scholar]
- 10.Payne G, Sancar A. Absolute action spectrum of E-FADH2 and E-FADH2-MTHF forms of Escherichia coli DNA photolyase. Biochemistry. 1990;29(33):7715–7727. doi: 10.1021/bi00485a021. [DOI] [PubMed] [Google Scholar]
- 11.Heelis PF. Photochemical properties of escherichia coll DNA photolyase: a flash photolysis study. Biochemistry. 1986;25(25):8163–8166. doi: 10.1021/bi00373a006. [DOI] [PubMed] [Google Scholar]
- 12.Ramsey AJ, Alderfer JL, Jorns MS. Energy transduction during catalysis by Escherichia coli DNA photolyase. Biochemistry. 1992;31(31):7134–7142. doi: 10.1021/bi00146a015. [DOI] [PubMed] [Google Scholar]
- 13.Kim S-T, Heelis PF, Sancar A. Energy transfer (deazaflavin→FADH2) and electron transfer (FADH2→T<>T) kinetics in Anacystis nidulans photolyase. Biochemistry. 1992;31(45):11244–11248. doi: 10.1021/bi00160a040. [DOI] [PubMed] [Google Scholar]
- 14.Kim ST, Hartman RF, Rose SD. Solvent dependence of pyrimidine dimer splitting in a covalently linked dimer-indole system. Photochemistry and Photobiology. 1990;52(4):789–794. doi: 10.1111/j.1751-1097.1990.tb08683.x. [DOI] [PubMed] [Google Scholar]
- 15.Epple R, Wallenborn E-U, Carell T. Investigation of flavin-containing DNA-repair model compounds. Journal of the American Chemical Society. 1997;119(32):7440–7451. [Google Scholar]
- 16.Song Q-H, Tang W-J, Hei X-M, Wang H-B, Guo Q-X, Yu S-Q. Efficient photosensitized splitting of thymine dimer by a covalently linked tryptophan in solvents of high polarity. European Journal of Organic Chemistry. 2005;(6):1097–1106. [Google Scholar]
- 17.Brash DE, Franklin WA, Sancar GB, Sancar A, Haseltine WA. Escherichia coli DNA photolyase reverses cyclobutane pyrimidine dimers but not pyrimidine-pyrimidone (6-4) photoproducts. Journal of Biological Chemistry. 1985;260(21):11438–11441. [PubMed] [Google Scholar]
- 18.Sancar A, Smith FW, Sancar GB. Purification of Escherichia coli DNA photolyase. Journal of Biological Chemistry. 1984;259(9):6028–6032. [PubMed] [Google Scholar]
- 19.Sancar GB, Smith FW, Sancar A. Identification and amplification of the E. coli phr gene product. Nucleic Acids Research. 1983;11(19):6667–6678. doi: 10.1093/nar/11.19.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sancar A, Sancar GB. Escherichia coli DNA photolyase is a flavoprotein. Journal of Molecular Biology. 1984;172(2):223–227. doi: 10.1016/s0022-2836(84)80040-6. [DOI] [PubMed] [Google Scholar]
- 21.Rupert CS, Goodgal SH, Herriott RM. Photoreactivation in vitro of ultraviolet-inactivated Hemophilus influenzae transforming factor. The Journal of General Physiology. 1958;41:451–471. doi: 10.1085/jgp.41.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Payne G, Heelis PF, Rohrs BR, Sancar A. The active form of Escherichia coli DNA photolyase contains a fully reduced flavin and not a flavin radical, both in vivo and in vitro. Biochemistry. 1987;26(22):7121–7127. doi: 10.1021/bi00396a038. [DOI] [PubMed] [Google Scholar]
- 23.Husain I, Sancar GB, Holbrook SR, Sancar A. Mechanism of damage recognition by Escherichia coli DNA photolyase. Journal of Biological Chemistry. 1987;262(27):13188–13197. [PubMed] [Google Scholar]
- 24.Vande Berg BJ, Sancar GB. Evidence for dinucleotide flipping by DNA photolyase. Journal of Biological Chemistry. 1998;273(32):20276–20284. doi: 10.1074/jbc.273.32.20276. [DOI] [PubMed] [Google Scholar]
- 25.Christine KS, MacFarlane AW, IV, Yang K, Stanley RJ. Cyclobutylpyrimidine dimer base flipping by DNA photolyase. Journal of Biological Chemistry. 2002;277(41):38339–38344. doi: 10.1074/jbc.M206531200. [DOI] [PubMed] [Google Scholar]
- 26.Yang K, Stanley RJ. Differential distortion of substrate occurs when it binds to DNA photolyase: a 2-aminopurine study. Biochemistry. 2006;45(37):11239–11245. doi: 10.1021/bi060408u. [DOI] [PubMed] [Google Scholar]
- 27.Sancar GB, Jorns MS, Payne G. Action mechanism of Escherichia coli DNA photolyase. III. Photolysis of the enzyme-substrate complex and the absolute action spectrum. Journal of Biological Chemistry. 1987;262(1):492–498. [PubMed] [Google Scholar]
- 28.Jorns MS, Baldwin ET, Sancar GB, Sancar A. Action mechanism of Escherichia coli DNA photolyase. II. Role of the chromophores in catalysis. Journal of Biological Chemistry. 1987;262(1):486–491. [PubMed] [Google Scholar]
- 29.Jorns MS, Wang B, Jordan SP. DNA repair catalyzed by Escherichia coli DNA photolyase containing only reduced flavin: elimination of the enzyme’s second chromophore by reduction with sodium borohydride. Biochemistry. 1987;26(21):6810–6816. doi: 10.1021/bi00395a034. [DOI] [PubMed] [Google Scholar]
- 30.Kim S-T, Heelis PF, Okamura T, Hirata Y, Mataga N, Sancar A. Determination of rates and yields of interchromophore (folate→flavin) energy transfer and intermolecular (flavin→DNA) electron transfer in Escherichia coli photolyase by time-resolved fluorescence and absorption spectroscopy. Biochemistry. 1991;30(47):11262–11270. doi: 10.1021/bi00111a011. [DOI] [PubMed] [Google Scholar]
- 31.Okamura T, Sancar A, Heelis PF, Begley TP, Hirata Y, Mataga N. Picosecond laser photolysis studies on the photorepair of pyrimidine dimers by DNA photolyase. 1. Laser photolysis of photolyase-2-deoxyuridine dinucleotide photodimer complex. Journal of the American Chemical Society. 1991;113(8):3143–3145. [Google Scholar]
- 32.Langenbacher T, Zhao X, Bieser G, Heelis PF, Sancar A, Michel-Beyerle ME. Substrate and temperature dependence of DNA photolyase repair activity examined with ultrafast spectroscopy. Journal of the American Chemical Society. 1997;119(43):10532–10536. [Google Scholar]
- 33.Kao Y-T, Saxena C, Wang L, Sancar A, Zhong D. Direct observation of thymine dimer repair in DNA by photolyase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(45):16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacFarlane AW, IV, Stanley RJ. Evidence of powerful substrate electric fields in DNA photolyase: implications for thymidine dimer repair. Biochemistry. 2001;40(50):15203–15214. doi: 10.1021/bi0114224. [DOI] [PubMed] [Google Scholar]
- 35.MacFarlane AW, IV, Stanley RJ. Cis-Syn thymidine dimer repair by DNA photolyase in real time. Biochemistry. 2003;42(28):8558–8568. doi: 10.1021/bi034015w. [DOI] [PubMed] [Google Scholar]
- 36.Kim ST, Sancar A, Essenmacher C, Babcock GT. Evidence from photoinduced EPR for a radical intermediate during photolysis of cyclobutane thymine dimer by DNA photolyase. Journal of the American Chemical Society. 1992;114(11):4442–4443. [Google Scholar]
- 37.Rustandi RR, Jorns MS. Photoinduced spin-polarized radical pair formation in a DNA photolyase·substrate complex at low temperature. Biochemistry. 1995;34(7):2284–2288. doi: 10.1021/bi00007a024. [DOI] [PubMed] [Google Scholar]
- 38.Li YF, Sancar A. Active site of Escherichia coli DNA photolyase: mutations at Trp277 alter the selectivity of the enzyme without affecting the quantum yield of photorepair. Biochemistry. 1990;29(24):5698–5706. doi: 10.1021/bi00476a009. [DOI] [PubMed] [Google Scholar]
- 39.Kim S-T, Li YF, Sancar A. The third chromophore of DNA photolyase: Trp-277 of Escherichia coli DNA photolyase repairs thymine dimers by direct electron transfer. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(3):900–904. doi: 10.1073/pnas.89.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park H-W, Kim S-T, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli . Science. 1995;268(5219):1866–1894. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 41.Mees A, Klar T, Gnau P, et al. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004;306(5702):1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 42.Komori H, Masui R, Kuramitsu S, et al. Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13560–13565. doi: 10.1073/pnas.241371398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan RS, Tatsch CE, Gushard RH, McAdon J, Warme PK. Chains of alternating sulfur and pi-bonded atoms in eight small proteins. International Journal of Peptide and Protein Research. 1978;11(3):209–217. doi: 10.1111/j.1399-3011.1978.tb02841.x. [DOI] [PubMed] [Google Scholar]
- 44.Morgan RS, McAdon JM. Predictor for sulfur-aromatic interactions in globular proteins. International Journal of Peptide and Protein Research. 1980;15(2):177–180. doi: 10.1111/j.1399-3011.1980.tb02566.x. [DOI] [PubMed] [Google Scholar]
- 45.Bodner BL, Jackman LM, Morgan RS. NMR study of 1:1 complexes between divalent sulfur and aromatic compounds: a model for interactions in globular proteins. Biochemical and Biophysical Research Communications. 1980;94(3):807–813. doi: 10.1016/0006-291x(80)91306-6. [DOI] [PubMed] [Google Scholar]
- 46.Némethy G, Scheraga HA. Strong interaction between disulfide derivatives and aromatic groups in peptides and proteins. Biochemical and Biophysical Research Communications. 1981;98(2):482–487. doi: 10.1016/0006-291x(81)90865-2. [DOI] [PubMed] [Google Scholar]
- 47.Reid KSC, Lindley PF, Thornton JM. Sulphur-aromatic interactions in proteins. FEBS Letters. 1985;190(2):209–213. [Google Scholar]
- 48.Lebl M, Sugg EE, Hruby VJ. Proton n.m.r. spectroscopic evidence for sulfur-aromatic interactions in peptides. International Journal of Peptide and Protein Research. 1987;29(1):40–45. doi: 10.1111/j.1399-3011.1987.tb02227.x. [DOI] [PubMed] [Google Scholar]
- 49.Viguera AR, Serrano L. Side-chain interactions between sulfur-containing amino acids and phenylalanine in α-helices. Biochemistry. 1995;34(27):8771–8779. doi: 10.1021/bi00027a028. [DOI] [PubMed] [Google Scholar]
- 50.Spencer DS, Stites WE. The M32L substitution of staphylococcal nuclease: disagreement between theoretical prediction and experimental protein stability. Journal of Molecular Biology. 1996;257(3):497–499. doi: 10.1006/jmbi.1996.0180. [DOI] [PubMed] [Google Scholar]
- 51.Zauhar RJ, Colbert CL, Morgan RS, Welsh WJ. Evidence for a strong sulfur-aromatic interaction derived from crystallographic data. Biopolymers. 2000;53(3):233–248. doi: 10.1002/(SICI)1097-0282(200003)53:3<233::AID-BIP3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 52.Druhan LJ, Swenson RP. Role of methionine 56 in the control of the oxidation-reduction potentials of the Clostridium beijerinckii flavodoxin: effects of substitutions by aliphatic amino acids and evidence for a role of sulfur-flavin interactions. Biochemistry. 1998;37(27):9668–9678. doi: 10.1021/bi980770b. [DOI] [PubMed] [Google Scholar]
- 53.Prytkova TR, Beratan DN, Skourtis SS. Photoselected electron transfer pathways in DNA photolyase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(3):802–807. doi: 10.1073/pnas.0605319104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Acocella A, Jones GA, Zerbetto F. What is adenine doing in photolyase? Journal of Physical Chemistry B. 2010;114(11):4101–4106. doi: 10.1021/jp101093z. [DOI] [PubMed] [Google Scholar]
- 55.Miyazawa Y, Nishioka H, Yura K, Yamato T. Discrimination of class i cyclobutane pyrimidine dimer photolyase from blue light photoreceptors by single methionine residue. Biophysical Journal. 2008;94(6):2194–2203. doi: 10.1529/biophysj.107.119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin C, Todo T. The cryptochromes. Genome Biology. 2005;6(5, article 220) doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liedvogel M, Maeda K, Henbest K, et al. Chemical magnetoreception: bird cryptochrome 1a is excited by blue light and forms long-lived radical-pairs. PLoS One. 2007;2(10) doi: 10.1371/journal.pone.0001106. Article ID e1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gegear RJ, Casselman A, Waddell S, Reppert SM. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature. 2008;454(7207):1014–1018. doi: 10.1038/nature07183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu H, Sauman I, Yuan Q, et al. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biology. 2008;6(1):p. e4. doi: 10.1371/journal.pbio.0060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodgers CT, Hore PJ. Chemical magnetoreception in birds: the radical pair mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(2):353–360. doi: 10.1073/pnas.0711968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biskup T, Schleicher E, Okafuji A, et al. Direct observation of a photoinduced radical pair in a cryptochrome blue-light photoreceptor. Angewandte Chemie. International Edition. 2009;48(2):404–407. doi: 10.1002/anie.200803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hitomi K, Nakamura H, Kim S-T, et al. Role of two histidines in the (6-4) photolyase reaction. Journal of Biological Chemistry. 2001;276(13):10103–10109. doi: 10.1074/jbc.M008828200. [DOI] [PubMed] [Google Scholar]
- 63.Jorns MS, Sancar GB, Sancar A. Identification of a neutral flavin radical and characterization of a second chromophore in Escherichia coli DNA photolyase. Biochemistry. 1984;23(12):2673–2679. doi: 10.1021/bi00307a021. [DOI] [PubMed] [Google Scholar]
- 64.Smith WW, Burnett RM, Darling GD, Ludwig ML. Structure of the semiquinone form of flavodoxin from Clostridium MP. Extension of 1.8 Å resolution and some comparisons with the oxidized state. Journal of Molecular Biology. 1977;117(1):195–225. doi: 10.1016/0022-2836(77)90031-6. [DOI] [PubMed] [Google Scholar]
- 65.Ludwig ML, Pattridge KA, Metzger AL, et al. Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes. Biochemistry. 1997;36(6):1259–1280. doi: 10.1021/bi962180o. [DOI] [PubMed] [Google Scholar]
- 66.Xu L, Mu W, Ding Y, et al. Active site of Escherichia coli DNA photolyase: Asn378 is crucial both for stabilizing the neutral flavin radical cofactor and for DNA repair. Biochemistry. 2008;47(33):8736–8743. doi: 10.1021/bi800391j. [DOI] [PubMed] [Google Scholar]
- 67.Heelis PF, Hartman RF, Rose SD. Photoenzymic repair of UV-damaged DNA: a chemist’s perspective. Chemical Society Reviews. 1995;24(4):289–297. [Google Scholar]
- 68.Goosen N, Moolenaar GF. Repair of UV damage in bacteria. DNA Repair. 2008;7(3):353–379. doi: 10.1016/j.dnarep.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 69.Berndt A, Kottke T, Breitkreuz H, et al. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. Journal of Biological Chemistry. 2007;282(17):13011–13021. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 70.Balland V, Byrdin M, Eker APM, Ahmad M, Brettel K. What makes the difference between a cryptochrome and DNA photolyase? A spectroelectrochemical comparison of the flavin redox transitions. Journal of the American Chemical Society. 2009;131(2):426–427. doi: 10.1021/ja806540j. [DOI] [PubMed] [Google Scholar]
- 71.Öztürk N, Song S-H, Selby CP, Sancar A. Animal type 1 cryptochromes: analysis of the redox state of the flavin cofactor by site-directed mutagenesis. Journal of Biological Chemistry. 2008;283(6):3256–3263. doi: 10.1074/jbc.M708612200. [DOI] [PubMed] [Google Scholar]
- 72.Okafuji A, Biskup T, Hitomi K, et al. Light-induced activation of class II cyclobutane pyrimidine dimer photolyases. DNA Repair. 2010;9(5):495–505. doi: 10.1016/j.dnarep.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 73.Pieper U, Eswar N, Braberg H, et al. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Research. 2004;32:D217–D222. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Damiani MJ, Yalloway GN, Lu J, McLeod NR, O'Neill MA. Kinetic stability of the flavin semiquinone in photolyase and cryptochrome-DASH. Biochemistry. 2009;48(48):11399–11411. doi: 10.1021/bi901371s. [DOI] [PubMed] [Google Scholar]
- 75.Song S-H, Öztürk N, Denaro TR, et al. Formation and function of flavin anion radical in cryptochrome 1 blue-light photoreceptor of monarch butterfly. Journal of Biological Chemistry. 2007;282(24):17608–17612. doi: 10.1074/jbc.M702874200. [DOI] [PubMed] [Google Scholar]
- 76.Hoang N, Schleicher E, Kacprzak S, et al. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biology. 2008;6(8) doi: 10.1371/journal.pbio.0060160. Article ID e160. [DOI] [PMC free article] [PubMed] [Google Scholar]