Abstract
Identification and isolation of adult stem cells are still challenging for stem cell biologists. For example, no consensus exists yet regarding definitive markers for corneal epithelial stem cells, which have been identified to reside in the limbus for two decades. This study characterized the molecular signatures and biological pathways of limbal epithelial progenitors, the rapid adherent cells (RAC) isolated by adhesion on collagen IV, using human genome microarrays, real-time PCR and immunofluorescent staining. The microarrays produced highly reproducible data not only for all gene transcripts, but also for significantly changed genes, although the total 12 samples of 3 cell populations in 2 arrays were isolated from 4 separate experiments at different time period. The hierarchical clustering heatmap visually revealed that RAC progenitor population displayed distinguishably characteristic gene expression profile. With verification of 27 important genes by quantitative real-time PCR, the microarray data not only confirm the expression patterns of 15 known genes as stem cell associated markers representing limbal stem cell phenotype, but also identified many significantly regulated genes expressed by limbal progenitor cells. Transcription factor TCF4 and cell surface protein SPRRs were identified as potentially positive or negative markers, respectively, for corneal epithelial progenitor cells. Using GenMAPP and MAPPFinder, we have identified three patterns of biological pathway profiles, overexpressed, underexpressed and balanced, by RAC progenitors based on Gene Ontology categories. These genes and related pathways are interesting targets for further identification and isolation of limbal stem cells as well as other tissue specific adult stem cells.
Keywords: Adult stem cells, progenitors, epithelium, gene expression, microarray
INTRODUCTION
Identification and isolation of adult stem cells are still challenging for stem cell biologists. For example, no consensus exists yet regarding definitive markers for corneal epithelial stem cells, which have been recognized to reside in a ring around the peripheral cornea called the limbus for two decades. The evidence through previous studies leaves little doubt that corneal epithelial stem cells reside in the limbus, and these cells exhibit the full complement of well-defined keratinocyte stem cell properties (see review articles by (Boulton and Albon, 2004; Dua and Azuara-Blanco, 2000; Lavker et al., 2004; Lavker and Sun, 2000; Tseng, 1989). Limbal tissue biopsies and cultivated limbal epithelial cells have been successfully transplanted to patients with limbal deficiency to reconstruct the diseased and damaged corneas (Koizumi et al., 2001; Pellegrini et al., 1997; Schwab et al., 2000; Tsai et al., 2000; Tsubota, 1999). However, the stem cells are only a small subpopulation, estimated as less as <1% of these limbal basal cells (Budak et al., 2005; Pajoohesh-Ganji and Stepp, 2005), perhaps as few as 100 cells/limbus are true stem cells (Collinson et al., 2002; Stepp and Zieske, 2005). The limbal basal epithelium also consists of transient amplifying cells (TAC) that are an intermediate population of progenitor cells, and perhaps terminally differentiated cells (TDC). Although many stem cell markers have been proposed, no molecular markers have been recognized that definitively identify the limbal stem cells to date (Budak et al., 2005; Li et al., 2007; Pajoohesh-Ganji and Stepp, 2005; Schlotzer-Schrehardt and Kruse, 2005; Stepp and Zieske, 2005).
In recent years, efforts have been made to characterize the phenotype of limbal stem cells. We have extensively evaluated these molecular markers and characterized that the basal cells at limbal epithelium are small primitive cells expressing three patterns of molecular markers (Chen et al., 2004): (1) exclusively positive for p63, ABCG2 and integrin α9 by a subset of basal cells; (2) relatively higher expression of integrin β1, EGFR, K19 and enolase-α1 by most basal cells, and (3) lack of expression of E-cadherin, connexin 43, involucrin, K3 and K12. Very recently, we have observed that two neurotrophic factors, nerve growth factor (NGF) and glial cell-derived neurotrophic factor (GDNF), and their corresponding receptors TrkA and GFRα-1 were exclusively localized to a subpopulation of basal limbal epithelial cells (Qi et al., 2008a; Qi et al., 2008b). Furthermore, keratin 15 (Figueira et al., 2007), N-cadherin (Hayashi et al., 2007) and CCAAT enhancer binding protein δ (C/EBP-δ, Barbaro et al., 2007) have been proposed as markers to identify limbal stem cells. All these markers that are positively or negatively expressed by limbal basal epithelial cells may serve as stem cell associated markers, and collectively, they represent a unique phenotype that identifies putative corneal epithelial stem cells (Chen et al., 2004; Hayashi et al., 2007). The presence of p63 in limbal basal cells appears to represent a higher proliferative potential, the presence of ABCG2 may be a feature of limbal stem cells that protects them from damage by drugs and toxins, and the NGF and GDNF with their receptors may serve as critical survival factors for these stem cells. The higher expression of integrins α9 and β1 may indicate their strong adhesion to extracellular matrix for limbal resistance to shear forces. The absence of connexin 43 and E-cadherin expression may be an inherent feature of stem cells, while the lack of expression of K3, K12 and involucrin indicates their poorly differentiated status.
The evidence for limbal stem cell concept has been largely derived from comparisons of properties and phenotypes between cornea and limbus. However, the stem cells are only a very small subpopulation located in the basal layer of limbal epithelium that contains larger number of surrounding TAC and TDC. The phenotype and properties of limbal stem cells may be diluted, mixed, or hidden by other cell types when only comparing the cornea and limbus,. Further characterization and identification could be performed if purified populations of limbal basal cells or progenitor cells enriched in putative stem cells are available. However isolation of limbal stem cells has not been accomplished to date due to the lack of a truly unique marker or a definitive method to identify these stem cells. We have successfully isolated 5 clonogenic cell populations from limbal epithelia and/or their cultures, based on stem cell phenotype, using different cell surface markers and properties including cell-sizing (de Paiva et al., 2006b), adherence to extracellular matrix (Li et al., 2005), sorting for side population or for expression of ABCG2 (de Paiva et al., 2005) or connexin 43 (Chen et al., 2006) cell surface markers. These 5 clonogenic populations isolated by different methods represent corneal epithelial progenitor cells with some properties that are characteristic of adult stem cells: (1) poorly differentiated: they expressed higher levels of stem cell associated markers (ABCG2, p63, or integrin β1) and negative levels of differentiation associated markers (K3, K12, involucrin or connexin 43) at both protein and mRNA levels; (2) high proliferative potential: they showed greater clonal forming efficiency (CFE) and growth capacity in culture; and (3) self-renewing: they also contained slow-cycling BrdU label-retaining cells in culture, an intrinsic characteristic of stem cells.
Among the methods used for isolating progenitor populations, adhesion to collagen IV has been demonstrated to be successful for isolating 3 cell populations from whole limbal epithelial cells (Li et al., 2005). The rapid adherent cell (RAC) population enriched for certain putative stem cell properties may represent limbal epithelial progenitor cells, while the slow adherent cells (SAC) and non-adherent cells (NAC) may represent TAC and TDC cells, respectively. To further characterize limbal stem cells, we compared gene expression profiles in the stem cell-containing RAC progenitor population with TAC-like SAC and TDC-like NAC populations using Affymetrix whole human genome microarrays. This study reports a variety of interesting findings that provide new insight on the unique characteristics of human corneal epithelial progenitor cells.
MATERIALS AND METHODS
Corneal limbal tissues and limbal epithelial cell isolation
Fresh human corneoscleral tissues (less than 72 hours post mortem), from donors aged 19–67 years, were obtained from the Lions Eye Bank of Texas (LEBT, Houston, TX) and from the National Disease Research Interchange (NDRI, Philadelphia, PA). They were cut through the horizontal meridian, frozen and sectioned for immunostaining. Limbal epithelial cells were isolated from multiple fresh limbal tissues and pooled for use in adhesion experiments by a previously described method (Kim et al., 2004; Li et al., 2005).
Isolation of progenitor cells by adhesion of limbal epithelial cells to collagen IV
Human limbal epithelial cells isolated from fresh limbal tissues were used for isolation of their progenitor cells using a previous described method (Li et al., 2005). For isolation of progenitor cells that are enriched with putative corneal epithelial stem cells, the cells were allowed to attach to a collagen IV coated dish at 37°C for 20 minutes. The attached limbal epithelial cells in 20 minutes were designated as rapid adherent cells (RAC). The unattached cells within the first 20 minutes were then transferred to another collagen IV coated dish and allowed to adhere for an additional 100 minutes. The attached limbal epithelial cells during 20 to 120 minutes were designated as slow adherent cells (SAC). The remaining unattached cells after 120 minutes were collected as non-adherent cells (NAC). These three selected cell populations were subjected to total RNA extraction for microarray analysis and real-time PCR.
Affymetrix GeneChip® Human Genome U133 Plus 2.0 arrays
Total RNA was isolated from these three cell populations (RAC, SAC and NAC) using a Qiagen RNeasy® Micro kit according to manufacturer’s protocol (Qiagen, Valencia, CA), quantified by NanoDrop® ND-1000 Spectrophotometer (Nanadrop technologies, Wilmington, DE) and stored at −80°C. The quality of these RNA samples was further checked using Agilent Bioanalyer 2100 and was confirmed by gel electrophoresis. Affymetrix GeneChip® Human Genome (HG) U133 Plus 2.0 arrays (Affymetrix, Santa Clara, CA) was performed by the Microarray Core Facility at Baylor College of Medicine (Houston, TX) using Affymetrix Two-Cycle Target Labeling and Control Reagent Kit according to the manufacturer’s protocols. The array was hybridized overnight at 45°C and stained with a strepavidin, R-phycoerythrin conjugate stain. Signal amplification was done using biotinylated antistreptavidin. The stained array was scanned on the Affymetrix GeneChip® Scanner 3000 (Affymetrix). The images were analyzed and quality control metrics recorded using Affymetrix GCOS software version 1.4.
Microarray data analysis
The human genome microarray data were analyzed using R software (R Development Core Team, 2008) from Bioconductor (http://www.bioconductor.org). Principal component analysis was performed using all the data to visualize the relationship among the individual samples. A linear model was fitted for each probe set using the R package limma (Smyth et al., 2003), with the cell types (RAC, SAC and NAC) as the predictors. Fold changes in gene expression were calculated by dividing the mean intensity signal from RAC samples by the mean intensity signal from the NAC or SAC samples. Hierarchical clustering of the selected probe sets was performed using Pearson correlation for distance matrix and the Ward’s linkage.
Gene Ontology (GO) and pathway profile analysis
GenMAPP (Gene Map Annotator and Pathway Profiler) and MAPPFinder (www.genmapp.org) were used to view whole human genome microarray data on biological pathways and Identify global trends in the data, giving a comprehensive picture of the gene expression changes associated with a particular GO term (Dahlquist, 2004; Doniger et al., 2003). The Ingenuity Pathway Analysis (IPA) suite (Ingenuity Systems, Inc. Redwood City, CA) was used for pathway analysis on selected probe sets that were regulated in the RAC progenitor population.
Reverse transcription (RT) and quantitative real-time PCR
As previously described (Luo et al., 2004; Yoon et al., 2007), the first strand cDNA was synthesized by RT from 1 μg of total RNA using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare Biosciences, Pittsburgh, PA), and the real-time PCR was performed in the Mx3005P™ system (Stratagene) with a 20μl reaction volume containing 5μl of cDNA, 1μl of TaqMan® Gene Expression Assay primers and probe (see 28 gene list in Table S1 in the section of Supplemental Data) and 10μl TaqMan® Gene Expression Master Mix (Applied Biosystems). The thermocycler parameters were 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. A non-template control was included to evaluate DNA contamination. The results were analyzed by the comparative threshold cycle (CT) method and normalized by a housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (de Paiva et al., 2006a; Yoon et al., 2007).
Immunofluorescent staining
Immunofluorescent staining for TCF4 and SPRR proteins on human corneal frozen sections was performed with goat anti-human TCF4 (1:100, Senta Cruz Biotechnology, Santa Cruz, CA), or rabbit antibodies against human SPRR1a (I:100, Abcam, Cambridge, MA) and SPRR2 (1:100, Alexis Biochemicals, San Diego, CA) using the methods previously described (Chen et al., 2004; de Paiva et al., 2005). Sections were examined and photographed with an epifluorescent microscope equipped with a digital camera (Eclipse E400 with a DS-Fi1, Nikon, Japan).
Statistical analysis
The Student’s t-test or Analysis of Variance (ANOVA) with Tukey’s post hoc testing was used for statistical comparisons. P ≤ 0.05 was considered statistically significant. All validation tests were performed using the GraphPad Prism 4.0 software (GraphPad Prism Software, Inc., San Diego, CA).
RESULTS
RNA quality and Affymetrix array data reproducibility
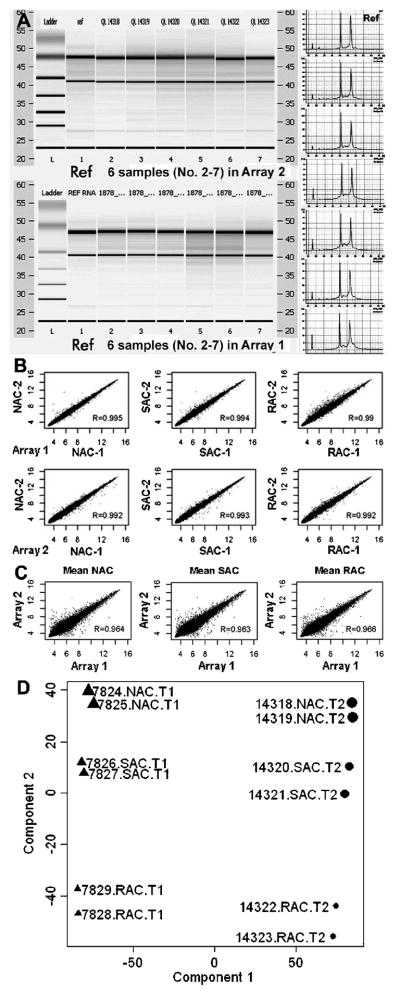
The quality of RNA samples was evaluated using the Nanodrop ND-1000 spectrophotometer and Agilent Bioanalyer 2100. All RNA samples showed 260/280 nm absorption ratios at 2.0 or above. As shown in Figure 1A, the profiles of total RNA samples isolated from limbal epithelial tissues were similar to the reference RNA, as analyzed by Agilent Biochip and gel electrophoresis.
Figure 1.
Quality of RNA samples and data reproducibility of Affymetrix microarrays. A. The total RNA samples were analyzed by Agilent Bioanalyer 2100 with Biochips and gel electrophoresis using a referent RNA as a quality control; B. The scatter plots showed that the expression values of all genes transcripts from each cell population (NAC, SAC or RAC) were highly correlated between the duplicate chips with high correlation coefficients (R>0.99); C. The scatter plots showed the data reproducibility between the two separate arrays (R>0.96); D. Principal components analysis (PCA) on the entire data sets (the 12 samples’ ID showed) from 2 arrays showed well separated gene expression patterns of 3 isolated cell populations, RAC, SAC, and NAC.
Affymetrix GeneChip® Human Genome U133 Plus 2.0 Arrays were performed twice from separate adhesion experiments. Each array analyzed three cell populations (RAC, SAC, and NAC) in duplicate, freshly isolated from limbal epithelial tissues in 2 separate experiments, using the Affymetrix GeneChip® microarray two-cycle protocol with 100ng of total RNA for each chip. Each array contained 6 chips for RAC, SAC, and NAC populations run in duplicate and a total of 12 chips were used for two microarrays. The data from 2 duplicate chips of each array as well as between 2 microarrays performed at a different time were highly reproducible even though each sample consisted of pooled cells from multiple limbal tissues obtained from different donors. The HG-U133 Plus 2.0 Array was comprised of more than 54,000 probe sets for more than 47,000 transcripts and variants, including 38,500 well-characterized human genes. Approximately 21,000 transcripts or 38% of the 54,675 probe sets on these arrays were present at a detectable level. The rates of present calls were very close in each pair of samples, as well as in the two arrays (Table S2 in Supplemental Data). The expression values of all genes from each cell population (NAC, SAC or RAC) were highly correlated between the duplicate chips with high correlation coefficients (Figure 1B, R>0.99) in each arrays, as well as between the two separate arrays (Figure 1C, R>0.96). To further identify the relationship of these GeneChip samples to each other, principal components analysis (PCA) was performed on the entire data sets. PCA was carried out on log-transformed data, using mean centering and scaling. As seen in Figure 1D, the PCA analysis has well divided these samples into 3 groups as RAC, SAC, and NAC, indicating the distinguishable expression profiles of these 3 cell populations.
Hierarchical clustering analysis of significantly changed genes in the RAC population
As reported in our previously published study (Li et al., 2005), the RAC, SAC and NAC three populations isolated from whole limbal epithelial cells may represent corneal epithelial progenitor cells, TAC and TDC, respectively. To further confirm these phenotypes, we compared the RAC gene expression profiles with NAC and SAC using Affymetrix whole genome microarrays. A hierarchical clustering was analyzed on the dataset consisting of the 776 significantly changed genes including 499 up- and 277 down-regulated genes, which were selected from data on 12 chips in 2 arrays through a filter criteria of at least 2-fold changes with P≤0.05. As shown in Figure 2, the hierarchical clustering heatmap for genes further displayed great reproducibility of the gene expression from these 3 cell populations between duplicate chips of each array, as well as between 2 arrays with RNA samples from 4 separate experiments performed at different times. The heatmap also showed a clear separation of the RAC group from NAC and SAC, indicating the RAC population has its distinguishably characteristic gene expression profile from NAC and SAC. Interestingly, the mRNA levels of most upregulated genes in RAC vs. NAC were also higher than SAC, showing the mRNA levels from the lowest levels (Blue) in NAC, to middle (yellow) in SAC and the highest levels (red) in RAC. In contrast, the expression levels of most down-regulated genes in RAC vs. NAC were also lower than SAC, showing the reversed color pattern from red to blue. It suggests that these 3 populations do represent 3 distinct cell types with levels of differentiation ranging from stem cell-containing progenitors to TAC and TDC. Since our studies were performed on quiescent cells isolated from unwounded limbal tissues, we recognize that the SAC RNA profiles may be different from the TACs that appear after an injury or after corneal stem cells are grown in culture.
Figure 2.
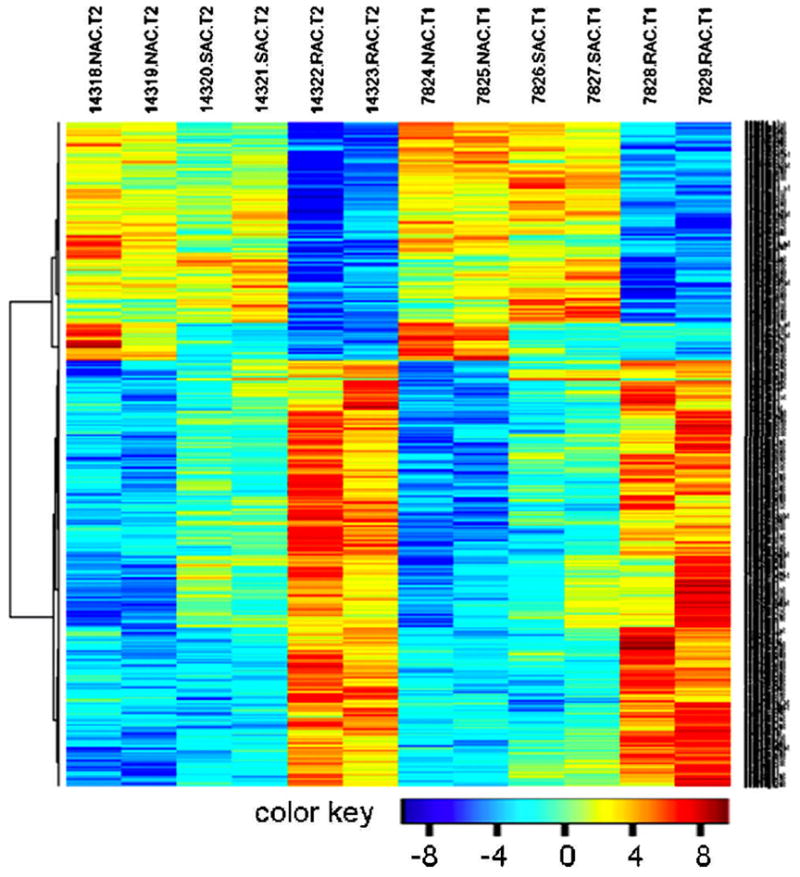
Heat map visualization of 776 significantly regulated probe sets. A hierarchical clustering was analyzed on the probe sets consisting of the 776 genes including 499 up- and 277 down-regulated gene transcripts, which were selected from 12 chips in 2 arrays (T1 and T2) through a filter criteria of at least 2-fold changes with P≤0.05 (F test). Columns: samples; Rows: genes; Color key indicates gene expression value, blue: Lowest, red: highest.
Analysis and validation of currently known genes that have been proposed as corneal epithelial stem cell associated markers in the RAC population
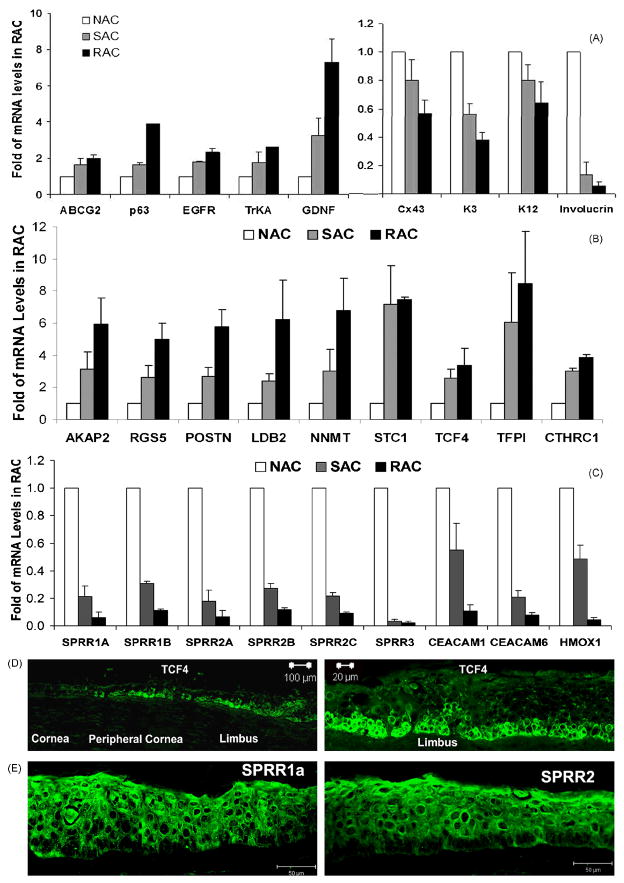
Based on the proposed stem cell makers in last 2 decades, limbal stem cell phenotype has been characterized with positive expression of stem cell associated markers including p63, ABCG2, integrin α9, integrin β1, EGFR, K15, enolase-α1, NGF, GDNF, TrkA, N-cadherin (Hayashi et al., 2007) and C/EBP-δ (Barbaro et al., 2007), but negative expression of differentiation markers such as E-cadherin, connexin 43, K12, K3 and involucrin (Barbaro et al., 2007; Chen et al., 2004; Hayashi et al., 2007; Pajoohesh-Ganji and Stepp, 2005; Qi et al., 2008b). In this study, we compared the gene expression profiles of RAC (representing progenitor cells) with NAC (representing TDC) and SAC (representing TAC). Interestingly, the microarray data reproducibly displayed a molecular profile of RAC that included a variety of previously proposed markers for putative stem cell phenotype. As shown in Table 1, all RAC chips of 2 arrays from 4 separate experiments expressed significantly higher levels of stem cell associated markers, including ABCG2, p63, integrin α9, integrin β1, EGFR, enolase α1, Keratin 15, TrkA, GDNF, N-cadherin and C/EBP-δ, but lower levels of differentiation associated markers, connexin 43, E-cadherin, K3, K12 and involucrin, than NAC (all with P values <0.05). A similar trend or pattern was observed when RAC and SAC were compared; although the differences of these genes between the two populations were not all significant (P values for RAC/SAC were not shown). Using RT and quantitative real-time PCR, the expression pattern of 5 stem cell associated genes and 4 differentiation genes were further verified in the 3 cell populations isolated from limbal epithelial tissues in separate adhesion experiments (Figure 3A). The patterns of these known stem cell associated markers expressed by RAC progenitors were consistent to previous reports, which supports our hypothesis that the limbal stem cell phenotype represented by a group of the markers is useful to identify the limbal stem cells despite the lack of a single definitive marker.
Table 1.
Gene expression levels of known limbal stem cell associated or differentiation markers in RAC population isolated from human limbal epithelium
| Limbal SC Markers | Gene Symbol | Gene Name Description | Fold Changes of mRNA |
||
|---|---|---|---|---|---|
| RAC/NAC | P | RAC/SAC | |||
| ABCG2 | ABCG2 | ATP-binding cassette, sub-family G, member 2 | 1.80 | 0.001 | 1.50 |
| p63 | TP73L | tumor protein p73-like | 2.25 | 0.000 | 1.64 |
| Integrin α9 | ITGA9 | integrin, alpha 9 | 1.97 | 0.000 | 1.62 |
| integrin β1 | ITGB1 | integrin, beta 1 (fibronectin receptor, CD29) | 1.50 | 0.036 | 1.37 |
| EGFR | EGFR | epidermal growth factor receptor | 2.48 | 0.007 | 1.49 |
| enolase α1 | ENO1 | enolase 1, (alpha) | 1.66 | 0.042 | 0.98 |
| Keratin 15 | KRT15 | keratin 15 | 2.08 | 0.000 | 1.95 |
| TrkA | NTRK1 | neurotrophic tyrosine kinase, receptor, type 1 | 3.85 | 0.000 | 2.03 |
| N-cadherin | CDH2 | cadherin 2, type 1, N-cadherin (neuronal) | 1.66 | 0.005 | 1.31 |
| C/EBP-δ | CEBPD | CCAAT/enhancer binding protein (C/EBP), delta | 1.776 | 0.012 | 1.531 |
| Connexin 43 | GJA1 | gap junction protein, alpha 1, 43kDa | 0.67 | 0.004 | 0.86 |
| E-cadherin | CDH2 | cadherin 1, type 1, E-cadherin (epithelial) | 0.61 | 0.001 | 0.79 |
| Keratin 12 | KRT12 | keratin 12 (Meesmann corneal dystrophy) | 0.42 | 0.041 | 0.71 |
| Keratin 3 | KRT3 | keratin 3 | 0.56 | 0.000 | 0.81 |
| Involucrin | IVL | involucrin | 0.17 | 0.006 | 0.64 |
The ratios of RAC/NAC and RAC/SAC represent the relative fold changes of mRNA levels in RAC versus NAC or SAC, respectively, by Affymetrix arrays; the P values are for RAC/NAC. The Affymetrix probe sets and gene accession numbers of these markers are listed in Table S3 in the Supplemental Data.
Figure 3.
Validation of 27 genes for stem cell phenotype. The expression patterns of known 5 stem cell associated and 4 differentiation markers (A), 9 highly up- (B) and 9 highly down- (C) regulated new genes were verified by RT and quantitative real-time PCR in the isolated RAC, SAC and NAC populations from limbal epithelial tissues obtained from separate adhesion experiments. The representative images of immunofluorescent staining on corneal limbal tissue frozen sections showing immunolocalozation of TCF4 that was positive only at basal cells of limbal and peripheral corneal epithelia (D) or SPRR1a and SPRR2 that were negative at limbal basal cells (E).
Analysis and validation of genes significantly up- or down-regulated in the RAC progenitor population
When the gene expression levels of RAC versus NAC and SAC were filtered with criteria that the fold changes of mRNA expression values of RAC/NAC were ≥2 or ≤0.5 fold with P value ≤ 0.05, 1362 or 1288 transcripts were significantly changed in these 2 arrays respectively. The mean expression values in the 3 cell populations, NAC, SAC and RAC, between 2 arrays from separate experiments were highly reproducible with significant correlation coefficients (R=0.937, 0.926, and 0.861, respectively; see Figure S1 in Supplemental Data).
Among the significantly changed genes in the RAC population, 814 genes, account for 59.8 or 63.2% respectively in 2 arrays, shared the same expression pattern, which included 551 up-regulated and 263 down-regulated transcripts. We further analyzed the top changed 42 genes in the RAC groups including 26 up- and 16 down-regulated genes that were expressed 6 fold up or down in the RAC compared to the NAC with P values ≤0.005 in the 2 arrays (Table 2). The expression pattern of 18 genes selected from the list were verified by RT and real-time PCR using RNA samples obtained from separate adhesion experiments. As shown in Figure 3B, the top 9 up-regulated genes in the RAC population detected by Affymetrix arrays were verified, including A kinase (PRKA) anchor protein 2 (AKAP2), regulator of G-protein signaling 5 (RGS5), periostin (POSTN), LIM domain binding 2 (LDB2), nicotinamide N-methyltransferase (NNMT), stanniocalcin 1 (STC1), transcription factor 4 (TCF4), tissue factor pathway inhibitor (TFPI), collagen triple helix repeat containing 1 (CTHRC1), all of which showed consistent expression patterns in the RAC, SAC and NAC populations. Among these upregulated molecules, the transcription factor TCF4 was further observed to be exclusively localized in the certain basal cells of limbal epithelium, as evaluated by immunofluorescent staining (Figure 3D). Interestingly, the most down-regulated genes in RAC progenitor cells were small proline-rich protein (SPRR) and carcinoembryonic antigen-related cell adhesion molecule (CEACAM) families. We further verified the 9 top down-regulated genes including SPRR genes (SPRR1A, SPRR1B, SPRR2A, SPRR2B, SPRR2C and SPRR3), CEACAM1, CEACAM6, and heme oxygenase (decycling) 1 (NMOX1), all of which displayed the same dramatic down-regulation pattern in the RAC population when compared with SAC and NAC groups (Figure 3C). Immunofluorescent staining using rabbit antibodies against human SPRR1a and SPRR2 showed both SPRR1a and SPRR2 proteins were positively localized in all layers of corneal epithelium and most limbal epithelial cells, but they were absent in clusters of basal epithelial cells in the human limbus (Figure 3E). This confirmation of the SPRR expression pattern at the protein level suggests that one or more members of SPRR family could be potential negative markers for limbal stem or progenitor cells. Further studies are necessary to test this hypothesis.
Table 2.
Top up- and down-regulated genes (≥6 fold, P≤0.005) in RAC limbal epithelial progenitors
| Gene Symbol | Gene Name Description | Fold Changes of mRNA |
||
|---|---|---|---|---|
| RAC/NAC | P | RAC/SAC | ||
| AKAP2 | A kinase (PRKA) anchor protein 2 | 11.46 | 0.0000 | 1.77 |
| C10orf10 | chromosome 10 open reading frame 10 | 10.65 | 0.0000 | 2.42 |
| RGS5 | regulator of G-protein signaling 5 | 10.29 | 0.0017 | 1.84 |
| DARC | Duffy blood group, chemokine receptor | 10.24 | 0.0000 | 2.69 |
| GIMAP6 | GTPase, IMAP family member 6 | 9.71 | 0.0000 | 2.46 |
| APOLD1 | apolipoprotein L domain containing 1 | 9.08 | 0.0000 | 2.45 |
| PDK4 | pyruvate dehydrogenase kinase, isozyme 4 | 9.01 | 0.0000 | 2.17 |
| POSTN | periostin, osteoblast specific factor | 8.63 | 0.0000 | 2.48 |
| LDB2 | LIM domain binding 2 | 8.59 | 0.0001 | 2.57 |
| NNMT | nicotinamide N-methyltransferase | 7.95 | 0.0000 | 1.71 |
| STC1 | stanniocalcin 1 | 7.74 | 0.0000 | 1.36 |
| RHOJ | ras homolog gene family, member J | 7.48 | 0.0000 | 2.54 |
| EMCN | endomucin | 7.36 | 0.0000 | 2.08 |
| CSF2RB | colony stimulating factor 2 receptor, beta | 7.07 | 0.0000 | 2.16 |
| AQP1 | aquaporin 1 (Colton blood group) | 7.05 | 0.0000 | 1.95 |
| COL15A1 | collagen, type XV, alpha 1 | 6.87 | 0.0000 | 2.66 |
| MSRB3 | methionine sulfoxide reductase B3 | 6.87 | 0.0001 | 2.24 |
| TSPAN7 | tetraspanin 7 | 6.72 | 0.0000 | 2.27 |
| SLC2A3 | solute carrier family 2 member 3 | 6.62 | 0.0000 | 1.92 |
| TCF4 | transcription factor 4 | 6.62 | 0.0021 | 2.62 |
| TFPI | tissue factor pathway inhibitor | 6.52 | 0.0000 | 2.02 |
| A2M | alpha-2-macroglobulin | 6.38 | 0.0000 | 2.41 |
| ITM2A | integral membrane protein 2A | 6.35 | 0.0000 | 2.35 |
| CTHRC1 | collagen triple helix repeat containing 1 | 6.34 | 0.0001 | 2.04 |
| C8orf4 | chromosome 8 open reading frame 4 | 6.31 | 0.0000 | 2.40 |
| GIMAP8 | GTPase, IMAP family member 8 | 6.30 | 0.0000 | 2.10 |
| KRTAP3-2 | keratin associated protein 3-2 carcinoembryonic antigen-related cell | 0.16 | 0.0014 | 0.29 |
| CEACAM1 | adhesion molecule 1 | 0.16 | 0.0015 | 0.33 |
| C15orf48 | chromosome 15 open reading frame 48 | 0.14 | 0.0000 | 0.54 |
| HMOX1 | heme oxygenase (decycling) 1 | 0.14 | 0.0000 | 0.17 |
| RNASE7 | ribonuclease, RNase A family, 7 | 0.13 | 0.0000 | 0.13 |
| C6orf128 | chromosome 6 open reading frame 128 | 0.12 | 0.0000 | 0.16 |
| KRT24 | keratin 24 | 0.12 | 0.0017 | 0.51 |
| SLC6A14 | solute carrier family 6 member 14 carcinoembryonic antigen-related cell | 0.11 | 0.0000 | 0.72 |
| CEACAM7 | adhesion molecule 7 | 0.11 | 0.0008 | 0.95 |
| IL1RL1 | interleukin 1 receptor-like 1 | 0.11 | 0.0000 | 0.07 |
| SPRR2A | small proline-rich protein 2B | 0.11 | 0.0015 | 0.51 |
| SPRR1B | small proline-rich protein 1B (cornifin) | 0.10 | 0.0015 | 0.43 |
| SPRR1A | small proline-rich protein 1A | 0.10 | 0.0015 | 0.39 |
| CEACAM6 | carcinoembryonic antigen-related cell adhesion molecule 6 carcinoembryonic antigen-related cell | 0.09 | 0.0000 | 0.32 |
| CEACAM5 | adhesion molecule 5 | 0.09 | 0.0024 | 0.72 |
| SPRR3 | small proline-rich protein 3 | 0.05 | 0.0041 | 0.75 |
The ratios of RAC/NAC and RAC/SAC represent the relative fold changes of mRNA levels in RAC versus NAC or SAC, respectively, by Affymetrix arrays; the P values are for RAC/NAC. The Affymetrix probe sets and accession numbers of these genes are listed in Table S4 in the Supplemental Data.
Analysis of Gene Ontology and pathway profiles associated with RAC progenitor population
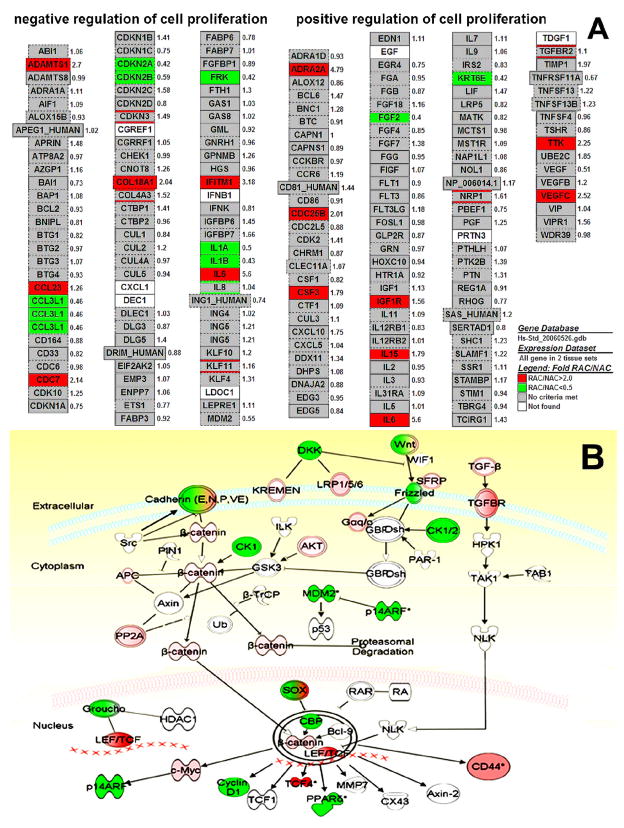
Beyond the identification of a series of individual genes whose expression was changed in RAC progenitor cells, we further determined groups of functionally related genes, based on the gene ontology system by GenMAPP and MAPPFinder software. Considering that these processes or functions may involve a number of genes, we did use less restrictive gene selection criteria that admitted any individual gene as long as its change in expression was significant (P≤0.05) and at least twofold. The Z-scores of the corresponding set of genes were calculated to identify the significant GO terms (Z-score ≥ 2.0, corresponding to P ≤ 0.05). Table 3 shows the top enriched GO categories with Z scores ≥ 3.0 and permutation P ≤ 0.005 according to the lists of ontology types, biological processes (P), molecular functions (F) and cellular components (C). Table 3 presents GO profiles enriched with up-regulated genes in the RAC progenitor population in comparison with NAC, which identified overexpressed biological processes related to regulation of cell adhesion, migration and growth, phosphate transport, proteoglycan biosynthesis, sulfur compound biosynthesis, calcium ion homeostasis, and G-protein coupled receptor protein signaling pathway, as well as transmembrane receptor tyrosine kinase activity, extracellular matrix and basement membrane. Table 4 presents those associated with down-regulated genes in RAC in comparison with NAC, which identified under-expressed biological processes related to hormone secretion, mRNA 3′-end processing, regulation of cyclin dependent protein kinase activity, chromatin assembly, interleukin-1, EGF and FGF receptor binding and regulator activity, oxygen binding, intermediate filament and apical junction complex. Table 5 presents balanced GO profiles enriched by both up- and down-regulated genes in RAC, including angiogenesis, vasculature development, cell-cell adhesion, endopeptidase, protease and enzyme inhibitor activity, chemokine and cytokine activity, and cell junction. The above 3 patterns of enriched GO profiles appear to represent main characteristics of limbal RAC progenitor population. The GenMAPP and MAPPFinder software generates maps showing the fold changes of expression values by genes associated with these characterized biological processes. For example, Figure 4A showed a gene map of cell proliferation profile by RAC progenitors. This map displayed numerous genes up- or down-regulated (fold changes of mRNA levels in RAC/NAC) in the RAC population with their potential role in positive or negative regulation pathways of cell proliferation. Using the Ingenuity Pathway Analysis suite, we further analyzed more signaling pathways that associated with the significantly regulated (up or down) genes (fold change more than 1.5 fold and P value ≤0.05) in RAC population. For example, Figure 4B showed a well characterized stem cell pathway, Wnt/β-catenin pathway, where numerous genes in RAC progenitors participated, including up-regulated genes like TCF4, TGFβ, TGFβ receptors, c-myc, CD44, etc, and down-regulated genes like E-cadherin, DKK, MDM2, p14, etc. The pathway analysis would explore the potential role of the significantly regulated genes in RAC progenitors.
Table 3.
Top over-expressed Gene Ontology biological pathways associated mainly with up-regulated genes in RAC limbal epithelial progenitors
| GO ID | Gene Ontology (GO) Description | GO Type | Number of genes |
Z Score | |||||
|---|---|---|---|---|---|---|---|---|---|
| Up | Measured | in GO | % Up | %Present | |||||
| 1 | 19884 | antigen presentation\, exogenous antigen | P | 6 | 14 | 21 | 42.9 | 66.7 | 7.535 |
| 2 | 6817 | phosphate transport | P | 15 | 81 | 89 | 18.5 | 91.0 | 6.819 |
| 3 | 6029 | proteoglycan metabolism | P | 6 | 21 | 25 | 28.6 | 84.0 | 5.846 |
| 4 | 6024 | glycosaminoglycan biosynthesis | P | 5 | 16 | 18 | 31.2 | 88.9 | 5.656 |
| 5 | 16477 | cell migration | P | 12 | 76 | 80 | 15.8 | 95.0 | 5.371 |
| 6 | 15698 | inorganic anion transport | P | 17 | 134 | 146 | 12.7 | 91.8 | 5.283 |
| 7 | 30166 | proteoglycan biosynthesis | P | 4 | 13 | 16 | 30.8 | 81.2 | 5.008 |
| 8 | 9887 | organ morphogenesis | P | 18 | 167 | 179 | 10.8 | 93.3 | 4.623 |
| 9 | 30334 | regulation of cell migration | P | 4 | 16 | 17 | 25.0 | 94.1 | 4.364 |
| 10 | 44272 | sulfur compound biosynthesis | P | 5 | 24 | 29 | 20.8 | 82.8 | 4.29 |
| 11 | 8277 | G-protein coupled receptor protein signaling pathway | P | 5 | 25 | 26 | 20.0 | 96.2 | 4.163 |
| 12 | 30155 | regulation of cell adhesion | P | 5 | 31 | 31 | 16.1 | 100.0 | 3.522 |
| 13 | 6874 | calcium ion homeostasis | P | 8 | 65 | 67 | 12.3 | 97.0 | 3.511 |
| 14 | 6790 | sulfur metabolism | P | 7 | 53 | 59 | 13.2 | 89.8 | 3.509 |
| 15 | 42060 | wound healing | P | 9 | 80 | 87 | 11.2 | 92.0 | 3.408 |
| 16 | 1558 | regulation of cell growth | P | 10 | 103 | 108 | 9.7 | 95.4 | 3.059 |
| 17 | 5201 | extracellular matrix structural constituent | F | 21 | 79 | 85 | 26.6 | 92.9 | 10.448 |
| 18 | 45012 | MHC class II receptor activity | F | 6 | 14 | 21 | 42.9 | 66.7 | 7.535 |
| 19 | 5520 | insulin-like growth factor binding | F | 5 | 18 | 20 | 27.8 | 90.0 | 5.238 |
| 20 | 30247 | polysaccharide binding | F | 12 | 84 | 85 | 14.3 | 98.8 | 4.934 |
| 21 | 5021 | vascular endothelial growth factor receptor activity | F | 4 | 14 | 15 | 28.6 | 93.3 | 4.772 |
| 22 | 5539 | glycosaminoglycan binding | F | 11 | 81 | 82 | 13.6 | 98.8 | 4.516 |
| 23 | 8201 | heparin binding | F | 9 | 61 | 62 | 14.8 | 98.4 | 4.391 |
| 24 | 4714 | transmembrane receptor tyrosine kinase activity | F | 9 | 62 | 67 | 14.5 | 92.5 | 4.33 |
| 25 | 19838 | growth factor binding | F | 7 | 50 | 53 | 14.0 | 94.3 | 3.697 |
| 26 | 31012 | extracellular matrix | C | 45 | 255 | 268 | 17.6 | 95.1 | 11.453 |
| 27 | 5581 | collagen | C | 11 | 32 | 32 | 34.4 | 100.0 | 8.918 |
| 28 | 5604 | basement membrane | C | 13 | 44 | 45 | 29.5 | 97.8 | 8.804 |
| 29 | 5583 | fibrillar collagen | C | 3 | 9 | 9 | 33.3 | 100.0 | 4.564 |
GO types: biological processes (P), molecular functions (F) and cellular components (C). The Z-scores: calculated to identify the significant GO terms (Z-score ≥ 2.0, corresponding to P ≤ 0.05)
Table 4.
Top under-expressed Gene Ontology biological pathways associated mainly with down-regulated genes in RAC limbal epithelial progenitors
| GO ID | Gene Ontology (GO) Description | GO Type | Number of genes |
Z Score | |||||
|---|---|---|---|---|---|---|---|---|---|
| Down | Measured | in GO | % Down | %Present | |||||
| 1 | 30073 | insulin secretion | P | 3 | 7 | 7 | 42.9 | 100.0 | 7.752 |
| 2 | 30072 | peptide hormone secretion | P | 3 | 8 | 8 | 37.5 | 100.0 | 7.201 |
| 3 | 31124 | mRNA 3′-end processing | P | 3 | 9 | 9 | 33.3 | 100.0 | 6.742 |
| 4 | 6334 | nucleosome assembly | P | 9 | 73 | 117 | 12.3 | 62.4 | 6.350 |
| 5 | 79 | cyclin dependent protein kinase activity | P | 6 | 38 | 39 | 15.8 | 97.4 | 6.107 |
| 6 | 31497 | chromatin assembly | P | 9 | 82 | 126 | 11.0 | 65.1 | 5.852 |
| 7 | 46879 | hormone secretion | P | 3 | 12 | 13 | 25.0 | 92.3 | 5.716 |
| 8 | 5149 | interleukin-1 receptor binding | F | 5 | 12 | 12 | 41.7 | 100.0 | 9.856 |
| 9 | 5154 | epidermal growth factor receptor binding | F | 3 | 5 | 6 | 60.0 | 83.3 | 9.299 |
| 10 | 30353 | FGF receptor antagonist activity | F | 3 | 8 | 8 | 37.5 | 100.0 | 7.201 |
| 11 | 5152 | interleukin-1 receptor antagonist activity | F | 3 | 8 | 8 | 37.5 | 100.0 | 7.201 |
| 12 | 5104 | fibroblast growth factor receptor binding | F | 3 | 9 | 9 | 33.3 | 100.0 | 6.742 |
| 13 | 48019 | receptor antagonist activity | F | 3 | 10 | 10 | 30.0 | 100.0 | 6.351 |
| 14 | 30545 | receptor regulator activity | F | 3 | 11 | 11 | 27.3 | 100.0 | 6.013 |
| 15 | 19825 | oxygen binding | F | 4 | 29 | 35 | 13.8 | 82.9 | 4.562 |
| 16 | 5882 | intermediate filament | C | 11 | 80 | 107 | 13.8 | 74.8 | 7.563 |
| 17 | 786 | nucleosome | C | 9 | 68 | 104 | 13.2 | 65.4 | 6.665 |
| 18 | 43296 | apical junction complex | C | 6 | 55 | 56 | 10.9 | 98.2 | 4.752 |
GO types: biological processes (P), molecular functions (F) and cellular components (C). The Z-scores: calculated to identify the significant GO terms (Z-score ≥ 2.0, corresponding to P ≤ 0.05)
Table 5.
Top balanced Gene Ontology biological pathways associated with both up- and down-regulated genes in RAC limbal epithelial progenitors
| GO ID | Gene Ontology (GO) Description | GO Type | Number of genes |
Percentages of genes |
Z Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Changed | Measured | in GO | % Up | % Down | % Changed | %Present | |||||
| 1 | 8544 | epidermis development | P | 7 | 7 | 14 | 62 | 68 | 11.3 | 11.3 | 22.6 | 91.2 | 5.263 |
| 2 | 1525 | angiogenesis | P | 9 | 5 | 14 | 54 | 55 | 16.7 | 9.3 | 26.0 | 98.2 | 4.858 |
| 3 | 1944 | vasculature development | P | 9 | 5 | 14 | 56 | 57 | 16.1 | 8.9 | 25.0 | 98.2 | 4.716 |
| 4 | 1568 | blood vessel development | P | 9 | 5 | 14 | 56 | 57 | 16.1 | 8.9 | 25.0 | 98.2 | 4.716 |
| 5 | 7626 | locomotory behavior | P | 13 | 8 | 21 | 113 | 126 | 11.5 | 7.1 | 18.6 | 89.7 | 4.195 |
| 6 | 6935 | chemotaxis | P | 12 | 8 | 20 | 108 | 121 | 11.1 | 7.4 | 18.5 | 89.3 | 4.054 |
| 7 | 7610 | behavior | P | 13 | 10 | 23 | 172 | 190 | 7.6 | 5.8 | 13.4 | 90.5 | 3.621 |
| 8 | 16337 | cell-cell adhesion | P | 14 | 7 | 21 | 156 | 170 | 9.0 | 4.5 | 13.5 | 91.8 | 3.295 |
| 9 | 4867 | serine-type endopeptidase inhibitor activity | F | 10 | 8 | 18 | 76 | 82 | 13.2 | 10.5 | 23.7 | 92.7 | 5.351 |
| 10 | 4866 | endopeptidase inhibitor activity | F | 14 | 10 | 24 | 121 | 130 | 11.6 | 8.3 | 19.9 | 93.1 | 4.972 |
| 11 | 30414 | protease inhibitor activity | F | 14 | 10 | 24 | 122 | 131 | 11.5 | 8.2 | 19.7 | 93.1 | 4.939 |
| 12 | 4857 | enzyme inhibitor activity | F | 16 | 14 | 30 | 213 | 231 | 7.5 | 6.6 | 14.1 | 92.2 | 4.836 |
| 13 | 8009 | chemokine activity | F | 7 | 4 | 11 | 40 | 49 | 17.5 | 10.0 | 27.5 | 81.6 | 4.451 |
| 14 | 5125 | cytokine activity | F | 13 | 11 | 24 | 187 | 212 | 7.0 | 5.9 | 12.9 | 88.2 | 3.845 |
| 15 | 1664 | G-protein-coupled receptor binding | F | 7 | 4 | 11 | 48 | 57 | 14.6 | 8.3 | 22.9 | 84.2 | 3.832 |
| 16 | 5923 | tight junction | C | 5 | 6 | 11 | 50 | 51 | 10.0 | 12.0 | 22.0 | 98.0 | 5.084 |
| 17 | 5911 | intercellular junction | C | 9 | 8 | 17 | 98 | 103 | 9.2 | 8.2 | 17.4 | 95.1 | 4.399 |
| 18 | 30054 | cell junction | C | 9 | 8 | 17 | 103 | 108 | 8.7 | 7.8 | 16.5 | 95.4 | 4.221 |
GO types: biological processes (P), molecular functions (F) and cellular components (C). The Z-scores: calculated to identify the significant GO terms (Z-score ≥ 2.0, corresponding to P ≤ 0.05)
Figure 4.
Gene map examples of enriched biological pathways by RAC progenitors. A. Gene map of cell proliferation profile, generated by GenMAPP and MAPPFinder software, displayed numerous genes up (red)- or down (green)-regulated (fold changes of mRNA levels of RAC/NAC) in RAC population with their potential role in positive or negative regulation of cell proliferation. Grey color indicates not significantly changed (fold change of RAC/NAC < 2 or > 0.5, or P>00.05). White indicates absent genes by RAC population. B. Gene map of Wnt/β-catenin pathway, analyzed by the Ingenuity Pathway Analysis suite, showed that the numerous significantly regulated, up (red) or down (green), genes (fold change more than 1.5 fold with P value ≤0.05) in RAC population were involved in Wnt/β-catenin pathway. White indicates absent or unchanged genes by RAC population.
DISCUSSION
Adult stem cells, the tissue-specific stem cells residing in many adult tissues, have been recognized to have ability to self renew and to intervene in maintaining the structural and functional integrity of their original tissue, and therefore they become an important cell source of choice for regenerative medicine and tissue engineering. The ocular surface is an ideal region to study epithelial stem cell biology, because of the unique spatial arrangement of stem cells and TACs (Dua and Azuara-Blanco, 2000; Tseng, 1989; Watt and Hogan, 2000). Zhou and colleagues have revealed differential transcriptional profiles of the stem cells-enriched limbal basal epithelial cells versus the corneal basal TACs in quiescent mouse tissues using laser capture microdissection technique (Zhou et al., 2006). However, identification and isolation is still challenging for stem cell biologists. No molecular markers have been recognized to be capable of definitively identifying the limbal stem cells to date although corneal epithelial stem cells have been identified to reside in limbus for 2 decades and many stem cell markers have been proposed (Budak et al., 2005; Li et al., 2007; Pajoohesh-Ganji and Stepp, 2005; Schlotzer-Schrehardt and Kruse, 2005; Stepp and Zieske, 2005). In recent years, great efforts have been made to characterize the phenotype of limbal stem cells. We have reported that the limbal basal cells are small primitive cells expressing three patterns of molecular markers (Chen et al., 2004). We have also isolated clonogenic progenitor cell populations from limbal epithelia and/or their cultures, based on stem cell phenotype, using different cell surface markers and properties including cell-sizing, adherence to extracellular matrix, sorting for side population or for expression of ABCG2 or connexin 43 cell surface markers (Chen et al., 2006; de Paiva et al., 2005; de Paiva et al., 2006b; Li et al., 2005). To further characterize limbal stem cells, we analyzed gene expression profiles of isolated stem cell-containing RAC progenitor population in comparison with TAC-like SAC and differentiated TDC-like NAC populations using Affymetrix whole human genome microarray.
This study has shown that the Affymetrix microarrays are a powerful and reliable technique that delivers highly reproducible gene expression data for the whole human genome from a small amount of total RNA (100ng). As shown in Figure 1, the data, not only for all gene transcripts, but also for significantly changed genes, from each pair of 2 chips in one array and between 2 arrays were highly reproducible, even though these total 12 samples of 3 isolated cell populations were from 4 separate experiments, and the 2 arrays were performed at different times over 1 year period. In particular, the heatmap generated by a hierarchical clustering analysis on the dataset consisting of the 776 significantly changed genes including 499 up- and 277 down-regulated genes (Figure 2) displayed great reproducibility of gene expression patterns for these 3 cell populations between duplicate chips of each array, as well as between 2 arrays. Interestingly, the mRNA levels of the most upregulated genes in RAC over NAC were also higher than SAC, while the transcripts of most down-regulated genes in RAC over NAC were also expressed lower than SAC. The heatmap visually showed a clear separation of the RAC group from the NAC and SAC groups, indicating the RAC population has its distinguishably characteristic gene expression profile from NAC and SAC.
Although the RAC was shown to be progenitor cells with certain stem-like properties (Li et al., 2005), we were unable to show all or most of known stem cell associated marker expressed by RAC using regular or routine methods. The microarray data allowed us to analyze all these known markers at one time. Interestingly, the expression patterns of 15 known genes, proposed as stem cell associated (ABCG2, p63, integrin α9, integrin β1, EGFR, enolase-α1, K15, TrkA, N-cadherin and C/EBP-δ) or differentiation associated (connexin 43, E-cadherin, K12, K3 and involucrin) markers, were consistent to the limbal stem cell phenotype as previous reports (Barbaro et al., 2007; Chen et al., 2004; Figueira et al., 2007; Hayashi et al., 2007; Qi et al., 2008a; Qi et al., 2008b). Among them, 9 genes were verified again by quantitative real-time PCR using samples from separate adhesion experiments. These findings further demonstrate that the RAC population indeed represents the progenitor cells. In addition, these data also verified that the Affymetrix arrays truly measure real patterns of gene expression.
With confidence that these array data were correct, we further analyzed the highly regulated genes in RAC population selected to meet criteria from 4 chips for each cell population in both arrays. The top 42 genes including 26 up- and 16 down-regulated genes were selected based on fold change ≥ 6 fold (up or down) with P value ≤ 0.005 (listed in Table 2). Among them, 18 genes including 9 up- and 9 down-regulated genes were verified for their expression pattern by RAC, SAC and NAC populations using RT and quantitative real-time PCR. Interestingly, the verified results were consistent to the microarray data. Although many genes of the list are not well characterized, they might be related to certain stem cell properties such as self renewal, proliferation, differentiation, etc. As further evaluated by immunofluorescent staining, the exclusive immnoreactivity of positive TCF4 and negative SPRRs at limbal basal layer indicates that they may potentially serve as positive or negative markers, respectively, for limbal progenitor cells. Further studies regarding the function of these molecules are needed to confirm our hypothesis.
Mining the treasures from huge database generated by microarrays is challenging. Not only are highly changed genes important, but many other genes related to stem cell properties, regardless of how high or low their mRNA levels. As shown in Table 1, changes in mRNA levels were 1.5 – 3.9 fold in known stem cell associated markers, and 0.67-0.17 (−1.5 to −5.9) fold in known differentiation markers. These genes would be overlooked if only the top regulated genes were investigated. To explore molecular signatures of progenitor cells, we further analyzed biological pathways and global trends that are associated to these significantly regulated genes in RAC. Using GenMAPP and MAPPFinder, we have identified three patterns of biological pathway profiles, overexpressed, underexpressed and balanced, by the RAC progenitor population based on Gene Ontology categories that include 3 major groups of terms, biological process, molecular function and cellular component (Tables 3–5). Many pathways and related genes are worth further investigation to discover new features and properties of adult stem cells.
In conclusions, this study characterized the molecular signatures and biological pathways of corneal epithelial progenitor cells, the RAC population isolated from limbal epithelial tissues by adhesion to collagen IV, using Affymetrix GeneChip whole human genome U133 Plus 2.0 microarrays. With verified expression patterns of 27 important genes, the reproducible genome microarray data not only confirm most currently known stem cell associated and differentiation markers that represent the limbal stem cell phenotype, but also revealed many highly significantly regulated genes and their associated biological pathways in limbal progenitor cells. These genes and related pathways are the potential targets for further identification and isolation of limbal stem cells.
Supplementary Material
Acknowledgments
We thank the Microarray Core Facility in Baylor College of Medicine (Houston, TX) for high quality performance of Affymetrix arrays. This work was supported by Department of Defense CDMRP PRMRP grant FY06 PR064719 (DQL), National Institutes of Health grant EY11915 (SCP), an unrestricted grant from Research to Prevent Blindness, the Oshman Foundation and the William Stamps Farish Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barbaro V, Testa A, Di IE, Mavilio F, Pellegrini G, De LM. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J Cell Biol. 2007;177:1037–1049. doi: 10.1083/jcb.200703003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton M, Albon J. Stem cells in the eye. Int J Biochem Cell Biol. 2004;36:643–657. doi: 10.1016/j.biocel.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Budak MT, Alpdogan OS, Zhou M, Lavker RM, Akinci MA, Wolosin JM. Ocular surface epithelia contain ABCG2-dependent side population cells exhibiting features associated with stem cells. J Cell Sci. 2005;118:1715–1724. doi: 10.1242/jcs.02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, de Paiva CS, Luo L, Kretzer FL, Pflugfelder SC, Li DQ. Characterization of putative stem cell phenotype in human limbal epithelia. Stem Cells. 2004;22:355–366. doi: 10.1634/stemcells.22-3-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Evans WH, Pflugfelder SC, Li DQ. Gap junction protein connexin 43 serves as a negative marker for a stem cell-containing population of human limbal epithelial cells. Stem Cells. 2006;24:1265–1273. doi: 10.1634/stemcells.2005-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson JM, Morris L, Reid AI, Ramaesh T, Keighren MA, Flockhart JH, et al. Clonal analysis of patterns of growth, stem cell activity, and cell movement during the development and maintenance of the murine corneal epithelium. Dev Dyn. 2002;224:432–440. doi: 10.1002/dvdy.10124. [DOI] [PubMed] [Google Scholar]
- Dahlquist KD. Using GenMAPP and MAPPFinder to view microarray data on biological pathways and identify global trends in the data. Curr Protoc Bioinformatics. 2004;Chapter 7(Unit) doi: 10.1002/0471250953.bi0705s05. [DOI] [PubMed] [Google Scholar]
- de Paiva CS, Chen Z, Corrales RM, Pflugfelder SC, Li D-Q. ABCG2 transporter identifies a population of clonogenic human limbal epithelial cells. Stem Cells. 2005;23:63–73. doi: 10.1634/stemcells.2004-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paiva CS, Corrales RM, Villarreal AL, Farley WJ, Li D-Q, Stern ME, et al. Corticosteroid and doxycycline suppress MMP-9 and inflammatory cytokine expression, MAPK activation in the corneal epithelium in experimental dry eye. Exp Eye Res. 2006a;83:526–535. doi: 10.1016/j.exer.2006.02.004. [DOI] [PubMed] [Google Scholar]
- de Paiva CS, Pflugfelder SC, Li DQ. Cell size correlates with phenotype and proliferative capacity in human corneal epithelial cells. Stem Cells. 2006b;24:368–375. doi: 10.1634/stemcells.2005-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua HS, Azuara-Blanco A. Limbal stem cells of the corneal epithelium. Surv Ophthalmol. 2000;44:415–425. doi: 10.1016/s0039-6257(00)00109-0. [DOI] [PubMed] [Google Scholar]
- Figueira EC, Di GN, Coroneo MT, Wakefield D. The phenotype of limbal epithelial stem cells. Invest Ophthalmol Vis Sci. 2007;48:144–156. doi: 10.1167/iovs.06-0346. [DOI] [PubMed] [Google Scholar]
- Hayashi R, Yamato M, Sugiyama H, Sumide T, Yang J, Okano T, et al. N-cadherin is expressed by putative stem/progenitor cells and melanocytes in the human limbal epithelial stem cell niche. Stem Cells. 2007;25:289–296. doi: 10.1634/stemcells.2006-0167. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Jun SX, de Paiva CS, Chen Z, Pflugfelder SC, Li D-Q. Phenotypic characterization of human corneal epithelial cells expanded ex vivo from limbal explant and single cell cultures. Exp Eye Res. 2004;79:41–49. doi: 10.1016/j.exer.2004.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi N, Inatomi T, Suzuki T, Sotozono C, Kinoshita S. Cultivated corneal epithelial stem cell transplantation in ocular surface disorders. Ophthalmology. 2001;108:1569–1574. doi: 10.1016/s0161-6420(01)00694-7. [DOI] [PubMed] [Google Scholar]
- Lavker RM, Sun TT. Epidermal stem cells: properties, markers, and location. Proc Natl Acad Sci U S A. 2000;97:13473–13475. doi: 10.1073/pnas.250380097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavker RM, Tseng SC, Sun TT. Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Exp Eye Res. 2004;78:433–446. doi: 10.1016/j.exer.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Li D-Q, Chen Z, Song XJ, de Paiva CS, Kim HS, Pflugfelder SC. Partial enrichment of a population of human limbal epithelial cells with putative stem cell properties based on collagen type IV adhesiveness. Exp Eye Res. 2005;80:581–590. doi: 10.1016/j.exer.2004.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hayashida Y, Chen YT, Tseng SC. Niche regulation of corneal epithelial stem cells at the limbus. Cell Res. 2007;17:26–36. doi: 10.1038/sj.cr.7310137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Li D-Q, Doshi A, Farley W, Corrales RM, Pflugfelder SC. Experimental dry eye stimulates production of inflammatory cytokines and MMP-9 and activates MAPK signaling pathways on the ocular surface. Invest Ophthalmol Vis Sci. 2004;45:4293–4301. doi: 10.1167/iovs.03-1145. [DOI] [PubMed] [Google Scholar]
- Pajoohesh-Ganji A, Stepp MA. In search of markers for the stem cells of the corneal epithelium. Biol Cell. 2005;97:265–276. doi: 10.1042/BC20040114. [DOI] [PubMed] [Google Scholar]
- Pellegrini G, Traverso CE, Franzi AT, Zingirian M, Cancedda R, De Luca M. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 1997;349:990–993. doi: 10.1016/S0140-6736(96)11188-0. [DOI] [PubMed] [Google Scholar]
- Qi H, Li DQ, Bian F, Chuang EY, Jones DB, Pflugfelder SC. Expression of glial cell-derived neurotrophic factor and its receptor in the stem-cell-containing human limbal epithelium. Br J Ophthalmol. 2008a;92:1269–1274. doi: 10.1136/bjo.2007.132431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Li DQ, Shine HD, Chen Z, Yoon KC, Jones DB, et al. Nerve growth factor and its receptor TrkA serve as potential markers for human corneal epithelial progenitor cells. Exp Eye Res. 2008b;86:34–40. doi: 10.1016/j.exer.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotzer-Schrehardt U, Kruse FE. Identification and characterization of limbal stem cells. Exp Eye Res. 2005;81:247–264. doi: 10.1016/j.exer.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Schwab IR, Reyes M, Isseroff RR. Successful transplantation of bioengineered tissue replacements in patients with ocular surface disease. Cornea. 2000;19:421–426. doi: 10.1097/00003226-200007000-00003. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Yang YH, Speed T. Statistical issues in cDNA microarray data analysis. Methods Mol Biol. 2003;224:111–136. doi: 10.1385/1-59259-364-X:111. [DOI] [PubMed] [Google Scholar]
- Stepp MA, Zieske JD. The corneal epithelial stem cell niche. Ocul Surf. 2005;3:15–26. doi: 10.1016/s1542-0124(12)70119-2. [DOI] [PubMed] [Google Scholar]
- Tsai RJ, Li LM, Chen JK. Reconstruction of damaged corneas by transplantation of autologous limbal epithelial cells. N Engl J Med. 2000;343:86–93. doi: 10.1056/NEJM200007133430202. [DOI] [PubMed] [Google Scholar]
- Tseng SC. Concept and application of limbal stem cells. Eye. 1989;3(Pt 2):141–157. doi: 10.1038/eye.1989.22. [DOI] [PubMed] [Google Scholar]
- Tsubota K. Ocular surface management in corneal transplantation, a review. Jpn J Ophthalmol. 1999;43:502–508. doi: 10.1016/s0021-5155(99)00140-9. [DOI] [PubMed] [Google Scholar]
- Watt FM, Hogan BL. Out of Eden: stem cells and their niches. Science. 2000;287:1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- Yoon KC, de Paiva CS, Qi H, Chen Z, Farley WJ, Li DQ, et al. Expression of Th-1 chemokines and chemokine receptors on the ocular surface of C57BL/6 mice: effects of desiccating stress. Invest Ophthalmol Vis Sci. 2007;48:2561–2569. doi: 10.1167/iovs.07-0002. [DOI] [PubMed] [Google Scholar]
- Zhou M, Li XM, Lavker RM. Transcriptional profiling of enriched populations of stem cells versus transient amplifying cells. A comparison of limbal and corneal epithelial basal cells. J Biol Chem. 2006;281:19600–19609. doi: 10.1074/jbc.M600777200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.