Abstract
20,23-Dihydroxyvitamin D3 [20,23(OH)2D3] is a biologically active metabolite produced by the action of cytochrome P450scc (CYP11A1) on vitamin D3. It inhibits keratinocyte proliferation, stimulates differentiation, and inhibits nuclear factor-κB activity, working as a vitamin D receptor agonist. We have tested the ability of purified mouse 25-hydroxyvitamin D3 1α-hydroxylase (CYP27B1) to add a 1α-hydroxyl group to this vitamin D analog and determined whether this altered its biological activity. 20,23(OH)2D3 incorporated into phospholipid vesicles was converted to a single product by CYP27B1, confirmed to be 1α,20,23-trihydroxyvitamin D3 [1,20,23(OH)3D3] by mass spectrometry and NMR. The 20,23(OH)2D3 was a relatively poor substrate for CYP27B1 compared with the normal substrate, 25-hydroxyvitamin D3, displaying a 5-fold higher Km and 8-fold lower kcat value. Both 20,23(OH)2D3 and 1,20,23(OH)3D3 decreased neonatal human epidermal keratinocyte proliferation, showing significant effects at a lower concentration (0.1 nM) than that seen for 1α,25-dihydroxyvitamin D3 [1,25(OH)2D3] at 24 h of treatment. Both compounds also decreased cell biomass relative to that of control cells, measured by staining with sulforhodamine B. They caused little stimulation of the expression of the vitamin D receptor at the mRNA level compared with the 30-fold induction observed with the same concentration (100 nM) of 1,25(OH)2D3 at 24 h. Addition of a 1α-hydroxyl group to 20,23(OH)2D3 greatly enhanced its ability to stimulate the expression of the CYP24 gene but not to the extent seen with 1,25(OH)2D3. This study shows that purified CYP27B1 can add a 1α-hydroxyl group to 20,23(OH)2D3 with the product showing altered biological activity, especially for the stimulation of CYP24 gene expression.
Introduction
1α,25-Dihydroxyvitamin D3 [1,25(OH)2D3] is an important secosteroid hormone that regulates calcium and phosphorus homeostasis through binding to the vitamin D receptor (Prosser and Jones, 2004; Masuda and Jones, 2006; Holick, 2007). In addition, 1,25(OH)2D3 also plays an important role in regulating the immune system and displays antiproliferative and anticancer properties (Prosser and Jones, 2004; Bouillon et al., 2006; Masuda and Jones, 2006; Takahashi and Morikawa, 2006; Holick, 2007; Bikle, 2009). However, the supraphysiological doses required for the anticancer effects result in toxic hypercalcemia, which limits the use of 1,25(OH)2D3 for therapeutic purposes. As a result, other vitamin D analogs are being tested that retain the antiproliferative and prodifferentiative properties of 1,25(OH)2D3 but are not calcemic (Masuda and Jones, 2006; Takahashi and Morikawa, 2006; Lee et al., 2008; Slominski et al., 2010). A new source of vitamin D analogs with such properties arises from the action of cytochrome P450scc on vitamin D3 (Slominski et al., 2006; Zbytek et al., 2008; Janjetovic et al., 2009; Slominski et al., 2010).
Cytochrome P450scc is well known for catalyzing the cleavage of the side chain of cholesterol, which is the first enzymatic step of steroid hormone synthesis (Tuckey, 2005). In the past few years, numerous studies have shown that P450scc is also active toward vitamin D3, producing 20-hydroxyvitamin D3 [20(OH)D3], 20,23-dihydroxyvitamin D3 [20,23(OH)2D3], 17,20,23-trihydroxyvitamin D3, and other minor hydroxyvitamin D3 metabolites (Guryev et al., 2003; Slominski et al., 2005; Tuckey et al., 2008b,c). P450scc can also metabolize 1α-hydroxyvitamin D3 to give 1α,20-dihydroxyvitamin D3 [1,20(OH)2D3] (Tuckey et al., 2008a). P450scc hydroxylates vitamin D2, producing 20-hydroxyvitamin D2, 17,20-dihydroxyvitamin D2, and 17,20,24-trihydroxyvitamin D2 as the major products (Slominski et al., 2006; Nguyen et al., 2009). Of these many products of vitamin D metabolism, 20(OH)D3 and 20,23(OH)2D3 have been most extensively studied for their biological effects. Both compounds inhibit keratinocyte proliferation, promote differentiation, and display anti-inflammatory activity by decreasing nuclear factor-κB activity, effects mediated via binding to the vitamin D receptor (Zbytek et al., 2008; Janjetovic et al., 2009; Slominski et al., 2010), and show antiproliferative and prodifferentiation effects on a range of leukemia cell lines (Slominski et al., 2010). More importantly, 20(OH)D3 has been shown to lack calcemic activity in rats, even at relatively high concentrations. However, the addition of a hydroxyl group at the 1α-position [1,20(OH)2D3] significantly increased its calcemic activity (Slominski et al., 2010).
CYP27B1 is the cytochrome P450 enzyme that catalyzes the final step of vitamin D activation, the 1α-hydroxylation of 25(OH)D3 to give 1,25(OH)2D3. This reaction mainly takes place in the kidneys, but CYP27B1 has also been found to be expressed in other tissues including skin, lungs, breasts, and prostate (Bikle et al., 1986; Holick, 2007; Anderson et al., 2008). The human enzyme has not yet been thoroughly characterized because of its low expression in vivo and instability once removed from its hydrophobic membrane environment. In contrast, the mouse enzyme is more stable and has been successfully expressed in Escherichia coli, extracted, and purified (Uchida et al., 2004; Tang et al., 2010). Using the mouse enzyme we have shown that CYP27B1 can 1α-hydroxylate a variety of other substrates besides 25(OH)D3, including 20(OH)D3 (Tang et al., 2010). Thus, CYP27B1 provides a potential enzymatic method for the 1α-hydroxylation of novel, biologically active vitamin D3 analogs, including those made by the action of P450scc. In the current study, we demonstrate the ability of CYP27B1 to 1α-hydroxylate 20,23(OH)2D3 and show that the 1α,20,23-trihydroxyvitamin D3 [1,20,23(OH)3D3] product has altered biological effects on keratinocytes, a well characterized target of 1,25(OH)2D3.
Materials and Methods
Synthesis of 20,23(OH)2D3.
20,23(OH)2D3, synthesized enzymatically from the action of cytochrome P450scc on vitamin D3, was purified by thin-layer chromatography and reverse-phase HPLC as published previously (Tuckey et al., 2008a).
Preparation of Enzymes.
Mouse adrenodoxin, human adrenodoxin reductase, and mouse 25-hydroxyvitamin D3 1α-hydroxylase (CYP27B1) were expressed in E. coli (Woods et al., 1998; Tuckey and Sadleir, 1999; Tang et al., 2010). Both the mouse CYP27B1 and human adrenodoxin reductase were coexpressed with the chaperonins GroEL/ES to facilitate correct protein folding. The cDNA for mouse CYP27B1 was modified to encode a 4-histidine tag at the C terminus of the CYP27B1, and the sequence encoding the mitochondrial-targeting sequence was removed. The expressed mouse CYP27B1 was then purified using nickel affinity and Octyl Sepharose chromatography as before (Tang et al., 2010).
Measurement of 20,23(OH)2D3 Metabolism in Phospholipid Vesicles.
Phospholipid vesicles containing 20,23(OH)2D3 were prepared by sonication. Dioleoyl phosphatidylcholine (1.08 μmol; Sigma-Aldrich, St. Louis, MO), bovine heart cardiolipin (0.19 μmol; Sigma-Aldrich), and 20,23(OH)2D3 (as required) were placed in glass tubes, and the ethanol solvent was removed under nitrogen. Buffer (0.5 ml) comprising 20 mM HEPES (pH 7.4), 100 mM NaCl, 0.1 mM dithiothreitol, and 0.1 mM EDTA was added, and the tubes were sonicated for 10 min in a bath-type sonicator (Lambeth et al., 1982). The incubation mixture comprised vesicles (510 μM phospholipid), CYP27B1 (0.15–0.5 μM), 15 μM adrenodoxin, 0.4 μM adrenodoxin reductase, 2 mM glucose 6-phosphate, 2 U/ml glucose-6-phosphate dehydrogenase, and 50 μM NADPH, in the same buffer used for vesicle preparation. Samples (typically 0.25–1.0 ml) were preincubated for 5 min at 37°C, and then the reaction was initiated by the addition of adrenodoxin. Samples were incubated at 37°C, with shaking, for various time intervals (see Results). The reactions were terminated by the addition of 2 ml of ice-cold dichloromethane and vortexing (Tuckey et al., 2008c). After phase separation, the lower organic phase was retained, and the upper aqueous phase was extracted twice more with 2-ml aliquots of dichloromethane as described. The dichloromethane was subsequently removed under nitrogen, and the residual sample was dissolved in a 64% methanol-36% water mixture for analysis. Reverse-phase HPLC (C18 column) was performed as described before (Tang et al., 2010) to measure product formation by CYP27B1. Kinetic parameters were determined by fitting the Michaelis-Menten equation to the experimental data using KaleidaGraph 3.6 (Abelbeck Synergy, Reading, PA).
Large-Scale Preparation of 1,20,23(OH)3D3.
Enzymatic production of 1,20,23(OH)3D3 was optimized for temperature, CYP27B1 concentration, and glycerol concentration using 20,23(OH)2D3 or other hydroxyvitamin D3 substrates (see Results). The trihydroxyvitamin D3 metabolite was prepared from a 40-ml incubation of 0.8 μM mouse CYP27B1with 20,23(OH)2D3 (0.025 mol/mol phospholipid) at 25°C, in a scaled-up version of the method described above for activity measurement. The 1,20,23(OH)3D3 was purified by preparative HPLC, using a Brownlee Aquapore column (25 cm × 10 mm, particle size 20 μm) and eluted under isocratic conditions with a mobile phase consisting of 80% methanol with water.
Mass Spectrometry.
Mass spectra of the trihydroxy metabolite were measured using a Bruker Esquire liquid chromatography-mass spectrometry system (Bruker Daltonics, Billerica, MA). The measurements were performed on both an electrospray ionization (ESI) source and an atmospheric pressure chemical ionization (APCI) source with nitrogen as the nebulizing gas. Data were transferred to an offline computer and processed with ACD software (Advanced Chemistry Development, Toronto, ON, Canada).
NMR.
Trihydroxyvitamin D3 and vitamin D3 were dissolved in 60 μl of methanol-d4 (99.8% d; Cambridge Isotope Laboratories, Inc., Andover, MA) and transferred to 3-mm Shigemi NMR tubes (Shigemi Inc., Allison Park, PA). NMR spectra were acquired on a Varian Inova-500 MHz NMR spectrometer equipped with a 3-mm inverse probe (Varian, Inc., Palo Alto, CA). Temperature was regulated at 296.5 K. All NMR data were processed with standard parameters. Chemical shifts were referenced to the residue solvent peak (proton at 3.31 ppm and carbon at 49.15 ppm). Positions of the three hydroxyl groups in the trihydroxyvitamin D3 were determined by analysis of the acquired NMR spectra and comparison with those of parent vitamin D3.
Cell Proliferation Assay Using MTT Reagent.
For determining the number of viable cells in proliferation MTT reagent (Promega, Madison, WI) was used. Pooled samples of normal neonatal human epidermal keratinocyte (HEKn) cells, isolated from the foreskins of African American donors, were used in their third passage and were seeded in 96-well plates. When cells attained 80% confluence, they were treated with 1,25(OH)2D3, 20,23(OH)2D3, or 1,20,23(OH)3D3 at concentrations ranging from 0.1 to 100 nM. The hydroxyvitamin D3 compounds were diluted from ethanol stocks in KGM supplemented with KGF medium (Lonza, Walkersville, MD) and 0.5% bovine serum albumin, with 100 μl being added to each well. Ethanol vehicle diluted similarly was added to control cells. After 20 or 44 h of incubation, 20 μl of MTT reagent (5 mg/ml) was added to the cells, and cells were further incubated for 4 h at 37°C. The medium was discarded, and cells were lysed in isopropanol-0.1 M HCl solution by shaking for 30 min at room temperature. The quantity of formazan product is directly proportional to the number of living cells in culture as measured by the amount of absorbance at 570 nm.
Cell Mass Estimated with Sulforhodamine B.
For determining total biomass, cellular proteins were stained with sulforhodamine B reagent (Sigma-Aldrich). HEKn cells were seeded in 96-well plates to reach 80% confluence, as for the MTT assay. Cells were then treated with 1,25(OH)2D3, 20,23(OH)2D3, or 1,20,23(OH)3D3 (or ethanol vehicle) also as described above. After 23 h of incubation, 20 μl of 50% trichloroacetic acid (Sigma-Aldrich) was added to the cells, and cells were further incubated for 1 h at 4°C. The medium was discarded, and cells were briefly washed, dried, and stained with 0.4% sulforhodamine B dye for 20 to 30 min. Excess stain was removed by rinsing cells quickly with 1% acetic acid solution. Cells were lysed in 10 mM Tris-HCl with shaking for 5 min at room temperature, to liberate the dye incorporated into the cells. Absorbance of the liberated dye was measured at 565 nm. An increase or decrease in the number of cells (total biomass) results in a concomitant change in the amount of dye incorporated by the cells during culture.
Reverse Transcription-Polymerase Chain Reaction.
RNA was extracted from HEKn cells treated with 1,25(OH)2D3, 20,23(OH)2D3 or 1,20,23(OH)3D3 for 6 or 24 h as above, using the Absolutely RNA RT-PCR Miniprep Kit (Stratagene, La Jolla, CA). Reverse transcription was performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Mannheim, Germany). The reaction was performed with LightCycler 480 Probes Master (Roche Applied Science). The primers and probes were designed with the Universal Probe Library (Roche Applied Science). The primer sequences for CYP24 and vitamin D receptor (VDR) were described previously (Janjetovic et al., 2010). Real-time PCR was performed using TaqMan PCR Master Mix at 95°C for 5 min, and then 45 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. The data were collected with a Roche Light Cycler 480. The amounts of mRNA were normalized by the comparative Ct method, using cyclophilin B as a housekeeping gene.
Statistical Analysis.
Data are presented as means ± S.E.M. and have been analyzed with Student's t test (for two groups) or one-way analysis of variance and an appropriate post hoc test (for more than two groups), using Microsoft Excel and Prism 4.00 (GraphPad Software Inc., San Diego, CA). Statistically significant differences are denoted in the figures and corresponding figure legends.
Results
Kinetics of 20,23(OH)2D3 Metabolism by Mouse CYP27B1.
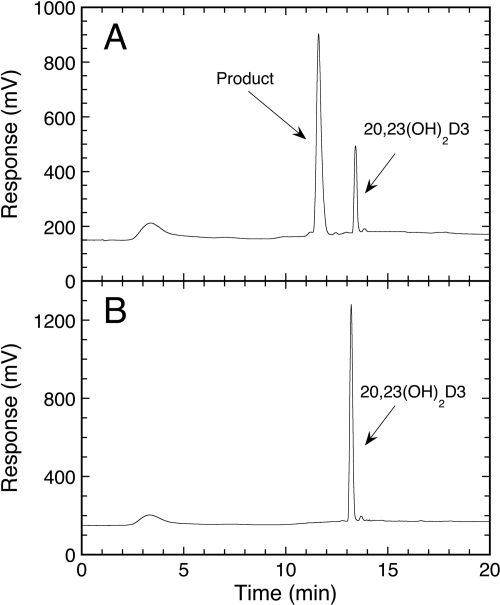
CYP27B1 activity was measured with substrate incorporated into phospholipid vesicles because this system mimics the native environment of the cytochrome in the inner mitochondrial membrane (Nakamura et al., 1997; Headlam et al., 2003) and CYP27B1 displays high activity in this system (Tang et al., 2010). Incubation of 20,23(OH)2D3 in phospholipid vesicles, with mouse CYP27B1, resulted in one product only (Fig. 1). This was subsequently confirmed to be 1,20,23(OH)3D3 (see mass spectra and NMR described later). A time course for CYP27B1 activity on 20,23(OH)2D3 was performed at 37°C and was linear for the first 2 min (data not shown), as previously observed with 25(OH)D3 as substrate (Tang et al., 2010). Hence, 2 min was used as the incubation time to measure the initial rate of product formation to determine the kinetics of metabolism of 20,23(OH)2D3 by CYP27B1. In phospholipid vesicles, 20,23(OH)2D3 gave a kcat of 4.49 ± 0.62 mol/(min · mol) CYP27B1 and a Km of 39.4 ± 10.6 mmol of substrate/mol of phospholipid. This compares to kcat and Km values for 1α-hydroxylation of 25(OH)D3 of 33.2 ± 0.8 mol/(min · mol) CYP27B1 and 7.16 ± 0.57 mmol of substrate/mol of phospholipid, respectively, under identical conditions.
Fig. 1.
Chromatogram showing the metabolism of 20,23(OH)2D3 by mouse CYP27B1. A, 20,23(OH)2D3 was incorporated into phospholipid vesicles at a ratio of 0.025 mol/mol phospholipid and was incubated with 0.6 μM CYP27B1 for 1 h in a reconstituted system containing adrenodoxin and adrenodoxin reductase. Samples were extracted using dichloromethane and analyzed by reverse-phase HPLC. B, control incubation without adrenodoxin showing the substrate but the absence of product.
Optimization of Metabolism of 20,23(OH)2D3 by CYP27B1 for Large-Scale Reactions.
We have previously reported that CYP27B1 is very labile with a 50% reduction in activity occurring during a 10-min incubation at 37°C with 25(OH)D3 as substrate (Tang et al., 2010). Because of this instability, we optimized the incubation condition in terms of temperature, time, and glycerol content to achieve the highest conversion of substrate to product. Time courses for product formation from 20,23(OH)2D3 revealed that despite a slower initial rate at 25°C, more product was produced by the end of a 1-h incubation at 25°C than at 37°C, reflecting the increased stability of the enzyme at 25°C. At both temperatures product formation had ceased by the end of the incubation. In a further optimization, we tested the effect of increasing the glycerol content from 2 to 10%. The 2% glycerol results from dilution of the stock enzyme, stored in 20% glycerol, into the incubation medium. The increase in glycerol in the incubation medium to 10% increased product formation at the end of a 1-h incubation by 5%. Therefore, we chose 25°C, 10% glycerol, and a CYP27B1 concentration of 0.8 μM as conditions to scale-up product formation from 20,23(OH)2D3 (see Materials and Methods). A 40-ml incubation of 20,23(OH)2D3 with mouse CYP27B1 under these conditions resulted in 78% conversion of substrate to product. After HPLC purification, 130 μg of pure product was obtained for NMR analysis and biological testing.
Identification of Metabolite Produced.
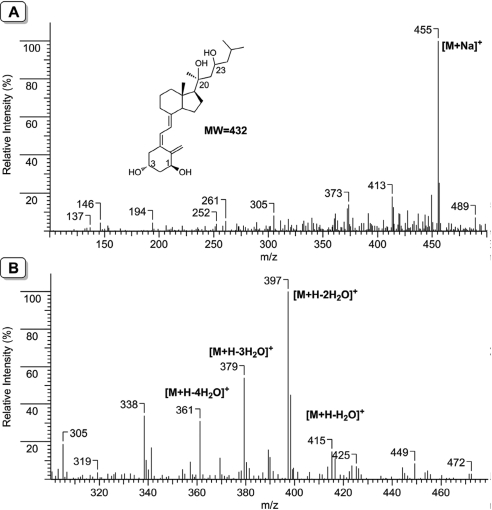
Mass spectrometry and NMR analysis confirmed that CYP27B1-catalyzed 1α-hydroxylation of 20,23(OH)2D3 with the product identified as 1α,20,23-trihydroxyvitamin D3. The dominant molecular ion was the sodium adduct with m/z = 455 ([M + Na]+, mol. wt. = 432) measured with an ESI source (soft ionization method) (Fig. 2A). When measured with an APCI source (hard ionization method), the molecular ion was not observed, but fragments corresponding to sequential neutral loss of water molecules were observed (Fig. 2B). These fragments include m/z = 415 ([M + H − H2O]+), m/z = 397 ([M + H − 2H2O]+), m/z = 379 ([M + H − 3H2O]+), and m/z = 361 ([M + H − 4H2O]+), consistent with the presence of a total of four hydroxyl groups in this metabolite.
Fig. 2.
Mass spectra of 1,20,23(OH)3D3. A, ESI source showing the molecular ion. B, APCI source showing the progressive loss of water molecules. Both measurements were done in the positive ion mode.
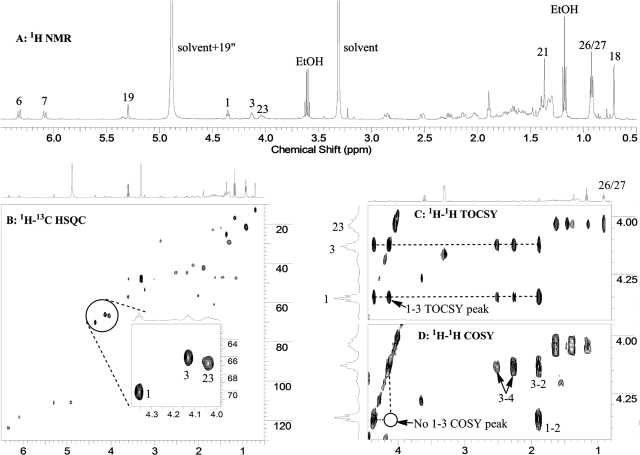
The new hydroxylation position was unambiguously established by analysis of the NMR spectra for this metabolite. As shown in Fig. 3, all four methyl groups (18-, 21-, 26-, and 27-Me) are intact (Fig. 3A), and there are three characteristic chemical shifts (4.04, 4.13, and 4.36 ppm) corresponding to three methine groups, each bearing a hydroxyl group. These three hydroxyl-bearing methines are further confirmed by the 1H-13C heteronuclear single quantum correlation (HSQC) spectrum (Fig. 3B and its inset). Unlike hydroxylation at C3 and C23, hydroxylation at C20 results in a tertiary carbon, which does not have a signal in either the one-dimensional proton or 2D 1H-13C HSQC spectrum; therefore, the new hydroxyl group must be on a methylene group in 20,23(OH)2D3. Examination of the 1H-1H total correlation spectroscopy (TOCSY) spectrum (Fig. 3C) clearly reveals that the proton at 4.04 ppm is in the side chain (i.e., 23-hydroxylation) because it belongs to the same spin network with 26 of 27 methyl groups, whereas the other two (4.13 and 4.36 ppm) are from the same ring because they have the same TOCSY correlation peaks. Because one of these two comes from the methine proton at C3, the new hydroxyl group has to be on the A-ring. Although these two protons belong to the same spin network as shown in TOCSY, they are not neighbors because they do not couple with each other as indicated by the 1H-1H correlation (correlation spectroscopy) NMR spectrum (Fig. 3D). This is only possible if the new hydroxylation has occurred at the C1 position. The methine proton at the C1 position is a triplet, with a coupling constant of 5.7 Hz to the two protons at C2. This small coupling constant indicates its equatorial (1α-hydroxyl configuration) rather than axial (1β-hydroxyl configuration) position, which would result in a much larger coupling constant. The above analysis unambiguously indicates that the metabolite is 1α,20,23-trihydroxyvitamin D3. Full assignments of proton and carbon chemical shifts for this metabolite are shown in Table 1. For comparison, chemical shifts for D3 are also presented in the table.
Fig. 3.
One-dimensional proton (A), 2D 1H-13C HSQC (B), 2D 1H-1H TOCSY (C), and 2D 1H-1H correlation spectroscopy (COSY) (D) NMR spectra of the metabolite. EtOH, ethanol.
TABLE 1.
NMR chemical shift assignments for 1,20,23(OH)3D3 and vitamin D3 from analysis of their NMR spectra, and carbon chemical shifts obtained from detected signals in HSQC and heteronuclear multiple bond correlation experiments
Solvent was CD3OD.
| Atom | 1,20,23(OH)3D3 |
D3 |
||
|---|---|---|---|---|
| 1H | 13C | 1H | 13C | |
| 1 | 4.36 β (t, J = 5.7Hz) | 70.1 | 2.12 α, 2.40 β | 33.8 |
| 2 | 1.89 α, 1.89 β | 42.3 | 1.98 α, 1.54 β | 36.8 |
| 3 | 4.13 | 66.1 | 3.76 | 70.7 |
| 4 | 2.52 α, 2.27 β | 44.9 | 2.53 α, 2.19 β | 47.2 |
| 5 | N.A. | 134.5 | N.A. | 137.5 |
| 6 | 6.33 | 123.7 | 6.22 | 122.8 |
| 7 | 6.09 | 118.1 | 6.04 | 119.1 |
| 8 | N.A. | —a | N.A. | 142.7 |
| 9 | 1.68 α, 2.87 β | 28.7 | 1.69 α, 2.86 β | 30.1 |
| 10 | N.A. | 148.6 | N.A. | 147.1 |
| 11 | — | — | 1.55 α, 1.67 β | 24.7 |
| 12 | 1.39 α, 2.13 β | 41.1 | 1.33 α, 2.03 β | 42.1 |
| 13 | N.A. | 45.9 | N.A. | 47.1 |
| 14 | 2.01 | 56.5 | 2.02 | 57.7 |
| 15 | 1.52 α, 1.52 β | 21.7 | 1.46 α, 1.46 β | 23.4 |
| 16 | 1.75 α, 1.83 β | 21.8 | 1.31 α, 1.91 β | 28.9 |
| 17 | 1.65 | 60.8 | 1.30 | 58.1 |
| 18 | 0.71 | 12.9 | 0.56 | 12.5 |
| 19 | 4.91, 5.30 | 110.8 | 4.75, 5.04 | 112.8 |
| 20 | N.A. | 75.9 | 1.40 | 37.6 |
| 21 | 1.37 | 25.3 | 0.95 | 19.6 |
| 22 | 1.64, 1.46 | 47.1 | 1.39, 1.04 | 37.5 |
| 23 | 4.04 | 66.8 | 1.39, 1.19 | 25.1 |
| 24 | 1.16, 1.39 | 47.6 | 1.16 | 40.8 |
| 25 | 1.73 | 24.1 | 1.54 | 29.3 |
| 26,27 | 0.92 | 21.5 | 0.89 | 23.2 |
N.A., not applicable (ternary carbons).
—, chemical shifts could not be unambiguously determined because of overlapping or weak signals at this position.
1,20,23(OH)3D3 Inhibits Proliferation of Keratinocytes.
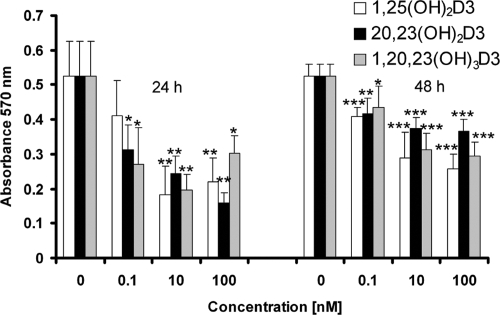
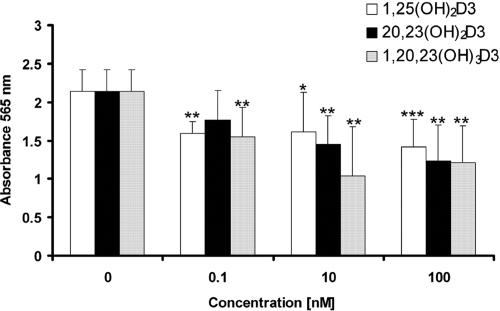
20,23(OH)2D3 and 1,20,23(OH)3D3 significantly inhibited proliferation of human epidermal keratinocytes in a time- and dose-dependent fashion (Fig. 4). Both compounds showed significant effects at a lower concentration (0.1 nM) than that seen for 1,25(OH)2D3 at 24 h of treatment. Both compounds also decreased cell biomass relative to that in control cells, measured by staining with sulforhodamine B (Fig. 5). The degree of inhibition was similar to that seen with 1,25(OH)2D3.
Fig. 4.
1,20,23(OH)3D3 inhibits HEKn proliferation. HEKn cells were seeded in 96-well plates and treated with a range of concentrations of 1,25(OH)2D3, 20,23(OH)2D3, or 1,20,23(OH)3D3 or with ethanol vehicle for 24 or 48 h. Uptake of MTT reagent was measured from the formazan product measured at 570 nm. n = 6. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
Fig. 5.
Effect of 1,20,23(OH)3D3 on HEKn biomass. HEKn cells were seeded in 96-well plates and were treated for 24 h with a range of concentrations of 1,25(OH)2D3, 20,23(OH)2D3, or 1,20,23(OH)3D3, or with ethanol vehicle. Cells were stained with sulforhodamine B and lysed, and the absorbance was measured at 565 nm. n = 6. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus control.
Effects of 1,20,23(OH)3D3 on the Expression of the Vitamin D Receptor and CYP24.
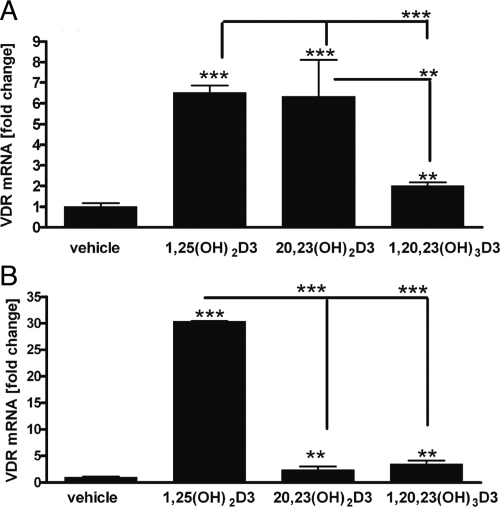
After 6 h of treatment of HEKn cells, both 1,25(OH)2D3 and 20,23(OH)2D3 caused a 6-fold increase in the expression of mRNA for the VDR, whereas 1,20,23(OH)3D3 caused only a doubling compared with vehicle-treated control cells (Fig. 6). After 24 h of treatment of cells with 1,25(OH)2D3, VDR expression at the mRNA level was increased 30-fold compared with that in control cells. In contrast, both 20,23(OH)2D3 and 1,20,23(OH)3D3 caused only a 2- to 3-fold stimulation; however, the stimulation was statistically significant. The data show that the effect of 20,23(OH)2D3 is not sustained from 6 to 24 h, possibly due to the metabolism of this compound by CYP24 or other vitamin D hydroxylases in keratinocytes (Bikle et al., 1986; Seifert et al., 2009).
Fig. 6.
Effect of 1,20,23(OH)3D3 on VDR expression. RNA from HEKn cells treated with 1,25(OH)2D3, 20,23(OH)2D3, or 1,20,23(OH)3D3 for 6 h (A) or 24 h (B) was extracted and reverse-transcribed. Real-time PCR was performed using TaqMan PCR Master Mix and primers specific for the VDR gene. The amounts of VDR mRNA were normalized by the comparative Ct method, using cyclophilin B as a housekeeping gene. n = 3. **, P < 0.01; ***, P < 0.001.
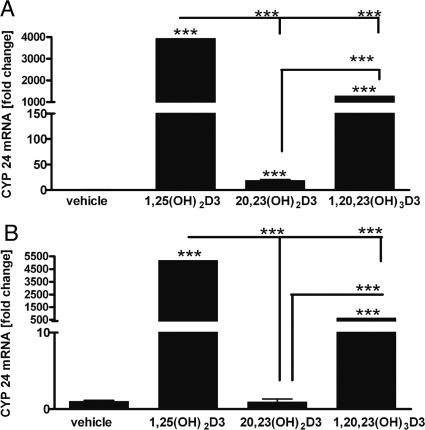
As shown in Fig. 7 and reported by us previously (Janjetovic et al., 2010), 20,23(OH)2D3 is a poor stimulator of the expression of the CYP24 gene, which encodes the 24-hydroxylase that inactivates 1,25(OH)2D3. In contrast, addition of the 1α-hydroxyl group to this compound greatly enhanced its ability to stimulate the expression of mRNA for CYP24, but not to the extent seen with 1,25(OH)2D3.
Fig. 7.
Effect of 1,20,23(OH)3D3 on CYP24 expression. HEKn cells were treated for 6 h (A) or 24 h (B) as for Fig. 6, and the RNA was extracted and reverse-transcribed. Real-time PCR was performed using TaqMan PCR Master Mix and primers specific for the CYP24 gene. The amounts of CYP24 mRNA were normalized by the comparative Ct method, using cyclophilin B as a housekeeping gene. n = 3. ***, P < 0.001.
Discussion
We have recently documented that the cytochrome P450scc-derived vitamin D analogs, 20(OH)D3 and 20,23(OH)2D3, possess many of the properties of 1,25(OH)2D3 such as the inhibition of proliferation, the promotion of differentiation, and suppression of inflammation, despite lacking the 1α-hydroxyl group usually seen in vitamin D receptor agonists (Zbytek et al., 2008; Janjetovic et al., 2009, 2010; Slominski et al., 2010). Of particular note is our finding that 20(OH)D3 lacks calcemic activity in rats but adding a 1α-hydroxyl group to it partially returns the calcemic activity (Slominski et al., 2010). To fully characterize these new P450scc-derived hydroxyvitamin D compounds, the effect of adding a 1α-hydroxyl group is therefore essential. Furthermore, because this addition may occur in vivo, it is of interest to know the activity of CYP27B1 on these new hydroxyvitamin D3 compounds. The expression of CYP27B1 in E. coli and its extraction and purification in the active form (Tang et al., 2010) has permitted an enzymatic approach to the synthesis of 1α-hydroxy derivatives of 20(OH)D3 and 20,23(OH)2D3. In the current study, we have successfully used expressed CYP27B1 to convert 20,23(OH)2D3 to 1,20,23(OH)3D3 in amounts sufficient for confirmation of its structure by mass spectrometry and NMR and biological testing on keratinocytes.
Kinetic analysis of the 1α-hydroxylation of 20,23(OH)2D3 incorporated into phospholipid vesicles, which mimic the inner mitochondrial membrane environment of the CYP27B1, has revealed that it is a relatively poor substrate for the enzyme. It has a kcat/Km value 40-fold lower than that for 25(OH)D3 resulting from both a higher Km and lower kcat. Therefore, we would predict relatively slow metabolism of 20,23(OH)2D3 in vivo compared with that of 25(OH)D3. The results of this study and previous ones looking at the substrate specificity of CYP27B1 clearly show that the activity of the enzyme is sensitive to the position of hydroxyl groups on the vitamin D3 side chain (Sakaki et al., 1999; Tang et al., 2010). In phospholipid vesicles, 20(OH)D3, 20,23(OH)2D3, and 24,25(OH)2D3 are all metabolized with kcat/Km values 3- to 40-fold lower than that for 25(OH)D3. In contrast, the presence of a double bond between carbons 22 and 23 on the side chain and a methyl group at C24, as seen in 25-hydroxyvitamin D2, have no significant effect on the kinetics of the 1α-hydroxylation (Tang et al., 2010). With all of the substrates mentioned above, none of the structural changes altered the site of hydroxylation, which was always the 1α-position.
Despite the relatively low activity of CYP27B1 toward 20,23(OH)2D3, greater than 75% conversion to product was obtained by using a high concentration of enzyme (0.8 μM) and a substrate concentration near its Km. The amount of expressed CYP27B1 required to produce the 130 μg of HPLC-purified product from a 40-ml incubation was obtained from approximately 2 liters of bacterial culture (Tang et al., 2010). Because of the instability of CYP27B1 during the 1-h incubation, we used 25°C rather than 37°C for the enzymatic synthesis of the 1,20,23(OH)3D3.
1,20,23(OH)3D3, retained some, but not all, of the biological properties exhibited by the parent compound on keratinocytes. Only minor differences in the inhibition of keratinocyte proliferation between 20,23(OH)2D3 and 1,20,23(OH)3D3 were observed. Both compounds significantly inhibited proliferation at a concentration as low as 0.1 nM with the effect being greater at 24 than at 48 h of treatment. A clear difference between the abilities of 20,23(OH)2D3 and 1,20,23(OH)3D3 to stimulate the expression of mRNA for VDR was seen at 6 h of treatment with the 1,20,23(OH)3D3 being a poor stimulator, but by 24 h of treatment both compounds caused minimal stimulation compared with that seen with 1,25(OH)2D3. A large difference in the abilities of 20,23(OH)2D3 and 1,20,23(OH)3D3 to stimulate the expression of CYP24 at the mRNA level was observed, with 20,23(OH)2D3 being a poor inducer compared with 1,20,23(OH)3D3 and 1,25(OH)2D3. We reported previously that 20(OH)D3 is a poor inducer of CYP24 expression compared with 1,20(OH)2D3 (Tuckey et al., 2008a; Zbytek et al., 2008). Taken together, these studies provide strong evidence that the 1α-hydroxyl group is required in vitamin D receptor agonists for robust stimulation of the expression of CYP24, the enzyme catalyzing 1,25(OH)2D3 inactivation. Partial receptor agonists lacking this functional group such as 20(OH)D3 and 20,23(OH)2D3 may therefore offer some advantage in therapeutic use by not rapidly inducing their own inactivation or that of 1,25(OH)2D3 either.
We have reported that 20(OH)D3 lacks calcemic activity in rats but 1,20(OH)2D3 does exhibit some calcemic activity, although less than that for 1,25(OH)2D3 (Slominski et al., 2010). Therefore, there appears to be a parallel requirement for the 1α-hydroxyl group for both calcemic activity and stimulation of CYP24 expression. The ability of expressed mouse CYP27B1 to add a 1α-hydroxyl group to vitamin D analogs lacking this functional group provides us with a tool to further explore this relationship in the future, particularly for 20,23(OH)2D3 and other P450scc-derived vitamin D metabolites.
This work was supported by the National Institutes of Health National Institute of Arthritis and Musculoskeletal and Skin Diseases [Grant R01-AR052190]; and by The University of Western Australia.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.034389.
- 1,25(OH)2D3
- 1α,25-dihydroxyvitamin D3
- 20(OH)D3
- 20-hydroxyvitamin D3
- 20,23(OH)2D3
- 20,23-dihydroxyvitamin D3
- P450
- cytochrome P450
- 1,20(OH)2D3
- 1α,20-dihydroxyvitamin D3
- 25(OH)D3
- 25-hydroxyvitamin D3
- 1,20,23(OH)3D3
- 1α,20,23-trihydroxyvitamin D3
- HPLC
- high-performance liquid chromatography
- ESI
- electrospray ionization
- APCI
- atmospheric pressure chemical ionization
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- HEKn
- neonatal human epidermal keratinocyte(s)
- PCR
- polymerase chain reaction
- VDR
- vitamin D receptor
- HSQC
- heteronuclear single quantum correlation
- 2D
- two-dimensional
- TOCSY
- total correlation spectroscopy
- Ct
- cycle threshold.
References
- Anderson PH, Hendrix I, Sawyer RK, Zarrinkalam R, Manavis J, Sarvestani GT, May BK, Morris HA. (2008) Co-expression of CYP27B1 enzyme with the 1.5 kb CYP27B1 promoter-luciferase transgene in the mouse. Mol Cell Endocrinol 285:1–9 [DOI] [PubMed] [Google Scholar]
- Bikle D. (2009) Nonclassic actions of vitamin D. J Clin Endocrinol Metab 94:26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikle DD, Nemanic MK, Gee E, Elias P. (1986) 1,25-Dihydroxyvitamin D3 production by human keratinocytes. Kinetics and regulation. J Clin Invest 78:557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillon R, Eelen G, Verlinden L, Mathieu C, Carmeliet G, Verstuyf A. (2006) Vitamin D and cancer. J Steroid Biochem Mol Biol 102:156–162 [DOI] [PubMed] [Google Scholar]
- Guryev O, Carvalho RA, Usanov S, Gilep A, Estabrook RW. (2003) A pathway for the metabolism of vitamin D3: unique hydroxylated metabolites formed during catalysis with cytochrome P450scc (CYP11A1). Proc Natl Acad Sci USA 100:14754–14759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headlam MJ, Wilce MC, Tuckey RC. (2003) The F-G loop region of cytochrome P450scc (CYP11A1) interacts with the phospholipid membrane. Biochim Biophys Acta 1617:96–108 [DOI] [PubMed] [Google Scholar]
- Holick MF. (2007) Vitamin D deficiency. N Engl J Med 357:266–281 [DOI] [PubMed] [Google Scholar]
- Janjetovic Z, Tuckey RC, Nguyen MN, Thorpe EM, Jr, Slominski AT. (2010) 20,23-Dihydroxyvitamin D3, novel P450scc product, stimulates differentiation and inhibits proliferation and NF-κB activity in human keratinocytes. J Cell Physiol 223:36–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjetovic Z, Zmijewski MA, Tuckey RC, DeLeon DA, Nguyen MN, Pfeffer LM, Slominski AT. (2009) 20-Hydroxycholecalciferol, product of vitamin D3 hydroxylation by P450scc, decreases NF-κB activity by increasing IκBα levels in human keratinocytes. PLoS ONE 4:e5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambeth JD, Kitchen SE, Farooqui AA, Tuckey R, Kamin H. (1982) Cytochrome P-450scc-substrate interactions. Studies of binding and catalytic activity using hydroxycholesterols. J Biol Chem 257:1876–1884 [PubMed] [Google Scholar]
- Lee HJ, Paul S, Atalla N, Thomas PE, Lin X, Yang I, Buckley B, Lu G, Zheng X, Lou Y-R, et al. (2008) Gemini vitamin D analogues inhibit estrogen receptor-positive and estrogen receptor-negative mammary tumorigenesis without hypercalcemic toxicity. Cancer Prev Res (Phila Pa) 1:476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Jones G. (2006) Promise of vitamin D analogues in the treatment of hyperproliferative conditions. Mol Cancer Ther 5:797–808 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Eto TA, Taniguchi T, Miyamoto K, Nagatomo J, Shiotsuki H, Sueta H, Higashi S, Okuda KI, Setoguchi T. (1997) Purification and characterization of 25-hydroxyvitamin D3 1α-hydroxylase from rat kidney mitochondria. FEBS Lett 419:45–48 [DOI] [PubMed] [Google Scholar]
- Nguyen MN, Slominski A, Li W, Ng YR, Tuckey RC. (2009) Metabolism of vitamin D2 to 17,20,24-trihydroxyvitamin D2 by cytochrome P450scc (CYP11A1). Drug Metab Dispos 37:761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DE, Jones G. (2004) Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci 29:664–673 [DOI] [PubMed] [Google Scholar]
- Sakaki T, Sawada N, Takeyama K, Kato S, Inouye K. (1999) Enzymatic properties of mouse 25-hydroxyvitamin D3 1α-hydroxylase expressed in Escherichia coli. Eur J Biochem 259:731–738 [DOI] [PubMed] [Google Scholar]
- Seifert M, Tilgen W, Reichrath J. (2009) Expression of 25-hydroxyvitamin D-1α-hydroxylase (1αOHase, CYP27B1) splice variants in HaCaT keratinocytes and other skin cells: modulation by culture conditions and UV-B treatment in vitro. Anticancer Res 29:3659–3667 [PubMed] [Google Scholar]
- Slominski A, Semak I, Wortsman J, Zjawiony J, Li W, Zbytek B, Tuckey RC. (2006) An alternative pathway of vitamin D2 metabolism. Cytochrome P450scc (CYP11A1)-mediated conversion to 20-hydroxyvitamin D2 and 17,20-dihydroxyvitamin D2. FEBS J 273:2891–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Semak I, Zjawiony J, Wortsman J, Li W, Szczesniewski A, Tuckey RC. (2005) The cytochrome P450scc system opens an alternate pathway of vitamin D3 metabolism. FEBS J 272:4080–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski AT, Janjetovic Z, Fuller BE, Zmijewski MA, Tuckey RC, Nguyen MN, Sweatman T, Li W, Zjawiony J, Miller D, et al. (2010) Products of vitamin D3 or 7-dehydrocholesterol metabolism by cytochrome P450scc show anti-leukemia effects, having low or absent calcemic activity. PLoS ONE 5:e9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Morikawa K. (2006) Vitamin D receptor agonists: opportunities and challenges in drug discovery. Curr Top Med Chem 6:1303–1316 [DOI] [PubMed] [Google Scholar]
- Tang EK, Voo KJ, Nguyen MN, Tuckey RC. (2010) Metabolism of substrates incorporated into phospholipid vesicles by mouse 25-hydroxyvitamin D3 1α-hydroxylase (CYP27B1). J Steroid Biochem Mol Biol 119:171–179 [DOI] [PubMed] [Google Scholar]
- Tuckey RC. (2005) Progesterone synthesis by the human placenta. Placenta 26:273–281 [DOI] [PubMed] [Google Scholar]
- Tuckey RC, Janjetovic Z, Li W, Nguyen MN, Zmijewski MA, Zjawiony J, Slominski A. (2008a) Metabolism of 1α-hydroxyvitamin D3 by cytochrome P450scc to biologically active 1α,20-dihydroxyvitamin D3. J Steroid Biochem Mol Biol 112:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckey RC, Li W, Zjawiony JK, Zmijewski MA, Nguyen MN, Sweatman T, Miller D, Slominski A. (2008b) Pathways and products for the metabolism of vitamin D3 by cytochrome P450scc. FEBS J 275:2585–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckey RC, Nguyen MN, Slominski A. (2008c) Kinetics of vitamin D3 metabolism by cytochrome P450scc (CYP11A1) in phospholipid vesicles and cyclodextrin. Int J Biochem Cell Biol 40:2619–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckey RC, Sadleir J. (1999) The concentration of adrenodoxin reductase limits cytochrome P450scc activity in the human placenta. Eur J Biochem 263:319–325 [DOI] [PubMed] [Google Scholar]
- Uchida E, Kagawa N, Sakaki T, Urushino N, Sawada N, Kamakura M, Ohta M, Kato S, Inouye K. (2004) Purification and characterization of mouse CYP27B1 overproduced by an Escherichia coli system coexpressing molecular chaperonins GroEL/ES. Biochem Biophys Res Commun 323:505–511 [DOI] [PubMed] [Google Scholar]
- Woods ST, Sadleir J, Downs T, Triantopoulos T, Headlam MJ, Tuckey RC. (1998) Expression of catalytically active human cytochrome P450scc in Escherichia coli and mutagenesis of isoleucine-462. Arch Biochem Biophys 353:109–115 [DOI] [PubMed] [Google Scholar]
- Zbytek B, Janjetovic Z, Tuckey RC, Zmijewski MA, Sweatman TW, Jones E, Nguyen MN, Slominski AT. (2008) 20-Hydroxyvitamin D3, a product of vitamin D3 hydroxylation by cytochrome P450scc, stimulates keratinocyte differentiation. J Invest Dermatol 128:2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]