Abstract
The human pregnane X receptor (hPXR) regulates the expression of CYP3A4, which plays a vital role in hepatic drug metabolism and has considerably reduced expression levels in proliferating hepatocytes. We have recently shown that cyclin-dependent kinase 2 (CDK2) negatively regulates hPXR-mediated CYP3A4 gene expression. CDK2 can be dephosphorylated and inactivated by protein phosphatase type 2C beta isoform long (PP2Cβl), a unique phosphatase that was originally cloned from human liver. In this study, we sought to determine whether PP2Cβl is involved in regulating hPXR's transactivation activity and whether PP2Cβl affects CDK2 regulation of this activity in HepG2 liver carcinoma cells. In transactivation assays, transiently coexpressed PP2Cβl significantly enhanced the hPXR-mediated CYP3A4 promoter activity and decreased the inhibitory effect of CDK2 on hPXR transactivation activity. In addition, shRNA-mediated down-regulation of endogenous PP2Cβl promoted cell proliferation, inhibited the interaction of hPXR with steroid receptor coactivator-1, and attenuated the hPXR transcriptional activity. The levels of PP2Cβl did not affect hPXR expression. Our results show for the first time that PP2Cβl is essential for hPXR activity and can positively regulate this activity by counteracting the inhibitory effect of CDK2. Our results implicate a novel and important role for PP2Cβl in regulating hPXR activity and CYP3A4 expression by inhibiting or desensitizing signaling pathways that negatively regulate the function of pregnane X receptor in liver cells and are consistent with the notion that both the activity of hPXR and the expression of CYP3A4 are regulated in a cell cycle-dependent and cell proliferation-dependent manner.
Introduction
The human pregnane X receptor (hPXR) plays a central role in activating the gene expression of cytochrome P450 (P450) enzymes in the human liver and other organs (Harmsen et al., 2007). CYP3A4, one of the most important P450s in humans, catalyzes the metabolism of more than 50% of clinically used drugs (Guengerich, 1999; Harmsen et al., 2007; Zhou, 2008). The master regulator of CYP3A4 gene expression, pregnane X receptor (PXR), is a member of the nuclear receptor (NR) superfamily of ligand-activated transcription factors and is activated by binding to various chemically and structurally distinct endobiotics and xenobiotics, including clinically used drugs (Kliewer et al., 1998; Lehmann et al., 1998; Harmsen et al., 2007).
The transcriptional activity of PXR is modulated not only by conventional ligand binding but also by cellular signaling pathways. Recent studies demonstrated a role for phosphorylation-dependent signaling events in regulating PXR-mediated CYP gene expression (Pondugula et al., 2009a). Kinases such as protein kinase A (Ding and Staudinger, 2005a; Lichti-Kaiser et al., 2009), protein kinase C (Ding and Staudinger, 2005b), cyclin-dependent kinase (CDK)2 (Lin et al., 2008), and p70 ribosomal S6 kinase (Pondugula et al., 2009b) phosphorylate PXR and regulate PXR-mediated CYP gene expression. Furthermore, CDK1, casein kinase II, and glycogen synthase kinase 3 also phosphorylate PXR (Lichti-Kaiser et al., 2009), although the functional significance of these phosphorylations is unknown.
Because phosphorylation regulates PXR function, it is logical to speculate that phosphatases are directly or indirectly involved in regulating PXR function by inhibiting the kinase pathways. However, in comparison to the understanding of the role of kinase signaling, there is only a meager understanding of the extent to which PXR is regulated by phosphatase signaling. For instance, okadaic acid, a nonspecific phosphatase inhibitor, affects PXR's transcriptional activity in cell-based gene reporter assays (Ding and Staudinger, 2005b), suggesting that okadaic acid-sensitive protein phosphatases (i.e., PP1 and PP2A) are involved in regulating PXR-mediated CYP gene expression, yet the exact mechanism remains unknown. It is important to more fully understand the contribution of phosphatases in regulating PXR function to comprehensively address the role of reversible phosphorylation in regulating PXR-mediated P450 expression.
Multiple research groups have established that CYP3A4 expression is significantly reduced in proliferating human liver cells (Pondugula et al., 2009a), strongly suggesting a link between cell cycle regulation and CYP3A4 expression. In fact, we have recently shown that CDK2 negatively regulates hPXR-mediated CYP3A4 gene expression in actively dividing HepG2 cells (Lin et al., 2008). However, protein phosphatase type 2C isoform beta long (PP2Cβl; a.k.a., PP2Cβ2 or PP2Cβx) can dephosphorylate phosphothreonine-160 and inactivate CDK2 (Cheng et al., 1999, 2000). Thus, in contrast to CDK2, which promotes cell proliferation, PP2Cβl arrests cell growth and promotes apoptosis (Seroussi et al., 2001; Klumpp et al., 2006; Tamura et al., 2006).
PP2Cβl's expression in the liver (Marley et al., 1998) and its inhibitory effect on CDK2, a negative regulator of PXR, led us to investigate the role of PP2Cβl in regulating the transcriptional activity of hPXR via CDK2 in actively proliferating liver cells. In this study, we sought to determine whether PP2Cβl is involved in regulating hPXR-mediated CYP3A4 gene expression and whether PP2Cβl counteracts the inhibitory effect of CDK2 on hPXR function in HepG2 cells.
Materials and Methods
Cell Culture, Plasmids, and Transient Transfections.
HepG2 human liver carcinoma cell line was propagated as described previously (Lin et al., 2008; Pondugula et al., 2009b) in growth media containing Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 100 U/ml penicillin (Invitrogen), 100 μg/ml streptomycin (Invitrogen), 2 mM l-glutamine (Invitrogen), and 1 mM sodium pyruvate (Invitrogen). The pcDNA3-hPXR, FLAG-pcDNA3-hPXR, and pGL3-CYP3A4-luc (CYP3A4 luciferase reporter) plasmids were generated as described previously (Lin et al., 2008). The pcDNA3-PP2Cβl plasmid was a gift from Dr. Sara Lavi (Department of Cell Research and Immunology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel). V5-CDK2 and V5-cyclin E constructs were gifts from Dr. Haojie Huang (Masonic Cancer Center and Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN). The CMV-Renilla luciferase plasmid was purchased from Promega (Madison, WI). Transient transfections were performed using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) as described previously (Lin et al., 2008; Pondugula et al., 2009b). In brief, transfections were performed in 6-well culture plates with 2 μg of total plasmid DNA per well. Cells to be used for hPXR transactivation and protein expression experiments were transfected with the following plasmids: hPXR (100 ng), CDK2 (200 ng), cyclin E (200 ng), PP2Cβl (400 ng), CMV-Renilla (100 ng), and pGL3-CYP3A4-luc (1000 ng). Additional pcDNA3 was added to ensure that the total plasmid DNA in each transfection reaction was 2 μg. Twenty-four hours after transfection in growth media, the cells to be used for luciferase assays or Western blot analyses were grown in assay media (Dulbecco's modified Eagle's medium with 5% charcoal/dextran-treated fetal bovine serum) and then treated with either 0.1% dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO) or 2 μM rifampicin (Sigma-Aldrich) for an additional 6 h before performing luciferase assays for transactivation analyses and harvesting of cell lysates for Western blot analyses.
PXR Transactivation Assay of Transiently Transfected HepG2 Cells.
After transfection, approximately 10,000 live cells were plated in each well of a 96-well culture plate (PerkinElmer Life and Analytical Sciences, Waltham, MA) and prepared for assaying as described above. The Dual-Glo Luciferase Assays (Promega) were performed to measure the luminescence using a PHERAstar microplate reader (BMG Labtech, Durham, NC). The activity of firefly luciferase was normalized to that of the Renilla and expressed in relative luminescence units.
Western Blot Analysis.
Transiently transfected HepG2 cells were lysed in radioimmunoprecipitation assay buffer, and then approximately 60 μg of total protein was resolved on NuPage 4 to 12% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes. The membranes were blocked for 2 h and then incubated overnight at 4°C with primary antibodies against PXR (rabbit polyclonal anti-hPXR serum (Saradhi et al., 2005) at a 1:10,000 dilution; PP2Cβl (rabbit polyclonal anti-PP2Cβ; Bethyl Laboratories, Montgomery, TX) at a 1:2000 dilution; or actin (mouse monoclonal anti-β-actin; Sigma-Aldrich) at a 1:5000 dilution. Infrared dye-conjugated secondary goat antibodies (anti-rabbit or anti-mouse) (LI-COR Biosciences, Lincoln, NE) were used at a 1:10,000 dilution, and antigen-antibody interactions were visualized using an Odyssey infrared imager (LI-COR Biosciences). The membranes that were probed for hPXR or PP2Cβl were stripped and reprobed using the anti-actin antibody to confirm equal loading of total protein in each lane.
Silencing PP2Cβl and Transactivation Assays.
HepG2 cells stably expressing hPXR and CYP3A4-luciferase (clone 1) (Lin et al., 2008) were transduced with either control nontarget lentiviral short hairpin RNA (shRNA) (Sigma-Aldrich) or PP2Cβl shRNA (Sigma-Aldrich; GenBank accession number NM_002706; The RNAi Consortium number TRCN0000002449; GenBank accession number NM_002706.x-1252s1c1) according to the manufacturer's instructions. Stably transduced cells were selected by using puromycin. Pools of the cells stably expressing either control (hereafter referred to as HepG2/Control shRNA) or PP2Cβl shRNA (hereafter referred to as HepG2/PP2Cβl shRNA) were used for transactivation assays, real-time reverse transcription-polymerase chain reaction (PCR) analyses, cell cycle analyses, mammalian two-hybrid assays, and real-time cell-based electronic sensing (RT-CES) assays.
For transactivation assays, approximately 10,000 live cells were seeded in each well of a 96-well culture plate (PerkinElmer Life and Analytical Sciences) containing either 0.1% DMSO or 2 μM rifampicin and grown for 24 h in assay media before measuring both the cell viability using the Alamar Blue assay system (Invitrogen) and the firefly luciferase activity using the Steady Lite Luciferase Assay System (PerkinElmer Life and Analytical Sciences). The firefly luciferase activity was normalized to the Alamar Blue cell viability to determine relative luciferase activity (RLA).
To determine whether overexpressing PP2Cβl can rescue the effect of PP2Cβl knockdown on hPXR, HepG2/PP2Cβl shRNA cells were transiently transfected with 100 ng CMV-Renilla and either 600 ng pcDNA3 or 600 ng pcDNA3-PP2Cβl. Additional pcDNA3 was added to ensure that the total plasmid DNA in each transfection reaction was 2 μg. After 24-h transfection, the cells were plated in assay medium and treated with 0.1% DMSO or 2 μM rifampicin for 24 h before performing the Dual-Glo Luciferase Assays.
Mammalian Two-Hybrid Assays.
The mammalian two-hybrid assays were performed as described previously to investigate the effect of PP2Cβl on the interaction between hPXR and steroid receptor coactivator-1 (SRC-1) (Pondugula et al., 2009b). In brief, pBIND-SRC-1 (Gal4-SRC-1) (500 ng) and a luciferase reporter pG5-luc (1000 ng), together with either pACT (empty vector) (200 ng), pACT-hPXR (VP16-hPXR) (200 ng), or pcDNA3-PP2Cβl (600 ng), as indicated, were cotransfected into HepG2/Control shRNA or HepG2/PP2Cβl shRNA cells for 24 h. pcDNA3 was added to ensure that the total plasmid DNA in each transfection reaction was 2.5 μg. The cells were then treated with DMSO or 5 μM rifampicin for 24 h before luciferase assays. pACT-hPXR contains the full-length hPXR fused to VP16. pBIND-SRC-1 contains the nuclear receptor interaction domain of SRC-1 (amino acids 621–765) fused to Gal4. pG5-luc contains the luciferase reporter gene controlled by a promoter containing Gal4 DNA binding sites. In the absence of pG5-luc, the luciferase signal decreased by >90% (data not shown), suggesting that the system is suitable for detecting the activity of pG5-luc. In this system, recruitment of VP16-hPXR to Gal4-SRC-1 enhances the expression of pG5-luc. The Gal4 vector (pBIND) also constitutively expresses Renilla luciferase to serve as an internal transfection control. Dual-Glo Luciferase Assay system was used to measure both the firefly and Renilla luciferase activities. Firefly luciferase activity from pG5-luc was normalized using Renilla luciferase activity. The relative rifampicin-induced pG5-luc activity was expressed as a fold induction over DMSO and was used to measure the rifampicin-induced hPXR/SRC-1 interaction. The data are shown as means and S.D.s obtained from eight independent observations. The p value was determined using Student's t test by comparing the relative rifampicin-induced pG5-luc activity in the absence or presence of hPXR cotransfection and also in the absence or presence of PP2Cβl when hPXR is cotransfected. Differences were considered significant for p < 0.05, 0.01, or 0.001, and nonsignificant for p > 0.05.
Real-Time Reverse Transcription-Polymerase Chain Reaction.
Total RNA was extracted from HepG2 cells by using the RNeasy Mini Kit (QIAGEN, Valencia, CA), and the quality and quantity of the total RNA were determined using a Nanodrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE). Reverse transcription was performed at 42°C for 30 min to synthesize cDNA from the mRNA from 50 ng of total RNA by using the QuantiTect Reverse Transcription Kit (QIAGEN). Real-time PCR was performed on this cDNA by using the QuantiTect SYBR Green Kit (QIAGEN) and iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Transcripts of the 18S small subunit ribosomal RNA (18S rRNA) housekeeping gene and hPXR were amplified using gene-specific primers. The 18S rRNA primers (forward: 5′-GAGGTTCGAAGACGATCAGA-3′ and reverse: 5′-TCGCTCCACCAACTAAGAAC-3′) were designed based on the sequence of GeneBank accession number BK000964 to generate a 315-base pair PCR product. hPXR primers (forward: 5′-TGTTCAAAGGCATCATCAGC-3′ and reverse: 5′-TCTGGGGAGAAGAGGGAGAT-3′) were designed based on the sequence of GenBank accession number AF061056 to generate a 307-base pair PCR product. Each of the 40 PCR cycles was comprised of 95°C for 15 s, 55°C for 30 s, and 72°C for 30 s. PCR was followed by a melt cycle at 65°C to 95°C. The sample's fluorescence was read at 2°C to 3°C below the melting temperature peak of that specific cDNA sequence to exclude contributions from nonspecific sources. To exclude the possibility of genomic DNA amplification during the PCR reaction, control reactions that had not been reverse transcribed (−RT) were performed and only accepted when the threshold value (Ct) was at least 20 cycles more than that of the +RT run. PCR products were electrophoresed on 2% agarose gels and detected by ethidium bromide. These products were then purified using the gel extraction kit (Sigma-Aldrich) and sequenced to verify their identities. Data are shown either as ΔCt [average Ct of hPXR − average Ct of 18S rRNA] or as gene expression fold change (2−ΔΔCt) relative to the no shRNA group. Values are means ± S.D. from triplicate samples.
Cell Cycle Analyses.
The cell cycle analyses were performed as reported previously (Lin et al., 2008). HepG2/Control shRNA or HepG2/PP2Cβl shRNA cells were plated in growth medium. The cells were then processed for fluorescence-activated cell sorting analyses to determine cell cycle distribution.
RT-CES Assay.
The RT-CES assay (ACEA Biosciences, San Diego, CA) was used to monitor real-time cell proliferation of HepG2 cells as described previously, and the cell index was derived from the cell status-based cell-electrode impedance (Zeng et al., 2009). In brief, approximately 1 × 104 cells were seeded in growth medium onto a 96x e-plate (ACEA Biosciences) and allowed to attach and spread for 12 h before real-time cell proliferation was monitored; the cell index was automatically determined every 60 min for 72 h.
Results and Discussion
PP2Cβl Enhances hPXR Transcriptional Activity.
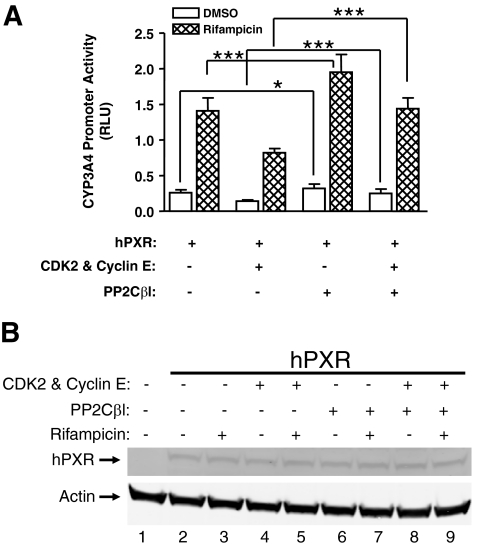
Hepatic CYP gene expression is affected by various signaling mechanisms, including reversible phosphorylation (Sidhu and Omiecinski, 1995; Marc et al., 2000; Pondugula et al., 2009a), and PXR-mediated CYP gene expression is modulated by both ligand binding and signaling cross-talk. We determined whether the phosphatase PP2Cβl affects hPXR transcriptional activity in HepG2 cells by measuring the activity of the PXR-regulated CYP3A4-luc reporter. As expected, the hPXR-agonist rifampicin (2 μM; 6 h) induced hPXR-mediated CYP3A4 promoter activity by approximately 5-fold (Fig. 1A). Interestingly, overexpressing PP2Cβl significantly (p < 0.05) enhanced both the basal and rifampicin-induced hPXR activity (Fig. 1A), suggesting that PP2Cβl is involved in positively regulating hPXR-mediated CYP3A4 gene expression. Multiple mechanisms, including reduction of hPXR protein stability or expression level or inhibition of signaling pathways that negatively regulate hPXR function, might contribute to this PP2Cβl effect; therefore, we sought to elucidate the responsible mechanisms.
Fig. 1.
PP2Cβl positively regulates hPXR function. A, PP2Cβl enhances hPXR-mediated CYP3A4 promoter activity and counteracts the inhibitory effect of CDK2 on hPXR. HepG2 cells were cotransfected with CYP3A4-luc− and hPXR−, as well as cyclin E−, CDK2−, or PP2Cβl− plasmids, as indicated, and CMV-Renilla luciferase plasmid (as a transfection control). Transactivation assays were performed as described under Materials and Methods. The values represent the means of six independent experiments, and the bars denote S.D. The p value was ascertained using the Student's t test, and differences were considered significant for p < 0.05 (*) or 0.001 (***) and nonsignificant for p > 0.05. B, cotransfection of PP2Cβl does not alter hPXR protein levels. Whole-cell lysates of the transiently transfected cells in A were subjected to Western blot analysis using rabbit polyclonal anti-hPXR serum. The pcDNA3 vector transfection was used as a negative control, and actin expression was analyzed as a loading control. Data shown are from a representative experiment.
PP2Cβl Counteracts CDK2-Mediated Inhibition of hPXR Transcriptional Activity.
We have shown that activating CDK2 decreases hPXR-mediated CYP3A4 expression and inhibiting CDK2 increases hPXR-mediated CYP3A4 expression in HepG2 cells (Lin et al., 2008). The activation of CDK2 requires the phosphorylation of threonine-160. Interestingly, PP2Cβl can dephosphorylate phosphothreonine-160 and inactivate CDK2 (Cheng et al., 1999, 2000). Therefore, it is possible that PP2Cβl enhances hPXR activity by dephosphorylating and inhibiting CDK2. We tested this possibility by determining whether overexpressing PP2Cβl can counteract the inhibitory effect of CDK2 on the transactivation activity of hPXR. As expected, overexpressing CDK2 and cyclin E inhibited both the basal and rifampicin-induced hPXR activity (Fig. 1A) (Lin et al., 2008). Interestingly, overexpressing PP2Cβl also significantly (p < 0.001) attenuated the inhibitory effect of CDK2 on both the basal and rifampicin-induced hPXR transactivation (Fig. 1A). These findings suggest that PP2Cβl enhances hPXR-mediated CYP3A4 gene expression by inhibiting CDK2 signaling in HepG2 cells.
Overexpressing PP2Cβl Does Not Affect hPXR Protein Level.
To explore the possibility that the apparent activating effect of PP2Cβl on hPXR transactivation is due to elevated protein levels of hPXR, we performed Western blot experiments. Analyses of Western blots showed that there was no measurable change in the protein levels of hPXR (Fig. 1B; data shown are from a representative experiment), indicating that the effect of PP2Cβl on hPXR transactivation was not caused by altered expression or stability of hPXR. Although immunoreactive hPXR protein was not detectable in HepG2 parental cells (Fig. 1B), hPXR mRNA was detectable (data not shown).
Knockdown of PP2Cβl Attenuates hPXR Transcriptional Activity.
Overexpressing PP2Cβl enhanced hPXR function, as shown in Fig. 1. To confirm the activating effect of PP2Cβl on hPXR's transactivation function, we determined whether silencing PP2Cβl in HepG2 cells, which stably express hPXR and the CYP3A4-luc reporter (clone 1), attenuates hPXR function. Western blot analysis showed that PP2Cβl shRNA, but not the nonsilencing control shRNA, selectively knocked down the protein levels of PP2Cβl by more than 80% (Fig. 2A). Consistent with our PP2Cβl overexpression results (Fig. 1A), silencing PP2Cβl significantly (p < 0.001) attenuated both the basal and rifampicin-induced transactivation function of hPXR (Fig. 2B). Real-time reverse transcription-polymerase chain reaction results showed that PP2Cβl knockdown did not significantly (p > 0.19) affect the mRNA level of hPXR. ΔCt values were 10.9 ± 0.5 and 11.4 ± 0.3 (p = 0.19) for HepG2/Control shRNA and HepG2/ PP2Cβl shRNA, respectively. When expressed as fold change (2−ΔΔCt) relative to the no shRNA group, the hPXR mRNA levels were 2.37 ± 2.12 and 1.08 ± 0.90 (p = 0.39) for HepG2/Control shRNA and HepG2/PP2Cβl shRNA, respectively. These results demonstrated that the attenuation of hPXR function in response to PP2Cβl knockdown is not due to reduced hPXR expression.
Fig. 2.
Knockdown of PP2Cβl attenuates hPXR-mediated CYP3A4 promoter activity. A, PP2Cβl shRNA knocks down the protein expression of endogenous PP2Cβl. HepG2 cells stably expressing hPXR and CYP3A4-luc (lane 1) were transduced with control nonsilencing shRNA (lane 2) or PP2Cβl shRNA (lane 3). Whole-cell lysates were collected and subjected to Western blot analysis using anti-PP2Cβl and anti-actin antibodies (as a loading control). Data shown are from a representative experiment. B, knockdown of PP2Cβl attenuates hPXR-mediated CYP3A4 promoter activity. HepG2 cells stably expressing hPXR and CYP3A4-luc were transduced with either control nonsilencing shRNA or PP2Cβl shRNA, and transactivation assays were performed as described under Materials and Methods. The RLA is shown as the mean value and S.D. from six independent observations. The Student's t test was used to determine statistical significance of unpaired samples by comparing the RLA obtained from the samples transduced with PP2Cβl shRNA to that of samples that were either untransduced or transduced with control shRNA. Differences were considered significant for p < 0.01 (**) or 0.001 (***) and nonsignificant (NS) for p > 0.05. C, overexpressing PP2Cβl rescues hPXR-mediated CYP3A4 promoter activity in PP2Cβl knockdown cells (HepG2/PP2Cβl shRNA). HepG2/PP2Cβl shRNA cells were transiently transfected with CMV-Renilla and either pcDNA3 or pcDNA3-PP2Cβl, and then the transactivation assays were performed as described under Materials and Methods section. The activity of firefly luciferase was normalized with that of the Renilla to determine relative luminescence units (RLU). The data represent the means and S.D.s of six independent experiments. The p value was determined using the Student's t test by comparing the samples transfected with pcDNA3 to PP2Cβl. Differences were considered significant for p < 0.05 (*) or 0.01 (**). D, PP2Cβl affects agonist-induced hPXR interaction with SRC-1. Mammalian two-hybrid assays were performed in HepG2/Control shRNA and HepG2/PP2Cβl shRNA cells as described under Materials and Methods. Induction of pG5-luc was used to measure the rifampicin-induced hPXR/SRC-1 interaction.
As expected, the attenuated hPXR transactivation in PP2Cβl knockdown cells was significantly (p < 0.01) rescued by overexpressing PP2Cβl (Fig. 2C), confirming that the attenuated hPXR function in HepG2/PP2Cβl shRNA cells was due to reduced PP2Cβl but not a reduced hPXR level. Taken together, these results suggest that PP2Cβl is essential for hPXR-mediated CYP3A4 gene expression in proliferating liver cells.
PP2Cβl Affects the Interaction between hPXR and SRC-1.
Phosphorylations regulate the activity of nuclear receptors by modifying their interaction with transcriptional coactivators such as SRC-1 and SRC-3 (Rochette-Egly, 2003; Pondugula et al., 2009a). The interaction between hPXR and SRC-1 is induced by hPXR agonists (Kliewer et al., 1998; Ding and Staudinger, 2005a,b; Johnson et al., 2006). We determined whether PP2Cβl affects agonist-induced hPXR interaction with SRC-1 by using the mammalian two-hybrid assays. As shown in Fig. 2D, rifampicin induced significant (p < 0.001) interaction between hPXR and SRC-1 in HepG2/Control shRNA cells. In contrast, no significant interaction between hPXR and SRC-1 was observed in HepG2/PP2Cβl shRNA cells. We were surprised to find that overexpressing PP2Cβl significantly (p < 0.001) restored the hPXR/SRC-1 interaction in HepG2/PP2Cβl shRNA cells, and it significantly (p < 0.01) enhanced the interaction in HepG2/Control shRNA cells (Fig. 2D). These results not only demonstrate that PP2Cβl is essential for agonist-dependent hPXR interaction with SRC-1 but also suggest a mechanism for PP2Cβl regulation of hPXR function.
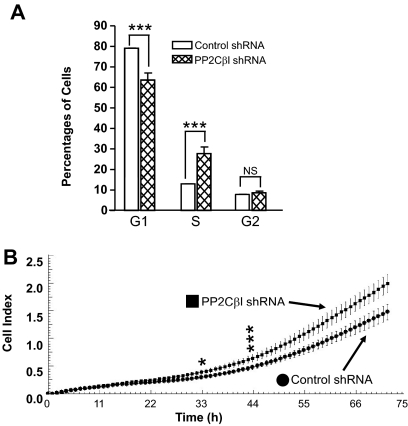
Knockdown of PP2Cβl Promotes Cell Cycle Progression of HepG2 Cells.
Stably overexpressing PP2Cβl in human embryonic 293 kidney cells arrests their cell growth (Seroussi et al., 2001). Therefore, we investigated whether PP2Cβl knockdown affects the cell cycle progression of HepG2 cells stably expressing hPXR. As shown in Fig. 3A, knockdown of PP2Cβl significantly (p < 0.001) increased the population of cells in the S phase of the cell cycle, yet it significantly (p < 0.001) decreased the population of cells in the G1 phase. These results indicate that down-regulating PP2Cβl protein expression promotes cell cycle progression from G1 to S phase in HepG2 cells.
Fig. 3.
Knockdown of PP2Cβl affects cell cycle distribution and cell proliferation. A, flow cytometry cell cycle analysis of HepG2/Control shRNA and HepG2/ PP2Cβl shRNA cells. The Student's t test was used to determine statistical significance of unpaired samples by comparing the percentage of cells transduced with control shRNA or PP2Cβl shRNA in different phases of the cell cycle. Differences were considered significant for p < 0.05 (*) or 0.001 (***) and nonsignificant (NS) for p > 0.05. B, RT-CES was used to monitor the cell proliferation of HepG2/Control shRNA and HepG2/PP2Cβl shRNA cells. The cell index represents the means of eight independent samples, and the bars indicate the S.D. The Student's t test was used to determine statistical significance of unpaired samples by comparing the cell index of HepG2/Control shRNA cells to that of HepG2/PP2Cβl shRNA cells. Differences were considered significant for p < 0.05 (*) or 0.001 (***). p values are shown for two time points (i.e., 33 and 44 h after monitoring was started) corresponding to 45 and 56 h after cell seeding.
Knockdown of PP2Cβl Promotes Proliferation of HepG2 Cells.
Because cell cycle analysis experiments suggested that PP2Cβl knockdown promotes cell cycle progression and may promote cell proliferation, we compared the cell proliferation of HepG2/Control shRNA and HepG2/PP2Cβl shRNA cells by using the RT-CES assay. As shown in Fig. 3B, knockdown of PP2Cβl started to significantly (p < 0.05) enhance cell proliferation approximately 45 h after cell seeding (i.e., 33 h after starting real-time cell index measurements), indicating that PP2Cβl can regulate the proliferation of HepG2 cells.
Several precautions were taken in planning our experiments to ensure that they yielded relevant results. For example, the transactivation assays and Western blot and coimmunoprecipitation analyses of transiently cotransfected cells were performed 24 h after transfection and subsequent 6-h DMSO or rifampicin treatment. These time points were chosen to allow appropriate ectopic protein expression and minimize the possibility of nonspecific actions from prolonged rifampicin treatment or protein overexpression. In addition, equal numbers of live cells were plated and treated with rifampicin for 6 h before the cell viability and luciferase activity were measured for the transactivation assays. Results of the Alamar Blue and CellTiter-Glo (Promega) cell viability assays did not show a significant change in the total number of live cells after 6-h rifampicin treatment (data not shown), indicating that neither the 2 μM rifampicin treatment nor the ectopic protein expression were toxic to HepG2 cells. Therefore, the observed changes in hPXR transactivation function were not due to cell number differences during the assay. Similar cell viability analyses of the cells used for the PP2Cβl knockdown experiments (i.e., no shRNA, control shRNA, or PP2Cβl shRNA) indicated no significant change in the total number of live cells in each cell sample after 24-h rifampicin treatment (data not shown), which suggests that the attenuated hPXR activity was not due to a cytotoxic effect of rifampicin during the 24-h assay period. As shown in Fig. 3B, knockdown of PP2Cβl did not significantly enhance cell proliferation within 24 h after cell seeding.
In this study, we show that elevated levels of PP2Cβl counteract the inhibitory effect of CDK2 on hPXR. Other signaling pathways that are inhibitory to PXR function exist. For example, nuclear factor-κB (NF-κB) signaling also negatively regulates the transcriptional activity of PXR as well as PXR-mediated P450 expression and activity (Gu et al., 2006; Xie and Tian, 2006; Zhou et al., 2006). PP2Cβl dephosphorylates and negatively regulates the activity of IκB kinase beta (IKKβ), a central intermediate signaling molecule in the activation of NF-κB (Prajapati et al., 2004; Sun et al., 2009). Therefore, it is possible that PP2Cβl enhances hPXR transactivation activity by attenuating NF-κB-mediated inhibition of PXR through dephosphorylating IKKβ. In addition, signal cross-talk through direct physical interactions between Ser/Thr protein phosphatases and NRs has been documented. For example, PP1 and PP2A physically associate with the vitamin D receptor (Bettoun et al., 2002), which belongs to the same subfamily of NRs as PXR does. However, coimmunoprecipitation experiments using mouse anti-FLAG M2-coated agarose beads and cell lysates that were harvested from HepG2 cells transiently transfected with either FLAG-PXR alone, or both PP2Cβl and FLAG-PXR revealed no specific physical interaction between hPXR and PP2Cβl (data not shown). Currently, it is not clear whether PP2Cβl directly dephosphorylates hPXR or modulates other signaling molecules or cofactors involved in hPXR transactivation.
CDK2 is a key regulator of cell cycle progression, and its activity fluctuates during the cell cycle, peaking at both the G1/S checkpoint and during S phase. We have previously shown that inhibition of hPXR activity occurs in thymidine-synchronized HepG2 cells during the S phase of the cell cycle, coinciding with the peak activity of CDK2, and that overexpression of CDK2 negatively regulates hPXR activity in unsynchronized HepG2 cells (Lin et al., 2008). In the current report, we show that down-regulating PP2Cβl significantly enriches the cells in the S phase of the cell cycle and promotes cell proliferation. It is possible that the cells were driven into S phase in response to PP2Cβl knockdown because PP2Cβl's normal inhibitory effect on CDK2 was released, and it is likely that CDK2 activity is increased in response to reduced levels of PP2Cβl. In addition, phosphorylation can affect the function of nuclear receptors by modulating either their location or interaction with coregulators such as SRC-1. hPXR was localized to the nucleus as we have shown previously (Pondugula et al., 2009b). Knockdown of PP2Cβl did not alter the localization of hPXR (data not shown). However, knockdown of PP2Cβl led to severe impairment of agonist-induced hPXR interaction with SRC-1, which was rescued by overexpressing PP2Cβl. These results suggest a responsible mechanism for PP2Cβl regulation of hPXR function, although other possible mechanisms cannot be ruled out.
During liver regeneration, the levels of growth factors such as human hepatocyte growth factor and augmenter of liver regeneration are up-regulated. These growth factors play important roles in liver regeneration by activating the proliferation of hepatocytes. Augmenter of liver regeneration induces hepatocyte proliferation (Ilowski et al., 2010), and remarkably represses both the basal and rifampicin-induced expression of CYP3A4, without changing the expressing levels of hPXR (Thasler et al., 2006). Human hepatocyte growth factor also activates the proliferation of hepatocytes and down-regulates the expression of P450s, including CYP3A4 (Donato et al., 1998), without changing the levels of hPXR (data not shown). These data suggest that in proliferating hepatocytes, the repression of CYP3A4 expression is not caused by a reduction in the levels of hPXR. Therefore, it is important to understand the mechanisms responsible for the reduction of CYP3A4 expression in proliferating hepatocytes; our data suggest a possible mechanism for CDK2 and PP2Cβl regulation of hPXR function. Because of the low endogenous level of hPXR, we have stably expressed hPXR in HepG2 (Lin et al., 2008). In this cell system, overexpression of either CDK2 or PP2Cβl affected the function of PXR without changing the levels of hPXR. However, whether the levels of PP2Cβl affect the expression of endogenous hPXR in proliferating hepatocytes has not been tested and needs to be investigated in future studies.
In conclusion, our results clearly demonstrate that increased levels of PP2Cβl enhance the function of hPXR in HepG2 cells and decreased levels of PP2Cβl that attenuate this function. More importantly, we have shown that PP2Cβl expression is essential for both the basal and inducible activity of hPXR in HepG2 cells. Furthermore, we show that elevated levels of PP2Cβl, which inactivates CDK2 (Cheng et al., 1999, 2000), counteract the inhibitory effect of CDK2 signaling on PXR function. It is possible that PP2Cβl plays an essential role to safeguard the function of PXR by inactivating or desensitizing the signaling pathways that negatively regulate the activity of PXR. Future studies using proliferating primary hepatocytes or in vivo liver regeneration animal models will improve our understanding of the role of PP2Cβl in regulating the function of hPXR.
Acknowledgments.
We thank Dr. Sara Lavi for sharing the PP2Cβl plasmid; Dr. Haojie Huang for sharing the CDK2 and cyclin E plasmids; Dr. Rakesh Tyagi (Special Centre for Molecular Medicine, Jawaharlal Nehru University, New Delhi, India) for sharing the hPXR antibody; Dr. Wenwei Lin, Dr. Dong Hanqing, and Fang Lei for sharing reagents and protocols; Dr. Kip Guy for reviewing the manuscript; and Dr. Cherise Guess for editing the manuscript.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant GM086415] (to T.C.); the National Institutes of Health National Cancer Institute [Grant P30-CA027165]; the American Lebanese Syrian Associated Charities; and St. Jude Children's Research Hospital.
Part of this work was previously presented as a poster as follows: Pondugula SR, Tong A, and Chen T (2009) Protein phosphatase 2Cβl is involved in positive regulation of human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. 16th North American Regional ISSX Meeting; 2009 Oct 18–22; Baltimore, MD. International Society for the Study of Xenobiotics, Washington, DC.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.032128.
- hPXR
- human pregnane X receptor
- P450
- cytochrome P450
- PXR
- pregnane X receptor
- NR
- nuclear receptor
- CDK
- cyclin-dependent kinase
- PP2Cβl
- protein phosphatase type 2C beta isoform long
- DMSO
- dimethyl sulfoxide
- shRNA
- short hairpin RNA
- PCR
- polymerase chain reaction
- RT-CES
- real-time cell-based electronic sensing
- RLA
- relative luciferase activity
- SRC-1
- steroid receptor coactivator-1
- 18S rRNA
- 18S small subunit ribosomal RNA
- Ct
- threshold cycle
- NF-κB
- nuclear factor-κB
- IKKβ
- IκB kinase beta.
References
- Bettoun DJ, Buck DW, 2nd, Lu J, Khalifa B, Chin WW, Nagpal S. (2002) A vitamin D receptor-Ser/Thr phosphatase-p70 S6 kinase complex and modulation of its enzymatic activities by the ligand. J Biol Chem 277:24847–24850 [DOI] [PubMed] [Google Scholar]
- Cheng A, Kaldis P, Solomon MJ. (2000) Dephosphorylation of human cyclin-dependent kinases by protein phosphatase type 2C alpha and beta 2 isoforms. J Biol Chem 275:34744–34749 [DOI] [PubMed] [Google Scholar]
- Cheng A, Ross KE, Kaldis P, Solomon MJ. (1999) Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev 13:2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Staudinger JL. (2005a) Induction of drug metabolism by forskolin: the role of the pregnane X receptor and the protein kinase a signal transduction pathway. J Pharmacol Exp Ther 312:849–856 [DOI] [PubMed] [Google Scholar]
- Ding X, Staudinger JL. (2005b) Repression of PXR-mediated induction of hepatic CYP3A gene expression by protein kinase C. Biochem Pharmacol 69:867–873 [DOI] [PubMed] [Google Scholar]
- Donato MT, Gómez-Lechón MJ, Jover R, Nakamura T, Castell JV. (1998) Human hepatocyte growth factor down-regulates the expression of cytochrome P450 isozymes in human hepatocytes in primary culture. J Pharmacol Exp Ther 284:760–767 [PubMed] [Google Scholar]
- Gu X, Ke S, Liu D, Sheng T, Thomas PE, Rabson AB, Gallo MA, Xie W, Tian Y. (2006) Role of NF-κB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J Biol Chem 281:17882–17889 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (1999) Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol 39:1–17 [DOI] [PubMed] [Google Scholar]
- Harmsen S, Meijerman I, Beijnen JH, Schellens JH. (2007) The role of nuclear receptors in pharmacokinetic drug-drug interactions in oncology. Cancer Treat Rev 33:369–380 [DOI] [PubMed] [Google Scholar]
- Ilowski M, Putz C, Weiss TS, Brand S, Jauch KW, Hengstler JG, Thasler WE. (2010) Augmenter of liver regeneration causes different kinetics of ERK1/2 and Akt/PKB phosphorylation than EGF and induces hepatocyte proliferation in an EGF receptor independent and liver specific manner. Biochem Biophys Res Commun 394:915–920 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Li CW, Chen LY, Ghosh JC, Chen JD. (2006) Regulation and binding of pregnane X receptor by nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone receptors (SMRT). Mol Pharmacol 69:99–108 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82 [DOI] [PubMed] [Google Scholar]
- Klumpp S, Thissen MC, Krieglstein J. (2006) Protein phosphatases types 2Calpha and 2Cbeta in apoptosis. Biochem Soc Trans 34:1370–1375 [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichti-Kaiser K, Xu C, Staudinger JL. (2009) Cyclic AMP-dependent protein kinase signaling modulates pregnane X receptor activity in a species-specific manner. J Biol Chem 284:6639–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Wu J, Dong H, Bouck D, Zeng FY, Chen T. (2008) Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J Biol Chem 283:30650–30657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc N, Galisteo M, Lagadic-Gossmann D, Fautrel A, Joannard F, Guillouzo A, Corcos L. (2000) Regulation of phenobarbital induction of the cytochrome P450 2b9/10 genes in primary mouse hepatocyte culture. Involvement of calcium- and cAMP-dependent pathways. Eur J Biochem 267:963–970 [DOI] [PubMed] [Google Scholar]
- Marley AE, Kline A, Crabtree G, Sullivan JE, Beri RK. (1998) The cloning expression and tissue distribution of human PP2Cbeta. FEBS Lett 431:121–124 [DOI] [PubMed] [Google Scholar]
- Pondugula SR, Dong H, Chen T. (2009a) Phosphorylation and protein-protein interactions in PXR-mediated CYP3A repression. Expert Opin Drug Metab Toxicol 5:861–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pondugula SR, Brimer-Cline C, Wu J, Schuetz EG, Tyagi RK, Chen T. (2009b) A phosphomimetic mutation at threonine-57 abolishes transactivation activity and alters nuclear localization pattern of human pregnane X receptor. Drug Metab Dispos 37:719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati S, Verma U, Yamamoto Y, Kwak YT, Gaynor RB. (2004) Protein phosphatase 2Cbeta association with the IkappaB kinase complex is involved in regulating NF-kappaB activity. J Biol Chem 279:1739–1746 [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C. (2003) Nuclear receptors: integration of multiple signalling pathways through phosphorylation. Cell Signal 15:355–366 [DOI] [PubMed] [Google Scholar]
- Saradhi M, Krishna B, Mukhopadhyay G, Tyagi RK. (2005) Purification of full-length human pregnane and xenobiotic receptor: polyclonal antibody preparation for immunological characterization. Cell Res 15:785–795 [DOI] [PubMed] [Google Scholar]
- Seroussi E, Shani N, Ben-Meir D, Chajut A, Divinski I, Faier S, Gery S, Karby S, Kariv-Inbal Z, Sella O, et al. (2001) Uniquely conserved non-translated regions are involved in generation of the two major transcripts of protein phosphatase 2Cbeta. J Mol Biol 312:439–451 [DOI] [PubMed] [Google Scholar]
- Sidhu JS, Omiecinski CJ. (1995) cAMP-associated inhibition of phenobarbital-inducible cytochrome P450 gene expression in primary rat hepatocyte cultures. J Biol Chem 270:12762–12773 [DOI] [PubMed] [Google Scholar]
- Sun W, Yu Y, Dotti G, Shen T, Tan X, Savoldo B, Pass AK, Chu M, Zhang D, Lu X, et al. (2009) PPM1A and PPM1B act as IKKbeta phosphatases to terminate TNFalpha-induced IKKbeta-NF-KappaB activation. Cell Signal 21:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Toriumi S, Saito J, Awano K, Kudo TA, Kobayashi T. (2006) PP2C family members play key roles in regulation of cell survival and apoptosis. Cancer Sci 97:563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thasler WE, Dayoub R, Mühlbauer M, Hellerbrand C, Singer T, Gräbe A, Jauch KW, Schlitt HJ, Weiss TS. (2006) Repression of cytochrome P450 activity in human hepatocytes in vitro by a novel hepatotrophic factor, augmenter of liver regeneration. J Pharmacol Exp Ther 316:822–829 [DOI] [PubMed] [Google Scholar]
- Xie W, Tian Y. (2006) Xenobiotic receptor meets NF-kappaB, a collision in the small bowel. Cell Metab 4:177–178 [DOI] [PubMed] [Google Scholar]
- Zeng FY, Cui J, Liu L, Chen T. (2009) PAX3-FKHR sensitizes human alveolar rhabdomyosarcoma cells to camptothecin-mediated growth inhibition and apoptosis. Cancer Lett 284:157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Tabb MM, Nelson EL, Grün F, Verma S, Sadatrafiei A, Lin M, Mallick S, Forman BM, Thummel KE, et al. (2006) Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J Clin Invest 116:2280–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SF. (2008) Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab 9:310–322 [DOI] [PubMed] [Google Scholar]