Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that mediates the biologic and toxic effects of its xenobiotic ligands. Previous cell culture studies have shown that, in addition to controlling the xenobiotic detoxification response, AHR activation leads to G0/G1 arrest, diminished capacity for DNA replication and inhibition of cell proliferation. In fact, recent work from our own and from other laboratories suggests that the AHR may function as a tumor suppressor gene that becomes silenced during the process of tumor formation. To test this hypothesis and determine whether the mouse Ahr gene acts as a tumor suppressor gene in vivo, we have examined the role of Ahr ablation on liver tumorigenesis induced by the genotoxic chemical diethylnitrosamine, a hepatic carcinogen that is not an AHR ligand. In mice administered a single i.p. injection of diethylnitrosamine, AHR antagonized liver tumor formation and growth by regulating cell proliferation, inflammatory cytokine expression, and DNA damage, parameters which were significantly elevated in the livers of control, and more so, of diethylnitrosamine-exposed Ahr−/− mice. Ahr−/− hepatocytes also showed significantly higher numbers of 4N cells, increased expression of proliferative markers and repression of tumor suppressor genes. These data support the concept that in its basal state in the absence of a xenobiotic ligand, the Ahr gene functions as a tumor suppressor gene and that its silencing may be associated with cancer progression.
Keywords: Ah receptor, DNA damage, tumor suppressor genes, hepatocellular carcinoma
INTRODUCTION
The AHR, a ligand-activated member of the bHLH/PAS family of transcription factors, controls a variety of developmental and physiological events, including induction of drug metabolizing enzymes, xenobiotic detoxification, neurogenesis, tracheal andsalivary duct formation, circadian rhythms, response to hypoxia, and hormone receptor function (1–3). More than 400 environmental toxicants and naturally occurring compounds are reported to bind and activate this cytosolic receptor (4) to form a nuclear heterodimeric transcription factor through dimerization with ARNT, also a bHLH/PAS protein. The transcriptionally active AHR/ARNT heterodimer recruits the basal transcriptional machinery, remodels chromatin and initiates transcription of genes coding for many phase I drug metabolizing enzymes, such as the cytochromes P450 CYP1A1, CYP1A2 and CYP1B1, and several phase II conjugating enzymes, such as ALDH3A1 and NQO1 (5–7). Following nuclear export, the AHR is degraded via the 26S proteasome pathway (8).
The AHR also functions as a cell cycle regulator (9–13) either by promoting cell cycle progression in the absence of an exogenous ligand (14–16), by promoting growth and immortalization of keratinocytes (17), or by inhibiting cell proliferation in cells exposed to the prototypical ligand, TCDD (18–21). From a mechanistic viewpoint, at least two different signaling pathways contribute to the AHR role in cell cycle regulation. On the one hand, AHR blocks TGFβ1-dependent inhibition of cell proliferation and promotion of apoptosis by repressing TGFβ1 expression (15, 22) and by accelerating TGFβ1 mRNA degradation (16). In addition, fibroblasts from AHR knockout mice overproduce TGFβ1 (15, 16) and their livers overproduce TGFβ1 and TGFβ3 (23), causing low proliferation rates and increased apoptosis. On the other hand, protein interactions between AHR and the RB/E2F axis further repress S-phase gene expression and prevent entry of the cells into S-phase (10, 11, 13, 20). Hence, depending on the cellular environment, the AHR could be considered a pro-proliferative gene in some cases and an anti-proliferative gene in others. Consistent with at least one aspect of this view, there is increasing evidence that AHR may function as a tumor suppressor in vivo. Epigenetic AHR silencing by promoter hypermethylation has recently been reported in a significant number of human acute lymphoblastic leukemia cases (24). Ahr repression was also substantial in DEN-induced liver tumors of mice, albeit in the absence of any significant promoter hypermethylation (25). Furthermore, AHR-null TRAMP mice show increased susceptibility to prostate tumorigenesis and develop prostate tumors with greater frequency than Ahr+/+ TRAMP mice, suggesting that the AHR possesses tumor suppressor properties (26).
To study the role of the Ahr gene in liver tumorigenesis, we have examined tumor incidence in DEN-induced liver tumors in wild type and Ahr−/− mice. DEN is metabolized by the cytochrome P450 CYP2E1 into a carcinogenic alkylating agent that causes DNA damage, but DEN is not an AHR ligand nor does it induce AHR responsive detoxification genes (27, 28). Using DEN allowed us to explore AHR functions that do not depend on xenobiotic AHR ligands or that result from AHR-mediated procarcinogen activation. The central question in these experiments was to determine whether loss of Ahr affected the formation and progression of liver tumors in DEN-induced mice and if it did, to characterize the molecular changes associated with the process. Our data are consistent with the hypothesis that during DEN-induced carcinogenesis, the Ahr functions as a tumor suppressor gene.
MATERIALS AND METHODS
Animals and hepatocarcinogen treatment
The Ahr−/− mice used in these experiments were derived from Ahr−/− mice bred into a C57BL/6J genetic background at the Jackson Labs. These mice were purchased 6 years ago and were thereafter maintained in our colony. Ahr−/− mice were bred with wild type Ahr+/+ mice and maintained as hemizygous Ahr+/− mice housed in a pathogen-free animal facility under standard 12-hour light/12-hour dark cycle with ad libitum water and chow. All experiments were conducted using the highest standards of humane care in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the University of Cincinnati Institutional Animal Care and Use Committee. For experimental treatment, sibling hemizygous mice were bred and the litters genotyped for loss or presence of the wild type allele in homozygosis. Groups (n = 5 – 9) of 15-day-old Ahr+/+ and Ahr−/− male and female littermates were administered a single i.p. injection of the genotoxic hepatocarcinogen DEN (Sigma) dissolved in saline at a dose of 20 mg/kg body weight. Littermates used as controls were injected with an equal volume of saline. Mice were sacrificed at 3 – 4 weeks or at 30 – 35 weeks post-DEN injection. To measure cell proliferation rates, mice were given a single i.p. injection of 150 mg/kg of BrdU 90 min before euthanasia. Immediately after euthanasia, livers were excised, weighed and photographed to facilitate scoring of surface liver lesions. One lobe was rapidly fixed in buffered 10% formalin for 16 hours for histologic analysis. Visible surface tumors were excised from both Ahr+/+ and Ahr−/− animals, immediately frozen in liquid N2 and stored at −80°C until use.
Tumor quantification
Grossly visible surface liver tumors were scored with the aid of digital photographs of the top and bottom of each liver. The size of surface nodules was quantified by measuring the diameter of nodules in millimeters. Liver histology and pathology were evaluated by microscopic evaluation of H&E-stained sections in a blinded fashion by a veterinary pathologist (G.P.B.).
Preparation of Liver tissues for histology, BrdU staining analyses, γH2A.X assays and ploidy determination
Formalin-fixed, paraffin-embedded liver tissues were sectioned at 4 μm. For immunohistochemical staining, sections were deparaffinized in xylenes and rehydrated through a graded series of ethanol/water solutions. For histology evaluation, H&E staining was performed by standard protocols. BrdU incorporation was analyzed using a Streptavidin-biotin BrdU detection kit (Zymed, South San Francisco, CA) exactly as recommended by the manufacturer. BrdU incorporation was scored blind, and 400 – 700 hepatocytes per field were counted from 6 – 8 random sections from each mouse liver and the mean ± SEM of at least 3 mice in each group was recorded. For γH2A.X immunostaining, sections were boiled for 10 min in Antigen Retrieval Solution (10 mM sodium citrate sodium buffer (pH 6.0) (DakoCytomation, Cambridge MA))and cooled to room temperature before proceeding with staining. Endogenous peroxidase activity was quenched by treatment in 3% H2O2 in methanol. Sections were blocked in 0.25% normal goat serum in TBS for 1 h at room temperature and incubated for 1 h at 37°C with monoclonal antibodies to histone H2AX-pSer139 (γH2A.X; Catalog no. 05-636, JBW301 from Millipore) at a 1:1500 dilution). Normal mouse serum was used as negative control. Sections were incubated for 1 h at room temperature with biotinylated goat anti-mouse secondary antibody (anti-mouse IgG staining systems Cat# sc-2050, Santa Cruz Biotechnology) at 1:200 dilution, counterstained with hematoxylin, dehydrated, mounted in permount medium and visualized under light microscopy. Positive cells were scored as per the BrdU staining protocol. Flow cytometric determination of γH2A.X was performed as previously described (13) by incubation with fluorescein isothiocyanate (FITC)-conjugated anti-phosphohistone H2A.X (p-Ser139) or negative control mouse IgG-FITC. Determination of ploidy levels was also as previously described by one of us (29).
Cytokine determination
Liver homogenates in PBS supplemented with a cocktail of protease inhibitors (Roche) were centrifuged at 2500×G to clarify the lysates and used to determine the levels of the two cytokines, Il6 and Tnfα using the Aushon Biosystems (Billerica, MA) Searchlight ELISA services.
RNA extraction, cDNA preparation and real-time PCR
RNA was isolated from at least three frozen DEN-induced liver tumors and three control livers of each genotype using Tri-reagent (Molecular Research Center Inc, Cincinnati, OH). Because tumor incidence was low in females, only livers from males were used for these experiments. cDNA was synthesized by reverse transcription of 30 μg total RNA in a total volume of 45 μl containing 1× reverse transcriptase buffer, 7 μM random hexamers primer, 0.5 mM dNTP mix, 10 mM dithiothreitol, 5 mM MgCl2, 20 U of RNase inhibitor (RNasin, Promega), and 100 U of SuperScript™ II RNase H− reverse transcriptase (Invitrogen). Samples were denatured and annealed to the primer for 10 min at 70°C, and reverse-transcribed for 3 h at 42oC. Before amplification, the reverse transcriptase was inactivated by heating to 70oC for 15 min and RNA was hydrolyzed by incubation with 0.05 N NaOH at 70°C for 10 min, neutralized with 0.05 N HCl and the cDNA precipitated with ethanol. The resulting cDNA products were dissolved in a final volume of 200 μl and a 2-μl aliquot was used as template for subsequent quantification by real-time PCR amplification. PCR reactions were conducted in duplicate in a total volume of 25 μl containing SYBR Green PCR Master Mix (Applied Biosystems), and 0.1 μM of each primer. Amplification was performed on an ABI 7500 (Applied Biosystems) where the reaction was heated to 95°C for 10 min followed by 40 cycles of denaturation at 95°C for 15 sec and annealing-elongation at 60°C for 60 sec. Detection of the fluorescent product was carried out during the elongation period, and emission data were quantified using threshold cycle (Ct) values. Ct values for each mouse sample were determined in duplicate, normalized to values for β-actin amplification of the same sample (ΔCt) and the mean of three mice was used to calculate the log2 of the ratio of the means ± SEM of each gene in the different Ahr−/− tissues (control, tumor, or tumor-adjacent normal) relative to their expression in the corresponding tissues of Ahr+/+ mice. Amplification of β-actin cDNA in the same samples was used as an internal control for all real-time PCR amplification reactions. PCR product specificity was confirmed using melting curve analyses and subsequent polyacrylamide gel electrophoresis. Primers for cDNA amplification are shown in Table S1.
Statistical analyses
Statistical analyses of RT-PCR data was performed using SigmaStat 3.1. Tumor incidence comparisons were made using chi-square analysis. Group comparisons were made using a two-tailed Student’s t-test. A p-value <0.05 was considered statistically significant.
RESULTS
Loss of Ahr promotes DEN-induced tumor formation
In the mouse liver, AHR is a master regulator of the detoxification of xenobiotic polycyclic aromatic hydrocarbons (30) and, through cooperative interactions with the RB/E2F axis, serves as an environmental checkpoint in the regulation of cell cycle progression (31). To investigate whether AHR plays a role in liver tumorigenesis, we analyzed the susceptibility of mice with a deletion of the Ahr gene to develop liver tumors after a single injection of the liver-specific carcinogen DEN. We found a significant enhancement of tumor incidence in the livers of Ahr−/− male mice compared with that in wild-type littermates of the same gender, characterized by significantly increased incidence, multiplicity of nodules, and size (Table 1). Specifically, the number of nodules per mouse was 4-fold higher and the nodule size was double in Ahr−/− mice compared to Ah+/+ mice. As is the case of liver cancer in humans, females showed fewer tumors than males, in agreement with previous observations in DEN-induced mouse liver cancer (32). Histological analysis of livers confirmed these findings. Liver nodules typically consisted of basophilic lesions with crowded nuclei and were classified as atypical foci, dysplastic adenomas or hepatocellular adenomas, characterized by clearly defined margins and compression of surrounding parenchyma. These lesions are believed to be precursors of hepatocellular carcinoma in humans (33). One single tumor in a DEN-exposed male presented with disorganized architecture and evidence of necrosis and was classified as hepatocellular carcinoma. These data support the idea that loss of the Ahr gene can be a critical event in liver cancer progression.
Table 1.
Tumor incidence at 9 months in DEN-treated mice
| Genotype | Gender | Treatment | Mice with tumors/total mice |
LW/BW (%) | Nodules per mouse | Nodule diameter (mm) | Histopathology | |
|---|---|---|---|---|---|---|---|---|
| HCC | Adenoma | |||||||
| Ahr+/+ | M | DEN | 0/6 | 3/6a | 4.6 ± 0.8 | 4 ± 1c | 1.5 ± 0.5d | |
| Control | 0/5 | 0/5 | 4.8 ± 0.6 | |||||
| F | DEN | 0/5 | 1/5 | 4.5 ± 0.4 | 5 | 3.2 ± 0.5 | ||
| Control | 0/8 | 0/8 | 4.0 ± 0.4 | |||||
| Ahr−/− | M | DEN | 1/7 | 7/7a | 9.3 ± 6.4b | 15 ± 4c | 3.4 ± 0.4d | Single HCC had necrosis, lymphocyte infiltration |
| Control | 0/3 | 0/3 | 3.6 ± 0.2b | |||||
| F | DEN | 0/9 | 4/9 | 3.9 ± 0.7 | 3 ± 1 | 3.1 ± 0.7 | ||
| Control | 0/7 | 0/7 | 3.6 ± 0.3 | |||||
Significantly different, Chi-square p=0.04.
Significantly different, t-test p=0.01.
Significantly different, t-test p=0.02.
Significantly different, t-test p=0.02
Ahr ablation deregulates proliferation and enhances DNA damage and apoptosis after DEN treatment
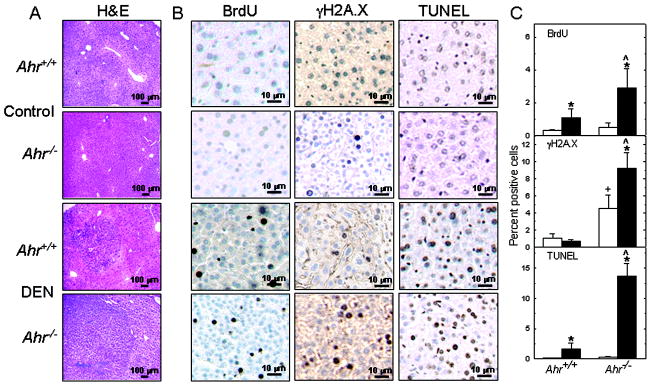
In previous cell culture work, we found that loss of Ahr in embryonic fibroblasts of Ahr−/− mice leads to a decrease in proliferation rate (16) and an increase in oxidative stress, DNA damage, and apoptosis (13). To determine what role AHR deficiency in the liver might play in tumor development, we used immunohistochemistry to examine livers from control and DEN-induced tumors in male mice of both genotypes. After 30 – 35 weeks of DEN treatment, less than 0.5% of the hepatocytes from unexposed mice of either genotype incorporated BrdU, whereas incorporation in livers from DEN-treated mice was significantly higher in Ahr−/− than in Ahr+/+ livers. BrdU staining was mainly circumscribed to the tumor, with little or no staining in the non-tumor areas. In contrast, Ahr−/− hepatocytes from livers of control untreated mice already showed a significant difference in phosphorylated H2A.X histone immune staining from their Ahr+/+ counterparts, a difference that was even greater in livers from DEN-treated mice. TUNEL analyses revealed no difference in the level of spontaneous apoptosis in untreated mice, but a large difference in DEN-induced apoptosis, with Ahr−/− livers showing 10-fold more apoptotic cells than Ahr+/+ livers. The histologic and immunohistochemistry data are shown in Fig. 1A–B and graphically summarized in Fig. 1C. Overall, the liver data regarding γH2A.X and TUNEL staining are consistent with our earlier cell culture findings, and show that even in the untreated mice, the Ahr−/− genotype confers susceptibility to DNA damage and is pro-apoptotic. Unlike in cultured cells, Ahr ablation does not deregulate proliferation, which is consistent with the fact that hepatocytes are quiescent in the liver of the intact animal (33).
Figure 1. AHR-deficient DEN-induced tumors show increased DNA proliferation, DNA damage and apoptosis.
(A) Four-micron-thick sections from formalin-fixed, paraffin-embedded left liver lobe of control and DEN-treated Ahr+/+ and Ahr−/− male mice were processed 30 – 35 weeks after i.p. inoculation for histological analysis by H&E staining, and (B), BrdU incorporation, and γH2A.X and TUNEL staining by immunohistochemistry. Corresponding micrographs from a single representative animal from each genotype are shown. Micrographs from at least 3 mice in each group were recorded blind by counting at least 400 hepatocytes per section chosen from several random fields of each micrograph. Open and closed bars represent means ± SEM of controls and DEN-treated mice, respectively. (*) denotes significantly different (p<0.05) from untreated controls of the same genotype; (+) denotes significantly different from untreated controls of different genotypes. (^) denotes significantly different between DEN-treated mice of different genotypes.
AHR deficiency impairs ploidy control
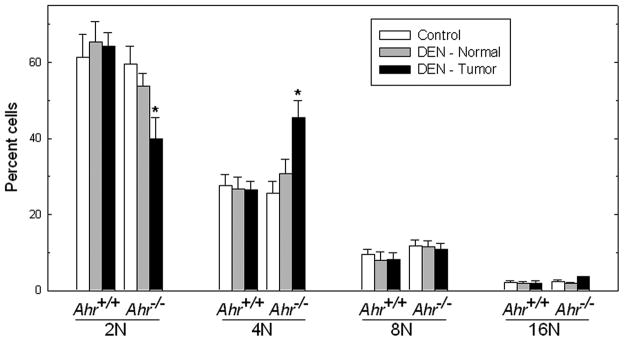
Genomic instability is intimately connected with the control of ploidy, with important implications for the pathophysiology of cancer (34). Oftentimes, exposure of cells in culture to genotoxic agents results in the acquisition of abnormal ploidy (35, 36). To assess the impact of DEN exposure on ploidy levels in AHR-proficient and AHR-deficient livers, we dissected tissues from livers of untreated Ahr+/+ and Ahr−/− mice and from liver tumors and normal surrounding tissues of DEN-exposed mice of both genotypes. We used flow cytometry to analyze the DNA content of individual nuclei from these tissues. Compared with untreated Ahr+/+ animals, exposure to DEN caused no changes in the ratios of 4N:2N hepatocytes, with approximately twice as many 2N than 4N hepatocytes in tumor or surrounding normal liver tissue of the same mice. In contrast, DEN induced a significant increase in the 4N:2N ratios in Ahr−/− mice, with approximately equal numbers of 4N and 2N hepatocytes in the tumor tissue of DEN-treated mice (Fig. 2). This increase in 4N cells affected only the tumor tissue, and not the normal surrounding tissues or the livers of untreated Ahr−/− mice. These data indicate that basal hepatocyte ploidy control is unaffected by loss of Ahr in the normal regions of livers from mice exposed to DEN, but that DEN exposure significantly impairs ploidy distribution in the preneoplastic tissue of AHR-deficient livers.
Figure 2. Aberrant ploidy in Ahr−/− hepatocytes from DEN-treated male mice.
Nuclei were isolated from hepatocytes from control livers and from tumor and tumor-adjacent normal tissue of DEN-treated Ahr+/+ and Ahr−/− male mice (n=3 – 5) and their DNA content was analyzed by flow cytometry. (*) Significantly different (p<0.05) between genotypes.
Loss of Ahr promotes increases in Il6 and Tnfα expression
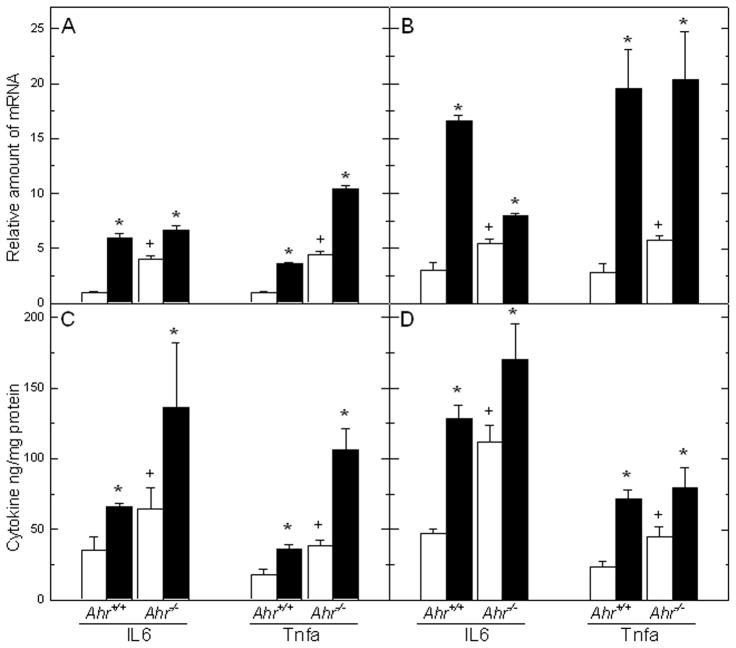
Inflammatory cells and proinflammatory cytokines found in tumors are major contributing factors to a state of chronic inflammation more likely to promote tumor growth, progression, and immunosuppression than they are to mount an effective host antitumor response (37, 38). Serum levels of interleukin-6 (IL6), a multifunctional cytokine largely responsible for the hepatic “acute phase response”, and tumor necrosis factor-alpha (TNFα), a major mediator of inflammation, are highly elevated in experimental models of HCC in mice (32, 39) and in a number of human conditions believed to be HCC precursors (40). To determine whether the Ahr genotype plays a significant role in the increase of inflammatory cytokine expression observed by others in DEN-treated mice (32), we used real time RT-PCR and ELISA to measure the levels of Il6 and Tnfα mRNAs and proteins as a function of genotype and treatment to the male mice. Expression of both cytokines was already significantly higher in Ahr−/− mice than in their Ahr+/+ counterparts, even in animals that had not been treated with DEN. Both Il6 and Tnfα mRNAs were significantly increased in mice of both genotypes already as early as 3 weeks after DEN injection (Fig. 3A), and more so, reaching nearly 5-fold increases over untreated controls by 36 weeks after treatment (Fig. 3B). Parallel determinations of the corresponding proteins in the same tissues showed a similar pattern of significant higher levels of IL6 and Tnfα in liver lysates of Ahr−/− at both 3 weeks and 36 weeks, and induction in livers of both genotypes after DEN treatment (Fig. 3C,D). These data suggest that AHR deficiency may promote a proinflammatory state by up-regulating the expression of these two cytokines and that this condition is exacerbated by DEN treatment.
Figure 3. Differential expression of pro-inflammatory cytokines Il6 and Tnfα in livers of control and DEN-treated Ahr+/+ and Ahr−/− male mice.
Il6 and Tnfα mRNA levels were quantified at 3 weeks (A) and 30 – 35 weeks (B) following control or DEN injection. Parallel determinations of the protein levels of the two cytokines were done at 3 weeks (C) and 30 – 35 weeks (D). Open and closed bars represent mean ± SEM of controls and DEN-treated mice, respectively. The symbols denoting significance are as detailed in the legend to Figure 1.
Loss of Ahr increases expression of proliferative markers and represses tumor suppressor genes
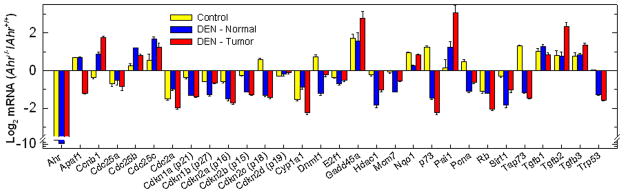
Direct comparison of the transcriptional characteristics of mouse and human HCC has allowed the identification of a limited number of genes that accurately predict the length of survival and provide new molecular insights into the pathogenesis of HCC (41). Comparative global gene expression profiling has provided important insights in the identification of genes associated with clinicopathological features of HCC, refining the diagnosis and prognostic predictions of HCC patients (41). Of particular interest in this regard is the expression of cell proliferation genes, which in DEN-induced mouse HCC is most similar to their expression in the poorer prognosis group of human HCC (42). To investigate whether AHR plays a significant role in the aberrant gene expression patterns associated with DEN-induced tumorigenesis, we examined the expression of survival and proliferation genes in hepatocytes from control, DEN-induced tumors, and normal tissues adjacent to the tumors. We found that most tumor suppressor genes and genes such as Cdc25b and Cdc25c, involved in the activation of cyclin-dependent kinases, were expressed at similar levels in control tissues of both mouse strains. In contrast, whereas the Cdc25 genes were up-regulated by DEN treatment, the tumor suppressor genes were repressed in normal and tumor hepatocytes of Ahr−/− mice to a larger extent than in Ahr+/+ mice. Among the pro-apoptotic genes, Apaf1 was up-regulated in control and normal tissues of DEN exposed-mice, but down-regulated in the tumors; p73 and TA-p73, its pro-apoptotic variant,, were up-regulated in control Ahr−/− hepatocytes, but were repressed in normal and tumor tissues of DEN-treated Ahr−/− relative to wild type mice. Sirt1 E2f1 and Rb were repressed in all three conditions tested, indicating that Ahr ablation is responsible for their down-regulation. On the other hand, Tp53 is up-regulated in hepatocytes of control mice and is repressed as consequence of DEN-treatment in Ahr−/− mice. The three Tgfb genes appear to be already up-regulated in control Ahr−/− hepatocytes; in DEN treated mice, Tgfβ2, but not Tgfβ1 or 3 is further induced in the tumor tissues of Ahr−/− mice. Gadd45a, a gene inducible by DNA damage, was highly expressed in control Ahr−/− hepatocytes, and its expression nearly doubled in normal and tumor tissues of DEN-treated Ahr−/− relative to Ahr+/+ mice (Fig. 4). With few exceptions, these data are consistent with the conclusion that the lack of AHR results in a type of tumor that more closely resembles the poor-prognosis phenotype of human HCC, suggesting that AHR may provide the protection characteristic of a tumor suppressor gene.
Figure 4. Analysis of the expression of pro-proliferative and pro-apoptotic markers in Ahr+/+ and Ahr−/− mice.
RNA extracted from hepatocytes of control livers and of tumor and tumor-adjacent normal tissue of DEN-treated Ahr+/+ and Ahr−/− male mice (n=3 – 5) was analyzed by real time RT-PCR for the accumulation of mRNA from the genes shown on the abscissa. The ordinate denotes the log2 of the ratio of the means ± SEM expression of those genes in the indicated Ahr−/− tissues (control, tumor, or tumor-adjacent normal) relative to their expression in the corresponding tissues of Ahr+/+ mice. The numerical expression value was determined from the change in threshold cycle number (ΔCt) of each mRNA relative to β-actin used as the normalization factor.
DISCUSSION
The results presented in this study show that ablation of the Ahr gene leads to a significant increase in DEN-dependent liver tumor incidence in male mice relative to wild type Ahr+/+ littermates of the same gender. There was no comparable change observed in female mice, in agreement with work in the literature (32). The increase in males results from higher incidence, higher nodule multiplicity and larger nodule size. The loss of the Ahr gene appears to sensitize liver hepatocytes to acquire a phenotype that is both pro-proliferative and anti-apoptotic when exposed to a DEN challenge, possibly due to a significantly higher constitutive level of DNA damage, as shown by γH2A.X immunostaining. Our previous cell culture work has shown that lack of AHR in fibroblasts creates a condition of heightened cellular oxidative stress that triggers significant increases in DNA damage (13). We surmise that a similar condition pertains to the hepatocytes of Ahr−/− mice, and that the higher prooxidant status of these mice becomes exacerbated by DEN, itself a DNA-damaging alkylating agent. In support of this contention is the observation that expression of the Gadd45a gene, a stress sensor inducible by DNA damage, is already high in control Ahr−/− hepatocytes and in tumor-adjacent tissues of the same mice, and becomes even higher in the tumor tissues themselves.
Exposure of cells in culture to genotoxic agents results in the development of abnormal ploidy (35, 36). Although in specific cases aneuploidy may act as a suppressor of tumorigenesis (43), genomic instability is more likely to drive, rather than suppress cancer (34, 43). It is likely that the sustained level of DNA damage found in Ahr deficient hepatocytes is responsible for the abnormally high level of tetraploid hepatocytes in these mice and the ensuing increase in liver tumors. There is now compelling evidence showing that tumors often contain a near tetraploid number of chromosomes, and that the uncontrolled proliferation of tetraploid cells can trigger tumor formation (44).
Expression of the pro-inflammatory cytokines Il6 and Tnfα was also higher in untreated AHR-deficient than in AHR-proficient mice. The difference was maintained at the same level over the whole experimental period of 35 weeks and was greatly increased by DEN treatment. Sustained oxidative stress and DNA damage are likely to contribute to a general state of cell damage and destruction, generating tissue necrosis and inducing the expression of pro-inflammatory cytokines. Greater expression of these cytokines in Ahr−/− hepatocytes might be a response involved in cell survival. DEN may aggravate this pre-existing cellular condition by creating a more extensive and long-lasting state of DNA damage. However, by 35 weeks after DEN treatment, mice of both genotypes show similar levels of these two cytokines, suggesting that the difference in tumorigenesis between the two genotypes may be independent of the damage caused by sustained endogenous cytokine expression.
Expression of typical cell proliferation markers, such as the cyclin-dependent kinase inhibitors and Rb, E2f1 and Tp53 is repressed in liver tumors and tumor-adjacent tissues of DEN-treated Ahr−/− mice. In the case of Rb and E2f1 repression is also evident in hepatocytes from control mice, suggesting that AHR modulates Rb and E2f1 expression and that Ahr ablation is sufficient to down-regulate these two genes, while DEN exposure further represses Rb expression. In contrast the genes coding for cyclin B1 and the Cdc25 phosphatases are up-regulated in those mice. These results are consistent with the classification of DEN-induced HCC in mice as having a biological phenotype similar to the poor-prognosis subclass of human HCC, characterized by the up-regulation of proliferation markers (41). As in that human subclass, pro-apoptotic markers are down-regulated in the tumors of Ahr−/− mice. The Tgfβ genes are also over-expressed in tumors and normal tissues of DEN-treated Ahr−/− mice and in untreated controls relative to the corresponding Ahr+/+ mice. This may be a unique feature of HCC in AHR-deficient mice, setting them aside from the human HCC-like class, being the result of the over-expression of Tgfβ genes in Ahr−/− mice (15). TGFβ over-expression may be at the root of the greater tumorigenesis of the Ahr−/−genotype. It appears that TGFβ has a dual role during tumorigenesis, functioning as a tumor suppressor in earlier tumor phases and turning into a tumor promoter in later phases of tumorigenesis (45). The switch occurs as a consequence of its being produced in high amounts in the tumor, which stimulates tumorigenesis by allowing tumor cells to escape immune surveillance (46) and to promote angiogenesis (47). In agreement with the regulatory role of TGFβ in Pai1 transcription (48), we find a concomitant increase in Pai1 expression. Pai1 encodes the plasminogen activator inhibitor serpin A1, whose expression increases migration and epithelial-mesenchymal transition and is highly elevated in many invasive tumors, including breast, brain and gastric cancers (49, 50).
In summary, our studies show that AHR functions as a tumor suppressor that modulates the tumorigenic effect of DEN and causes a lower incidence of tumors than that found in mice lacking this protein. This AHR function is radically different from the pro-oncogenic role that AHR plays as an activator of the carcinogenic potential of its procarcinogenic ligands. Because, as discussed earlier, DEN is not an AHR ligand or a substrate for AHR-regulated cytochrome P450s, this effect is not the result of AHR-mediated DEN detoxification. Rather, it is more likely that the combination of a higher level of oxidative stress and sustained TGFβ expression---two molecular events that may be causally connected---is responsible for the heightened tumorigenicity of the Ahr−/− genotype. High constitutive levels of oxidative stress and TGFβ expression might be a further phenotypic characteristic to be taken into consideration when assessing the prognosis of humans with HCC. This phenotype need not be solely associated with AHR deficiency, but may result from activation of different oncogenic pathways.
Supplementary Material
Acknowledgments
We thank Michael Danneman for his help in scoring immunocytochemistry micrographs.
This research was supported by NIEHS grants R01 ES06273, R01 ES10807 and the NIEHS Center for Environmental Genetics grant P30 ES06096.
ABBREVIATIONS
- ALDH3A1
aldehyde dehydrogenase-3A1
- AHR
aryl hydrocarbon receptor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- bHLH
basic-region helix-loop-helix
- DEN
diethylnitrosamine
- HCC
hepatocellular carcinoma
- NQO1
NAD(P)H-dependent quinone oxidoreductase
- PAI-1
plasminogen activator inhibitor type-1
- PAS
Period-Aryl hydrocarbon nuclear translocator-Simple-minded
- RB
retinoblastoma protein
- TRAMP
Transgenic adenocarcinoma of the mouse prostate
- TCDD
2, 3, 7, 8-tetrachlorodibenzo-p-dioxin
- TGFβ
transforming growth factor-beta
Footnotes
The authors declare no conflicts of interest.
References
- 1.Pocar P, Fischer B, Klonisch T, Hombach-Klonisch S. Molecular interactions of the aryl hydrocarbon receptor and its biological and toxicological relevance for reproduction. Reproduction. 2005;129:379–89. doi: 10.1530/rep.1.00294. [DOI] [PubMed] [Google Scholar]
- 2.Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr Opin Genet Dev. 1999;9:580–7. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 4.Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chem Biol Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 5.Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–40. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 6.Whitlock JP., Jr Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–25. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 7.Schnekenburger M, Peng L, Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim Biophys Acta. 2007;1769:569–78. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollenz RS. The mechanism of AH receptor protein down-regulation (degradation) and its impact on AH receptor-mediated gene regulation. Chem Biol Interact. 2002;141:41–61. doi: 10.1016/s0009-2797(02)00065-0. [DOI] [PubMed] [Google Scholar]
- 9.Kolluri SK, Weiss C, Koff A, Gottlicher M. p27kip1 induction and inhibition of proliferation by the intracellular Ah receptor in developing thymus and hepatoma cells. Genes Dev. 1999;13:1742–53. doi: 10.1101/gad.13.13.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strobeck MW, Fribourg AF, Puga A, Knudsen ES. Restoration of retinoblastoma mediated signaling to Cdk2 results in cell cycle arrest. Oncogene. 2000;19:1857–67. doi: 10.1038/sj.onc.1203510. [DOI] [PubMed] [Google Scholar]
- 11.Puga A, Barnes SJ, Dalton TP, Chang C, Knudsen ES, Maier MA. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J Biol Chem. 2000;275:2943–50. doi: 10.1074/jbc.275.4.2943. [DOI] [PubMed] [Google Scholar]
- 12.Weiss C, Faust D, Schreck I, Ruff A, Farwerck T, Melenberg A, et al. TCDD deregulates contact inhibition in rat liver oval cells via Ah receptor, JunD and cyclin A. Oncogene. 2008;27:2198–207. doi: 10.1038/sj.onc.1210859. [DOI] [PubMed] [Google Scholar]
- 13.Marlowe JL, Fan Y, Chang X, Peng L, Knudsen ES, Xia Y, et al. The Ah Receptor Binds to E2F1 and Inhibits E2F1-induced Apoptosis. Mol Biol Cell. 2008;19:3263–71. doi: 10.1091/mbc.E08-04-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Q, Whitlock JPJ. The aromatic hydrocarbon receptor modulates the Hepa 1c1c7 cell cycle and differentiated state independently of dioxin. Molecular & Cellular Biology. 1996;16:2144–50. doi: 10.1128/mcb.16.5.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elizondo G, Fernandez-Salguero P, Sheikh MS, Kim GY, Fornace AJ, Lee KS, et al. Altered cell cycle control at the G(2)/M phases in aryl hydrocarbon receptor-null embryo fibroblast. Mol Pharmacol. 2000;57:1056–63. [PubMed] [Google Scholar]
- 16.Chang X, Fan Y, Karyala S, Schwemberger S, Tomlinson CR, Sartor MA, et al. Ligand-independent regulation of transforming growth factor beta1 expression and cell cycle progression by the aryl hydrocarbon receptor. Mol Cell Biol. 2007;27:6127–39. doi: 10.1128/MCB.00323-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray SS, Swanson HI. Dioxin-induced immortalization of normal human keratinocytes and silencing of p53 and p16INK4a. J Biol Chem. 2004;279:27187–93. doi: 10.1074/jbc.M402771200. [DOI] [PubMed] [Google Scholar]
- 18.Hushka DR, Greenlee WF. 2,3,7,8-Tetrachlorodibenzo-p-dioxin inhibits DNA synthesis in rat primary hepatocytes. Mutat Res. 1995;333:89–99. doi: 10.1016/0027-5107(95)00135-2. [DOI] [PubMed] [Google Scholar]
- 19.Bauman JW, Goldsworthy TL, Dunn CS, Fox TR. Inhibitory effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on rat hepatocyte proliferation induced by 2/3 partial hepatectomy. Cell Prolif. 1995;28:437–51. doi: 10.1111/j.1365-2184.1995.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 20.Marlowe JL, Knudsen ES, Schwemberger S, Puga A. The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S-phase specific gene expression. J Biol Chem. 2004;279:29013–22. doi: 10.1074/jbc.M404315200. [DOI] [PubMed] [Google Scholar]
- 21.Levine-Fridman A, Chen L, Elferink CJ. Cytochrome P4501A1 promotes G1 phase cell cycle progression by controlling aryl hydrocarbon receptor activity. Mol Pharmacol. 2004;65:461–9. doi: 10.1124/mol.65.2.461. [DOI] [PubMed] [Google Scholar]
- 22.Carvajal-Gonzalez JM, Roman AC, Cerezo-Guisado MI, Rico-Leo EM, Martin-Partido G, Fernandez-Salguero PM. Loss of dioxin-receptor expression accelerates wound healing in vivo by a mechanism involving TGFbeta. J Cell Sci. 2009;122:1823–33. doi: 10.1242/jcs.047274. [DOI] [PubMed] [Google Scholar]
- 23.Zaher H, Fernandez-Salguero PM, Letterio J, Sheikh MS, Fornace AJ, Jr, Roberts AB, et al. The involvement of aryl hydrocarbon receptor in the activation of transforming growth factor-beta and apoptosis. Mol Pharmacol. 1998;54:313–21. doi: 10.1124/mol.54.2.313. [DOI] [PubMed] [Google Scholar]
- 24.Mulero-Navarro S, Carvajal-Gonzalez JM, Herranz M, Ballestar E, Fraga MF, Ropero S, et al. The dioxin receptor is silenced by promoter hypermethylation in human acute lymphoblastic leukemia through inhibition of Sp1 binding. Carcinogenesis. 2006;27:1099–104. doi: 10.1093/carcin/bgi344. [DOI] [PubMed] [Google Scholar]
- 25.Peng L, Mayhew CN, Schnekenburger M, Knudsen ES, Puga A. Repression of Ah receptor and induction of transforming growth factor-beta genes in DEN-induced mouse liver tumors. Toxicology. 2008;246:242–7. doi: 10.1016/j.tox.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fritz WA, Lin TM, Safe S, Moore RW, Peterson RE. The selective aryl hydrocarbon receptor modulator 6-methyl-1,3,8-trichlorodibenzofuran inhibits prostate tumor metastasis in TRAMP mice. Biochem Pharmacol. 2009;77:1151–60. doi: 10.1016/j.bcp.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guengerich FP, Kim D-H, Iwasaki M. Role oh human P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol. 1991;4:168–79. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 28.Chen ZY, Eaton DL. Differential regulation of cytochrome(s) P450 2B1/2 by phenobarbital in hepatic hyperplastic nodules induced by aflatoxin B1 or diethylnitrosamine plus 2-acetylaminofluorene in male F344 rats. Toxicol Appl Pharmacol. 1991;111:132–44. doi: 10.1016/0041-008x(91)90142-2. [DOI] [PubMed] [Google Scholar]
- 29.Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, et al. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–77. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 30.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–50. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 31.Puga A, Ma C, Marlowe JL. The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways. Biochem Pharmacol. 2009;77:713–22. doi: 10.1016/j.bcp.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–4. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 33.Fausto N. Mouse liver tumorigenesis: models, mechanisms, and relevance to human disease. Semin Liver Dis. 1999;19:243–52. doi: 10.1055/s-2007-1007114. [DOI] [PubMed] [Google Scholar]
- 34.Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nat Rev Mol Cell Biol. 2004;5:752–62. doi: 10.1038/nrm1443. [DOI] [PubMed] [Google Scholar]
- 35.Lan Z, Sever-Chroneos Z, Strobeck MW, Park CH, Baskaran R, Edelmann W, et al. DNA damage invokes mismatch repair-dependent cyclin D1 attenuation and retinoblastoma signaling pathways to inhibit CDK2. J Biol Chem. 2002;277:8372–81. doi: 10.1074/jbc.M108906200. [DOI] [PubMed] [Google Scholar]
- 36.Mayhew CN, Carter SL, Fox SR, Sexton CR, Reed CA, Srinivasan SV, et al. RB loss abrogates cell cycle control and genome integrity to promote liver tumorigenesis. Gastroenterology. 2007;133:976–84. doi: 10.1053/j.gastro.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 37.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–45. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 38.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 39.Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–6. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 40.Abiru S, Migita K, Maeda Y, Daikoku M, Ito M, Ohata K, et al. Serum cytokine and soluble cytokine receptor levels in patients with non-alcoholic steatohepatitis. Liver Int. 2006;26:39–45. doi: 10.1111/j.1478-3231.2005.01191.x. [DOI] [PubMed] [Google Scholar]
- 41.Lee JS, Thorgeirsson SS. Comparative and integrative functional genomics of HCC. Oncogene. 2006;25:3801–9. doi: 10.1038/sj.onc.1209561. [DOI] [PubMed] [Google Scholar]
- 42.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–6. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 43.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–87. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weaver BA, Cleveland DW. The role of aneuploidy in promoting and suppressing tumors. J Cell Biol. 2009;185:935–7. doi: 10.1083/jcb.200905098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ten Dijke P, Goumans MJ, Itoh F, Itoh S. Regulation of cell proliferation by Smad proteins. J Cell Physiol. 2002;191:1–16. doi: 10.1002/jcp.10066. [DOI] [PubMed] [Google Scholar]
- 46.Hazelbag S, Gorter A, Kenter GG, van den BL, Fleuren G. Transforming growth factor-beta1 induces tumor stroma and reduces tumor infiltrate in cervical cancer. Hum Pathol. 2002;33:1193–9. doi: 10.1053/hupa.2002.130109. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, et al. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–12. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 48.Kutz SM, Higgins CE, Samarakoon R, Higgins SP, Allen RR, Qi L, et al. TGF-beta 1-induced PAI-1 expression is E box/USF-dependent and requires EGFR signaling. Exp Cell Res. 2006;312:1093–105. doi: 10.1016/j.yexcr.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 49.Chazaud B, Ricoux R, Christov C, Plonquet A, Gherardi RK, Barlovatz-Meimon G. Promigratory effect of plasminogen activator inhibitor-1 on invasive breast cancer cell populations. Am J Pathol. 2002;160:237–46. doi: 10.1016/S0002-9440(10)64367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei H, Hemminki K, Johansson R, Altieri A, Enquist K, Henriksson R, et al. PAI-1 -675 4G/5G polymorphism as a prognostic biomarker in breast cancer. Breast Cancer Res Treat. 2008;109:165–75. doi: 10.1007/s10549-007-9635-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.