Abstract
Background
In insects, hemocyanin superfamily proteins accumulate apparently to serve as sources of amino acids during metamorphosis, reproduction and development. Storage hexamerins are important members of the hemocyanin superfamily. Although insects possess storage hexamerins, very little is known about the character and specific functions of hexamerin 1 and storage protein 1 in insect development.
Results
To gain insight into the function of storage proteins in insects, cDNAs for two storage proteins were cloned from the fat body of Spodoptera exigua. S. exigua hexamerin 1 (SeHex) cDNA contained an open reading frame of 2124 nucleotides encoding a protein of 707 amino acids with a predicted molecular weight of 82.12 kDa. S. exigua storage protein 1 (SeSP1) cDNA contained an open reading frame of 2256 bp encoding a protein of 751 amino acids with a predicted molecular weight of ~88.84 kDa. Northern blotting analyses revealed that SeHex mRNA is expressed in the fat body, cuticle, midgut and Malpighian tubules and SeSP1 in fat body, Malpighian tubules and tracheae. SeHex and SeSP1 mRNAs were expressed in fat body at different levels from first instar larvae to pupae, with expression was much lower from first instar larvae to first-day fifth instar larvae. SeHex transcript expression was high in fat body of wandering larvae (pre-pupae) and steadily decreased to the seventh pupal day. SeSP1 transcript expression was high in fat body of wandering larvae, 2-day-old fifth instar larvae and 2-, 4- and 7-day-old pupae. SeHex and SeSP1 mRNAs levels were expressed lower than control on the condition of starvation at 12 h. Of insects injected with SeHex and SeSP1 dsRNA, 38.7% and 24.3% survived to 204 h after treatment, respectively. This was significantly lower than in the controls groups.
Conclusions
These findings provide new data on the tissue distribution, expression patterns and the function in starvation of storage proteins. RNA interference results revealed that storage protein genes are key in metamorphosis, reproduction and insect development. The results for SeHex and SeSP1 interference reveal that a potential method to control this pest is to disrupt the regulation of storage proteins.
Background
In insects, hemocyanin superfamily proteins accumulate apparently to serve as sources of amino acids during metamorphosis, reproduction and periods when food is unavailable and the demand for amino acids is high [1,2]. Before the initiation of insect metamorphosis and reproduction, these proteins are progressively stored in larval hemolymph. Hexamerins are mainly synthesized by the fat body during larval development, stored in the hemolymph, and sequestered in the fat body, where they serve as sources of nitrogen and amino acids for pupae and adults during metamorphosis and reproduction [3-5].
The insect tracheal system has many respiratory proteins that transport oxygen in the hemolymph and, in preparing for metamorphosis, insect larvae store a huge amount of protein in hemolymph [6-9]. These specialized oxygen-transport proteins evolved from the copper-containing hemocyanins [10]. In most Hexapoda, gas exchange is mediated by the tracheal system, a network of tubules that open to the atmosphere on the cuticle and radiate to all parts of the body. Oxygen is delivered through the trachea and tracheoles in the gaseous phase [11]..
Insect hexamerins share sequence similarities with other proteins of divergent functions [12] and belong to a protein superfamily that also comprises arthropod phenoloxidases, crustacean pseudohemocyanins, insect storage hexamerins and dipteran hexamerin receptors [10,12-15]. The most abundant and widely distributed storage proteins that accumulate in the hemolymph or fat body are composed of six identical or similar subunits of ~80 kDa, and thus are also called hexamerins [4,13,16-18]. Storage hexamerins include the hexamerins, juvenile hormone-related protein, riboflavin-binding hexamerin precursor, methionine-rich storage protein (storage protein 1, SP1), very-high-density lipoprotein, tyrosine-rich proteins and hemocyanin-related proteins [4,19].
Genes for storage hexamerins have been cloned from plants, bacteria and fungi. Gordadze et al. cloned two hexamerin cDNAs from the mosquito Aedes aegypti [20]. So far, SPs and hexamerins have been cloned and studied in Musca domestica [21], Apriona germari [22,23], Apis mellifera [5], Drosophila melanogaster [24] and Reticulitermes flavipes [25]. In Lepidoptera, SPs and hexamerins have been cloned and studied in Spodoptera litura [26], Plodia interpunctella [2], Corcyra cephalonica [27], Manduca sexta [28], Amsacta albistriga [29-31], Plutella xylostella [32], Helicoverpa zea [33], Omphisa fuscidentalis [34], and Sesamia nonagrioides [35,36].
Spodoptera exigua, commonly called the beet armyworm, is a worldwide agricultural pest that has developed resistance to many chemical pesticides. In the present study, we cloned cDNA for two hexamerin genes from S. exigua (EF646282 and EU259816). We observed expression of these two genes not only in fat body, but also in Malpighian tubules and other tissues. Furthermore, the genes exhibited differential expression patterns in fat body and they play a role in starvation. We used RNA interference (RNAi) to investigate the functions of the genes.
Results
SeHex and SeSP1 cDNA sequence analysis
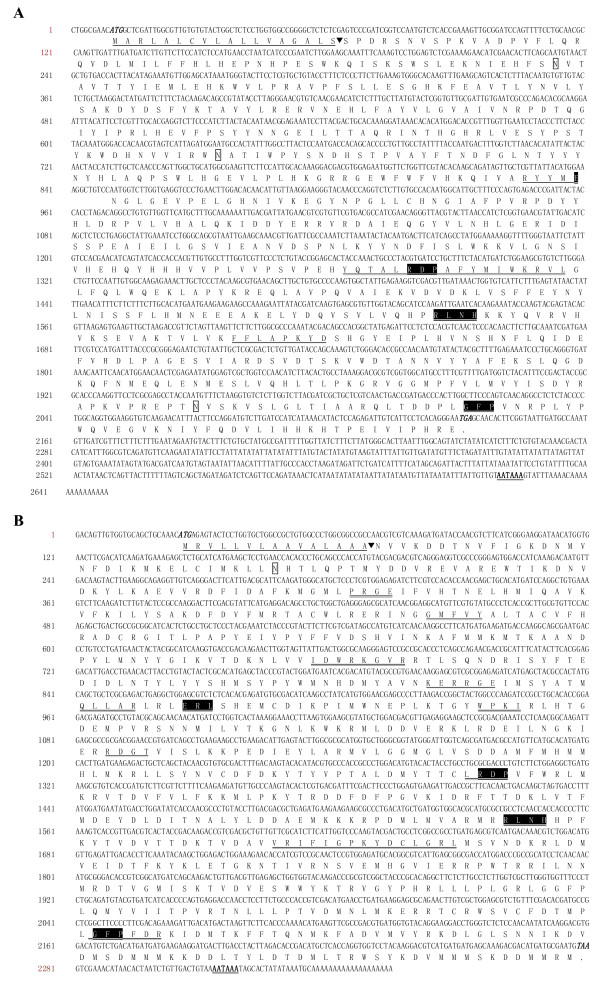
SeHex had an open reading frame of 2124 bp, encoding a protein of 707 amino acids (Figure 1A) with a predicted molecular weight (MW) of approximately 82.12 kDa and an isoelectric point (pI) of 6.48. SP1 had an open reading frame of 2256 bp, encoding a protein of 751 amino acids (Figure 1B) with a predicted MW of approximately 88.84 kDa and pI of 9.10.
Figure 1.
Nucleotide and deduced amino acid sequences for S. exigua Hex and SP1 cDNAs. Italic and bold nucleotides indicate the start and stop codons, respectively. The termination signal AATAAA is bold and underlined. The hemocyanin group of motifs (or signature motifs) (amino acid residues ERL, RDP, RLNH and GFP) are shaded in black. (A) SeHex cDNA sequence analysis. Underlined amino acid residues (1-18) and the arrowhead represent the signal peptide and putative cleavage site, respectively. Regions that are highly conserved in lepidopteran Hex genes are double underlined. Potential N-glycosylation sites (residues 75, 209, 479 and 647) are boxed. The nucleotide sequence reported in this paper has been submitted to GenBank (accession number EF646282). (B) SeSP1 cDNA sequence analysis. Underlined amino acid residues (1-15) and the arrowhead represent the signal peptide and putative cleavage site, respectively. Regions that are highly conserved in lepidopteran SP1 genes are double underlined. The potential N-glycosylation site (residue 47) is boxed. The nucleotide sequence reported in this paper has been submitted to GenBank (accession number EU259816).
SeHex cDNA (GenBank accession no. EF646282) has 43-74% identity to other known Hex genes (Additional file 1). SeHex is most similar to Hex from the lepidopteran H. armigera (74% identity). It is also similar to Hex genes from Trichoplusia ni, Hyalophora cecropia, Galleria mellonella and O. fuscidentalis (Figure 2). SeSP1 cDNA (GenBank no. EU259816) has 28-96% identity to other known SP1 genes (Additional file 1). SeSP1 is most similar to SP1 from the lepidopteran S. litura (96% identity). It is also similar to SP1 genes from T. ni, S. nonagrioides, H. cecropia, Hyphantria cunea, M. sexta, Samia cynthia, Heliconius erato, Bombyx mori, Chilo suppressalis, P. interpunctella, A. aegypti, Culex quinquefasciatus, Anopheles gambiae, Periplaneta americana, Perla marginata, Thermobia domestica, Reticulitermes speratus, A. germari, Sinella curviseta, Tribolium castaneum, Tenebrio molitor and R. flavipes (Figure 2).
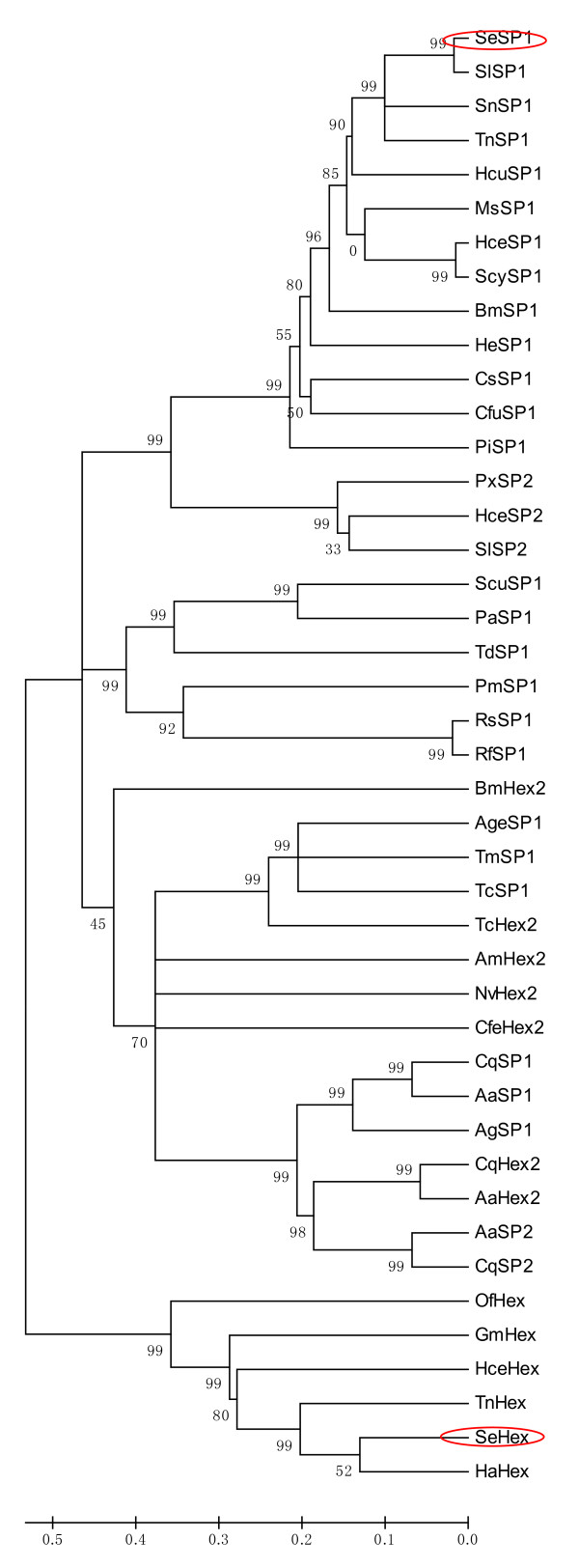
Figure 2.
Phylogenetic analysis of insect hemocyanins based on their amino acid sequences. Full-length amino acid sequences were aligned using Mega 3.1 to generate a phylogenetic tree (Hex, hexamerin; Hex2, class 2 hexamerin gene; SP1, storage protein 1; SP2, storage protein 2). A bootstrap analysis was carried out and the robustness of each cluster was verified in 1000 replications. The hemocyanins were from Aedes aegypti (AaHex2, GALLHP82 and AaSP1, XM_001659481 and AaSP2, XM_001659481), Anopheles gambiae (AgSP1, XM_321800), Apis mellifera (AmHex2, NM_001011600), Apriona germari (AgeSP1, AF509880), Bombyx mori (BmHex2, NM_001044125 and BmSP1, NM_001113276), Camponotus festinates (CfeHex2, AJ251271), Chilo suppressalis (CsSP1, AB248057), Choristoneura fumiferana (CfuSP1, AF007768), Culex quinquefasciatus (CqSP1, XM_001843442 and CqSP2, XM_001843444), Galleria mellonella (GmHex, GALLHP82X;), Heliconius erato (HeSP1, EU711403), Helicoverpa armigera (HaHex, AY661710), Hyalophora cecropia (HceHex, AF032397 and HceSP1, AF032399 and HceSP2, AF032398), Hyphantria cunea (HcuSP1, U60988), Manduca sexta (MsSP1, L07609), Nasonia vitripennis (NvHex2, XM_001606979), Omphisa fuscidentalis (OfHex, EF429085), Periplaneta americana (PaSP1, FM242648), Perla marginata (PmSP1, AM690365), Plodia interpunctella (PiSP1, AF356843), Plutella xylostella (PxSP2, AB266596), Reticulitermes flavipes (RfSP1, AY572858), Reticulitermes speratus (RsSP1, AB371986), Samia cynthia (ScySP1, AB288051), Sesamia nonagrioides (SnSP1, DQ147770), Sinella curviseta (ScuSP1, FM242638), Spodoptera exigua (SeHex, EF646282 and SeSP1, EU259816), Spodoptera litura (SlSP1, AJ249470; SlSP2, AJ249468), Tenebrio molitor (TmSP1, AF395329), Thermobia domestica (TdSP1, FM165290), Tribolium castaneum (TcHex2, XM_968706 and TcSP1, XM_967951) and Trichoplusia ni (TnHex, CBLJHSP and TnSP1, L03280).
Protein sequence analysis
In Figure 1, underlined amino acid residues 1-18 and 1-15 represent the signal peptides for SeHex and SeSP1, respectively. The deduced amino acid sequences for SeHex and SeSP1 contain not only four signature motifs of the hemocyanin family, such as ERL (residues 277-279 and 281-283), RDP (424-426), RLNH (506-509) and GFP (668-670 and 630-632), but also other conserved motifs. Sequence alignment for lepidopteran Hex proteins revealed three highly conserved regions (Additional file 1A). Alignment of lepidopteran SP1 proteins revealed ten highly conserved motifs (Additional file 1B). Four potential N-glycosylation sites (amino acids 75, 209, 479 and 647) were found in SeHex, but only one potential site (amino acid 47) in SeSP1 (Figure 1, Additional file 1).
Results for two hexamerin genes from the termite R. flavipes (RfHex) revealed conserved hexamerin signature motifs (ADKP/QFKM/QK/RQ, YFTEDVGL and TALRDPAY/NQ) [24]. The protein sequence encoded by SeHex had only two signature motifs (YFTNDFGL and TALRDPAFY) in common with RfHex. Sequence alignment revealed four conserved signature motifs (ERL, RDP, RLNH and GFP) for insect hexamerins and storage proteins. Three conserved motifs (RYYMRRLS, YQTALR/SDPAY/FYMIM/WKRVL and FFLAPKYD) were observed for lepidopteran Hex genes (Figure 1 and Additional file 1). Moreover, lepidopteran SP1 genes had ten highly conserved regions (PRGE, GMFV/LY, I/VDW/SRKGV/LR/P, KERR/QGE, QLLAR, WPKI, RDGT, LRDP, VRI/VFL/IGPKY/FDCM/LGR/LL and GFPFDR) (Figure 1 and Additional file 1).
Tissue distribution of Hex and SP1 in S. exigua
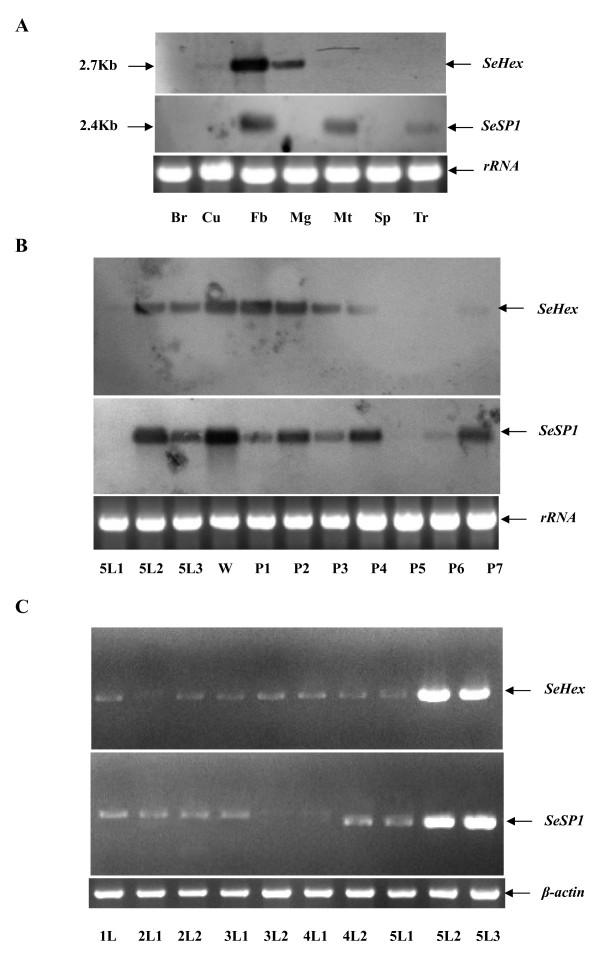
Northern blotting revealed strong SeHex bands for fat body and midgut and weak bands for cuticle and Malpighian tubules, but no transcripts were detected for brain, spermary and trachea samples from S. exigua larvae (Figure 3A). The results suggest that SeHex is specifically expressed in fat body, midgut, cuticle and Malpighian tubules. Northern blotting results for SeSP1 transcripts revealed expression in fat body, Malpighian tubules and tracheae, but not in brain, cuticle, midgut and spermary (Figure 3A).
Figure 3.
Northern blotting analyses of SeHex and SeSP1 transcript expression in fat body. (A) For Northern blotting analysis, total RNA of day-2 fifth instar S. exigua larvae was extracted from various tissues: brain (Br), cuticle (Cu), fat body (Fb), midgut (Mg), Malpighian tubules (Mt), spermary (Sp) and tracheae (Tr). (B) Developmental expression of SeHex and SeSP1 was analyzed by Northern blotting of fat body from fifth instar larvae to pupae of S. exigua. RNA was extracted from fat body of fifth instar larvae (5L), wandering (pre-pupae) larvae (W) and pupae (P). 5L1-5L3 denoted day-1 to day-3 fifth instar larvae and P1-P7 denote day-1 to day-7 pupae. rRNA was used as a control in all Northern blotting analysis. (C) Developmental expression of SeHex and SeSP1 in young larvae was analyzed by RT-PCR in fat body of S. exigua. RNA was extracted from the fat body of first (1L), second (2L), third (3L), fourth (4L) and fifth instar larvae (5L). L1 means the first day of young larvae, and so on. β-Actin was used as a control in all RT-PCTR analyses.
Developmental expression of SeHex and SeSP1
SeHex and SeSP1 mRNAs were expressed in fat body at different levels from fifth instar larvae to pupae. SeHex transcripts were highly expressed in fat body in wandering larvae (pre-pupae), as well as in day-1 and day-2 pupae. Transcripts were present at lower levels in fat body of day-2 and day-3 fifth instar larvae and the day-3 and day-4 pupae. SeHex transcripts were very low in fat body of day-1 and day-7 pupae (Figure 3B). Moreover, SeHex expression steadily decreased from wandering larvae to 6-day-old pupae. The results suggest that SeHex mRNA is constitutively expressed at a rather high level in fat body from the second day of larva stages to the fourth day of pupa stages. SeSP1 transcripts were highly expressed in fat body in wandering larvae (pre-pupae), as well as in day-2 fifth instar larvae and day-2, day-4 and day-7 pupae. It was present at lower levels on day-3 fifth instar larvae and day-1, day-3 and day-6 pupae (Figure 3B).
Comparative RT-PCR results showed that SeHex and SeSP1 mRNAs were expressed at very low levels in fat body from first instar larvae to day-1 fifth instar larvae. SeHex and SeSP1 mRNAs expression was higher in day-2 fifth instar larvae (Figure 3C).
SeHex and SeSP1 proteins' function identified on the condition of starvation
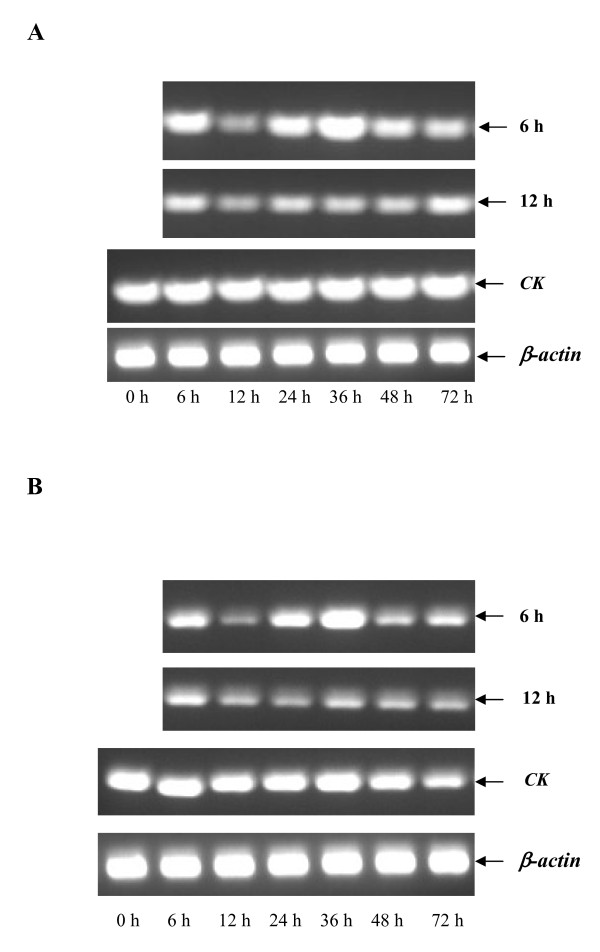
RT-PCR analysis showed that SeHex and SeSP1 transcripts were mainly lowly expressed when the insects were been on the condition of starvation. And SeHex and SeSP1 transcripts expressed in the same mode or tread in the different starvation treatment (Figure 4A, 4B). In the group of starvation with 6 h, once stress after finals, SeHex and SeSP1 mRNA expressional levels lower than control, but higher than the group of starvation with 12 h at 6 h, 24 h and 36 h (Figure 4A, 4B). In the group of starvation with 12 h, once stress was finished, SeHex and SeSP1 mRNA expressed decreased and keep the lower expressional levels.
Figure 4.
SeHex and SeSP1 mRNA expression on the condition of starvation. The day-2 fifth instar larvae were used as experimental insects in the treatment of starvation. In the experiment of starvation, two experimental group insects were hungered for 6 h and 12 h, respectively. Followed these insect continue to rear at 25 ± 1°C with an L14:D10 photoperiod using an artificial diet. The insects were observed at 0 h (before treatment), 6 h, 12 h, 24 h, 36 h, 48 h and 72 h after treatment. Every three to five lively larvae were randomly selected and stored at -80°C for subsequent RNA extraction. (A) for SeHex in starvation, (B) for SeSP1 in starvation. The housekeeping gene β-actin was used as a reference.
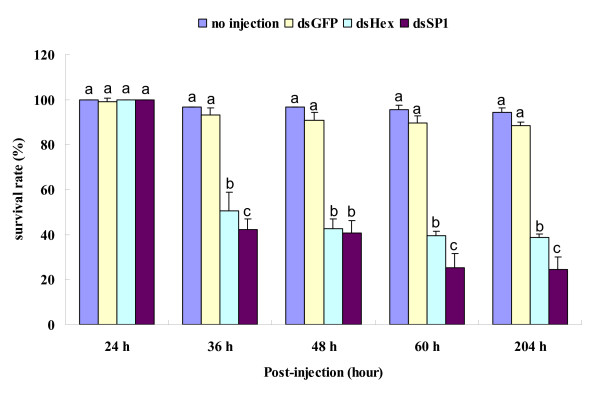
Survival rate and SeHex and SeSP1 transcript analysis after double-stranded RNA injection
Double-stranded RNA for SeHex and SeSP1 (dsSeHex and dsSeSP1) was injected into day-2 fifth instar larvae. The survival rate of insects injected with dsSeHex was 50.55%, 42.60%, 39.42% and 38.73% at 36, 48, 60 and 204 h after injection, respectively, which is significantly lower than the survival of insects in the two control groups (Figure 5). A sharp decrease in survival rate was observed between 24 and 36 h after dsSeHex injection. The survival rate of insects injected with dsSeSP1 was 42.18%, 40.54%, 25.07% and 24.29% at 36, 48, 60 and 204 h after injection, respectively, which is significantly lower than the survival of insects in the two control groups (Figure 5). Two sharp decreases in survival rate were observed from 24 to 36 h and from 48 to 60 h after dsSeSP1 injection. Prior to death, these larvae usually became less vital and smaller in size.
Figure 5.
Survival after injection of SeHex and SeSP1 dsRNA. Insect survival rates at different times after injection of dsSeHex, dsSeSP1 and dsGFP. The survival rate was assessed at key developmental stages of 24 h (fifth instar, 3-day-old larvae), 36 h, 48 h (pre-pupae), 60 h (pupae) and 204 h (adult) after injection. Results were arcsine square-root transformed before analysis to correct for the non-normal distribution of percentage values. Different letters at the same detection time indicate a significant difference in survival rate (p < 0.05, Duncan's test). No significant difference was found by ANOVA (p > 0.05). All error bars represent standard deviation (n = 3).
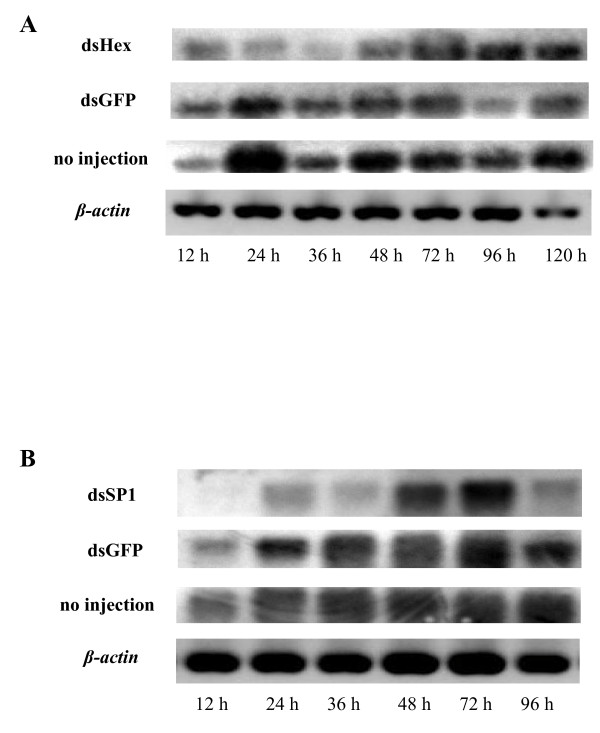
To test the RNAi efficiency, semi-quantitative RT-PCR was performed to detect SeHex and SeSP1 transcripts. SeHex and SeSP1 mRNA levels substantially decreased at 12, 24 and 36 h after injection compared to the negative controls (dsGFP and no injection) (Figure 6A, B). However, transcription of both genes recovered to some extent 48 h after injection.
Figure 6.
(A) SeHex and (B) SeSP1 transcript analysis by RT-PCR amplification after dsRNA injection. Three larval states (before death, still living and less vital) were randomly selected at each time point after injection. Total RNA was extracted and reversed to cDNA using AMV reverse transcriptase (Takara). SeHex and SeSP1 specific probes were radiolabeled with [α-32P]-dCTP. The specific primers SeHex-FP and SeHex-RP or SeSP1-FP and SeSP1-RP were used to amplify cDNAs in the same PCR reactions. The PCR products were separated on 2% agarose gel and transferred to a Hybond-N+ nylon membrane. Hybridization, washing and signal detection of the blots were carried out as described previously. (A) Lanes marked 12 h (5L2), 24 h (5L3), 36 h (5L3), 48 h (W), 72 h (P1), 96 h (P2) and120 h (P3) indicate the times (and developmental stage) after injection. (B) Lanes marked 12 h (5L2), 24 h (5L3), 36 h (5L3), 48 h (W), 72 h (P1) and 96 h (P2) indicate the times (and developmental stage) after injection. The housekeeping gene β-actin was used as a reference.
Discussion
Many different storage proteins are found in larvae and pupae at different stages, but not all species have all the storage proteins. Ryan et al. were the first to identify a 74-kDa hexamerin subunit in A. mellifera larval hemolymph [37]. Two storage proteins have also been investigated in the honeybee A. mellifera [38], the ant Camponotus festinatus [39,40], and several other ant species [41]. In Lepidoptera, two different types of methionine-rich hexamerins can be classified according to their common amino acid composition [16,42,43], and three or four SPs are found in some species. Storage proteins first reported in T. ni in final-stage larvae [44] and two SPs in M. sexta have been separated and cloned [45,46]. Six storage proteins of four different types have been found in Caplodes ethlius [19]. In the present study, two storage proteins were cloned from S. exigua (Figure 1). The results in Figure 2 show that three storage proteins (SP1, SP2 and Hex2) are found in the Hymenoptera A. aegypti and C. quinquefasciatus. Moreover, no more than three different storage proteins have been found in Lepidoptera (Figure 2).
Storage proteins serve as a source of amino acids for tissue metamorphosis during pupal development and have been shown to be a component of the sclerotizing system of the cuticle [47]. They also serve as an ecdysteroid carrier in the hemolymph and function in nutrient uptake and storage; some are also capable of binding the insect morphogenetic hormone juvenile hormone [48-50]. Storage proteins are mainly synthesized in fat body during larval development and stored in the hemolymph [9]. Corcyra cephalonica hexamerin protein 2 was expressed in fat body and carcass tissue, but not in salivary gland, midgut or Malpighian tubules [27]. Kim et al. reported that A. germari hexamerin was expressed not only in fat body, but also in midgut, in agreement with our results for SeHex1 [22]. We found that SeHex was expressed not only in fat body and midgut, but also in cuticle and Malpighian tubules (Figure 3A). SP1 in Amsacta albistriga was detected in fat body and pupal ovary [31]. However, the results for SeSP1 reveal that it is expressed in fat body, Malpighian tubules and tracheae (Figure 3A). Storage proteins do not bind copper or transport oxygen to the hemolymph [51,52], and lower expression levels were detected in Malpighian tubules and tracheae (Figure 3A). This is consistent with the theory of hemocyanin superfamily evolution.
Storage proteins accumulate in large quantities in hemolymph during final instar larvae, are taken up by fat body cells and serve as a reservoir for subsequent development [4,43,53]. CcHex2 mRNA was present at all stages of larval development of C. cephalonica and reached a maximum in fat body of final instar larvae [27]. AalSP1 expression gradually increased from day-1 to day-7 final instar larvae [30]. It has been reported that expression levels of insect storage proteins (including Hex1, Hex2, SP1 and SP2) reach a maximum in the last stage of final instar larvae [31,54]. SeHex and SeSP1 mRNA expression also reached a maximum in the final stage (pre-pupae) of last instar larvae (Figure 3B). However, SeHex and SeSP1 mRNA expression was much lower before the first day of fifth instar larvae, so it was not detected in the 25 μg of total RNA used for Northern blotting. In order to get the exact expression patterns of SeHex and SeSP1 mRNA in the larvae developmental stages, the more sensitive way of RT-PCR is used. And RT-PCR results confirmed that expression of these storage proteins is high in final instar larvae (Figure 3C). This result showed that storage protein can be accumulated on the good nutritional status, just as the last larvae in S. exigua.
The function of storage proteins is clear. In M. sexta, starvation is effective in reducing mRNA levels of both hexamerin genes [55]. In the group of starvation with 12 h, SeHex and SeSP1 mRNA expression was much lower than control (Figure 4A, 4B), that showed storage proteins are key in the stress of starvation. Hakim and colleagues reported that a monomeric α-arylphorin storage protein can stimulate stem cell proliferation in the lepidopterans M. sexta and Spodoptera littoralis and in the beetle Leptinotarsa decemlineata. In addition, feeding an artificial diet containing arylphorin increased the growth rate of several insect species [56]. It is known that storage proteins are crucial for insect development and disruption has a negative effect on the natural development and movement of an insect, which may eventually die. RNAi was used to investigate the function of SeHex and SeSP1 in the present study and resulted in adult survival rates of 38.73% and 24.29%, respectively (Figure 5). Moreover, SeHex and SeSP1 mRNA was substantially lower at 36 h after injection compared to the negative control (Figure 6). And the expression levels of some control larvae are weird may result in the side effect of inject. The results confirm that SeHex and SeSP1 mRNA expression is crucial for insect life. Abnormal morphological development was not observed after RNAi treatment. It is also found that SeHex and SeSP1 mRNA are lower on the condition of starvation. Hence, a lack of nutritional reserves or developmental failure may be key reasons for the lethality. Nevertheless, the SeHex and SeSP1 RNAi results suggest that a potential method for control of this pests is to disrupt the regulation of storage proteins.
Conclusions
We demonstrated that genes for two storage hexamerins exist in S. exigua. SeHex transcripts were expressed not only in fat body, but also in cuticle, midgut and Malpighian tubules. SeSP1 mRNA was expressed in fat body, Malpighian tubules and tracheae. Furthermore, SeHex and SeSP1 have differential expression patterns in fat body. The results of starvation treatment suggest that SeHex and SeSP1 have important functions in S. exigua. RNAi results showed that these proteins are critical for S. exigua metamorphosis, indicating that a potential method for control of this pest is to disrupt the regulation of storage proteins.
Methods
Insect culture
S. exigua larvae were reared at 25 ± 1°C with an L14:D10 photoperiod on an artificial diet [37]. The developmental stages were synchronized at each molt by collecting new larvae or pupae. The brain, midgut, fat body, cuticle, Malpighian tubules, spermary and tracheae from different stages from fifth instar larvae and the fat body from day-1 fifth instar to day-7 pupae were dissected in saline containing 0.75% NaCl and stored at -80°C until further use.
RNA isolation and cDNA synthesis
Total RNA was isolated from the fat body of S. exigua pupae using the acid guanidinium thiocyanate-phenol-chloroform method [57,58]. The fat body (100 mg) was homogenized in solution D (4 M guanidinium thiocyanate; 25 mM sodium citrate, pH 7; 0.5% sarcosyl; 0.1 M 2-mercaptoethanol), placed on ice for 5 min, and then sodium acetate, phenol (pH 7.0) and chloroform/isoamyl alcohol (49:1) were added. The mixture was centrifuged at 10,000×g at 4°C for 20 min. The supernatant was transferred to a new tube and then isopropanol was added. After centrifugation, the RNA pellet was washed with 75% ethanol and then dissolved in ddH2O. A sample of 1 μg of total RNA was reverse transcribed at 42°C for 1 h in a 25- μl reaction solution containing reaction buffer, 10 mM DTT, 0.5 mM dNTPs, 0.5 μg of oligo-dT18 and reverse transcriptase from avian myeloblastosis virus (AMV, Takara, Japan) [59].
Polymerase chain reaction and rapid amplification of cDNA ends
Two pairs of degenerate primers were designed based on the conserved amino acid sequences of known Hex and SP1 proteins. The first PCR reaction was performed with primers DF1 and DR1 using the following conditions: three cycles of 30 s at 94°C, 30 s at 45°C and 90 s at 72°C, followed by 30 cycles of 30 s at 94°C, 30 s at 48°C and 90 s at 72°C. A second PCR was carried out using the nested primers DF2 and DR2 using the same cycle conditions [60]. The products were subjected to agarose electrophoresis and the DNA bands were then excised from the gel and purified using a DNA gel extraction kit (Omega, USA). The PCR products were cloned into the pMD18-T vector (Takara) and sequenced by the dideoxynucleotide method (Invitrogen).
cDNA was synthesized according to the manufacturer's protocol (SMART™ kit, Clontech/Takara). Specific primers, Hex-5R1, Hex-5R2, SP1-5R1 and SP1-5R2 for 5'-RACE and Hex-3F1, Hex-3F2, SP1-3F1 and SP1-3F2 for 3'-RACE (Table 1), were designed and synthesized based on the cDNA sequence of the known PCR fragment. 5'-RACE was performed using 2.5 μl of 5'-ready-cDNA with Universal Primer Mix (UPM, Clontech/Takara) and Hex-5R1 or SP1-5R1. Then nested PCR was carried out using Nested Universal Primer A (NUP, Clontech/Takara) and Hex-5R2 or SP1-5R1. 3'-RACE was performed using 2.5 μl of 3'-ready-cDNA with UPM and Hex-3F1 or SP1-3F1, then with NUP and Hex-3F2 or SP1-3F2. The PCR conditions were as follows: 10 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 60°C and 150 s at 72°C, then 10 min at 72°C. Samples were then kept at 12°C.
Table 1.
Primers for cDNA cloning, Northern blotting, semi-quantitative RT-PCR, dsRNA synthesis and detection of SeHex and SeSP1 RNAi
| PCR Fragme |
Primers | |||
|---|---|---|---|---|
| Name | Directiona | Typeb | Nucleotide Sequence (5'-3') | |
| 1 | Hex-DF1 | F | D | TGG ARM GHC TGT CYA ACG |
| Hex-DF2 | F | D | ACA AKG GMA TBH HST TCC | |
| SP1-DF1 | F | D | GGC ATG TTC DTV TAT GC | |
| SP1-DF2 | F | D | TAC CCN TAC TWC TTC GTC | |
| Hex-DR1 | R | D | GAA BGG CAT RCC DCC GAC | |
| Hex-DR2 | R | D | GTC CAG CTS SAW GAA GTT | |
| SP1-DR1 | R | D | CGT ACT TGG GNC CAN DGA AG | |
| SP1-DR2 | R | D | TGA ANG GVT GRT GRT TGA G | |
| 2 RACE |
3-CDS | R | O | AAG CAG TGG TAT CAA CGC AGA GTA C(T)30VN |
| NUP | F | A | AAG CAG TGG TAT CAA CGC AGA GT | |
| Hex-5R1 | R | G | GCT GAT GTC AAT ACG TTC ACC GAG | |
| Hex-5R2 | R | G | CGT AAC CCT GTT CGA TGG CGT CAC | |
| SP1-5R1 | R | G | GAT GAC ATG GCT ATC GAC GA | |
| SP1-5R2 | R | G | CGT AGG GAG CAG GCA GAG TG | |
| Hex-3F1 | F | G | GTG AGC GTG TTG GTA CAG CAT CC | |
| Hex-3F2 | F | G | GAG TGA AGT TGC TAA GAC CGT TC | |
| SP1-3F1 | F | G | CAA CTG ACA AGC TAG TGA CC | |
| SP1-3F2 | F | G | CGC TGA GAT GAA GAA GAA GC | |
| 3 probe | Hex-FP | F | G | CGT TTG CAC GAG GTC TTC C |
| Hex-RP | R | G | GGC TTC TTC TTC ATT CAT GTG C | |
| SP1-FP | F | G | GAC CAA GGC AGC GAA TGA CC | |
| SP1-RP | R | G | CAG GGC GTT GGT GAT ATC CA | |
| 4 dsRNA | dsHex-F | F | G | GAC AGC CAC GGC TAT GAG ATT CC |
| dsHex-FT | F | G | GGA TCC TAA TAC GAC TCA CTA TAG GNG ACA GCC ACG GCT ATG AGA TTC C | |
| dsHex-R | R | G | CCT TAG AAA CAT TGG TAG GCT CG | |
| dsHex-RT | R | G | GGA TCC TAA TAC GAC TCA CTA TAG GNC CTT AGA AAC ATT GGT AGG CTC G | |
| dsSP1-F | F | G | GAC TGG AAA GAA CAC CAT CGT C | |
| dsSP1-FT | F | G | GGA TCC TAA TAC GAC TCA CTA TAG GNG ACT GGA AAG AAC ACC ATC GTC | |
| dsSP1-R | R | G | GGT CCT TCC TGT ACA CCA TCA C | |
| dsSP1-RT | R | G | GGA TCC TAA TAC GAC TCA CTA TAG GNG GTC CTT CCT GTA CAC CAT CAC | |
a F, forward, R, reverse; b D, degenerate primer; G, gene-specific primer; A, nested universal primer; O, 3'-RACE CDS primer.
Cloning of full-length SeHex and SeSP1 cDNA
Based on the conserved amino acid and nucleotide sequences of hexamerin and storage protein 1 from B. mori, A. aegypti, A. gambiae, G. mellonella, Helicoverpa armigera and M. sexta, we designed eight degenerate primers (Hex-DF1, Hex-DF2, Hex-DR1, Hex-DR2, SP1-DF1, SP1-DF2, SP1-DR1and SP1-DR2) for PCR reactions. Fragments of 800 bp (SeHex-DF2 and SeHex-DR2) and 1100 bp (SeSP1-DF2 and SeSP1-DR2) were first obtained from fat-body cDNA by a second PCR. After sequencing, the deduced amino acid sequence was found to have a high degree of similarity to insect hexamerin classes. We then performed 5' and 3' rapid amplification of cDNA ends (RACE) using several specific primers (Table 1) designed based on the fragment sequences, as well as universal primers (Clontech). The 600- and 1000-bp PCR products for SP1 and two 1100-bp PCR products for Hex were amplified by 5' and 3'-RACE, respectively. After assembling the overlapping fragments, full-length cDNAs of 2651 bp for Hex and 2254 for SP1 were obtained.
Analysis of SeHex and SeSP1 cDNA sequences
The neighbor-joining method was used to construct a phylogenetic tree with MEGA 3.1 software based on the amino acid sequences of known insect hexamerins. SeHex and SeSP1 cDNA sequences were compared with those for other Hex, SP1 or hexamerin proteins deposited in GenBank using the BLAST-N or BLAST-X tools at the National Center for Biotechnology Information (NCBI) website. The amino acid sequences encoded by SeHex and SeSP1 were deduced from the corresponding cDNA sequences using the translation tool at the ExPASy Proteomics website http://expasy.org/tools/dna.html. A bootstrap analysis was carried out and the robustness of each cluster was verified using 1000 replicates. Other protein sequence analysis tools used in this study, including MW, pI and N-glycosylation sites, were obtained from the ExPASy Proteomics website http://expasy.org/. Multiple sequence alignment of insect hemocyanin family protein sequences was performed using the tool at the multiple sequence alignment website http://bioinfo.genotoul.fr/multalin/multalin.html.
Probe labeling and Northern blotting analysis
Tissue-specific expression of SeHex and SeSP1 was determined by Northern blotting. cDNA fragments of 990 bp (primers Hex-FP and Hex-RP) and 895 bp (primers SP1-FP and SP1-RP) were labeled with [α-32P]dCTP using a random primer DNA labeling kit (Takara, Japan) as the hybridization probe [59]. Samples of 25 μg of total RNA isolated from brain, midgut, fat body, cuticle, Malpighian tubules, spermary and tracheae of day-3 fifth instar larvae were separated on a formaldehyde agarose gel containing ethidium bromide [59]. The RNA was subsequently blotted onto a Hybond-N+ membrane (Amersham) and pre-hybridized at 42°C for 3-6 h. The [α-32P]-labeled SeHex and SeSP1 probes were added and incubated at 42°C for up to 12 h in 5× SSPE (0.75 M NaCl, 0.05 M NaH2PO4, 0.005 M EDTA, pH 7.4) containing 50% formamide, 5× Denhardt's solution, 0.1% SDS and 100 mg/ml salmon sperm DNA [61,62]. After hybridization, the membrane was washed with 0.2× SSPE (0.03 M NaCl, 0.002 M NaH2PO4, 0.0002 M EDTA, pH 7.4) at 45°C and exposed to X-ray film at -70°C for 24 h [59].
Developmental expression of SeHex and SeSP1 by Northern blotting and RT-PCR amplification
Developmental expression of SeHex and SeSP1 from fifth instar larvae to pupae was determined by Northern blotting. The fat body was dissected from day-1 fifth instar larvae to day-7 pupae. Total RNA was isolated from the fat body and 25 μg was separated on a formaldehyde agarose gel containing ethidium bromide. Northern blotting was carried out as described above. A sample of 25 μg of total RNA was also used to detect the expression of SeHex and SeSP1 in young larvae, but the results were poor. Thus, more sensitive RT-PCR was used to detect the expression of SeHex and SeSP1 in larval stages. The PCR reaction was performed with primers Hex-FP and Hex-RP (or SP1-FP and SP1-RP) using the following conditions: 10 min at 94°C, followed by 37 cycles of 30 s at 94°C, 30 s at 60°C and 70 s at 72°C, then 10 min at 72°C. Samples were then kept at 12°C. The PCR products were detected by electrophoresis on agarose gel containing ethidium bromide.
The expression of SeHex and SeSP1 in starvation treatment by RT-PCR amplification
The day-2 fifth instar larvae were used as experimental insects in the treatment of starvation. In the experiment of starvation, all of the insects were divided into two processed groups and a control group. Two experimental group insects were starvation with 6 h and 12 h, respectively. Followed by these insect continue to rear at 25 ± 1°C with an L14:D10 photoperiod using an artificial diet. The insects were observed at 0 h (before treatment), 6 h, 12 h, 24 h, 36 h, 48 h and 72 h after treatment. Every three to five lively larvae were randomly selected and stored at -80°C for subsequent RNA extraction. The way of PCR reaction just as the previous.
Injection of dsRNA into S. exigua larvae
dsRNA for SeHex and SeSP1 genes was prepared according to methods established in the State Key Laboratory of Biocontrol of Sun Yat-sen University [60]. The T7 RiboMAX™ Express RNAi system (Promega, USA) was used for synthesis. Day-2 fifth instar larvae were used for injection experiments. For each treatment, 5 μg of dsRNA dissolved in 5 μl of DEPC water was injected into the side of the thorax of a larva using a 10- μl syringe (Hamilton) and the injection point was sealed immediately with wax. Control larvae were injected with an equivalent volume of dsRNA corresponding to a GFP gene or were not injected. Each group comprised 30-40 individual larvae and semi-quantitative RT-PCR and survival analyses were carried out in triplicate.
Data analysis and insect survival
Insect survival and morphological changes were recorded every 12 h. The insects were observed at 24 h (day-2 5th instar larvae), 36 h (day-3 5th instar larvae), 48 h (pre-pupae), 60 h (pupae) and 204 h (adult) after injection. To assess the effect of a treatment, ANOVA was performed using the cumulative percentage of abnormal and dead larvae as the dependent variable and treatment (dsSeHex, dsSeSP1, dsGFP or no injection) as the independent variable. Post hoc Duncan's tests were used to determine differences among groups when treatment effects were detected. Results were arcsine square-root transformed before analysis to correct for the non-normal distribution of percentage values.
Semi-quantitative RT-PCR analysis of gene silencing
Insects were observed at 12, 24, 36, 48, 72, 96, 120 and 168 h (pupae) after injection. Larvae in three states (before death, still living and less vital) were randomly selected at each time point and stored at -80°C for subsequent RNA extraction. Total RNA was extracted from individual larvae using the acid guanidinium thiocyanate-phenol-chloroform method [61,62] and converted to cDNA using AMV reverse transcriptase (Takara). The Hex-FP/Hex-RP or SP1-FP/SP1-RP primers were used to amplify all three cDNAs in the same PCR reactions. Pilot experiments demonstrated that 22-24 cycles were optimal for linear amplification of the PCR products, and this protocol was then used in subsequent experiments. PCR amplification was performed in a 25- μl reaction mixture under the following conditions: 10 min at 94°C; 22-24 cycles of 1 min at 94°C, 1 min at 60°C and 1 min at 72°C; and a final 10 min at 72°C. The PCR products were separated on a 2% agarose gel and transferred to a Hybond-N+ nylon membrane. Hybridization, washing and signal detection of the blots were similar to methods previously described [59].
Authors' contributions
BT carried out all experiments and wrote the manuscript. SGW was involved in helping to draft the manuscript and revising it critically for important intellectual content. FZ conceived the project and supervised the experiments. All authors read and approved the final manuscript.
Supplementary Material
Figure S1 Alignment of the amino acid sequences deduced for (A) Hex and (B) SP1 genes in insects. The amino acid sequences deduced for insect Hex and SP1 genes were aligned using Vector NTI 9.0 software. Highly conserved regions are yellow and sky-blue.
Contributor Information
Bin Tang, Email: tbzm611@yahoo.com.
Shigui Wang, Email: sgwang@mail.hz.zj.cn.
Fan Zhang, Email: zf6131@263.net.
Acknowledgements
This work was supported by the National Basic Research Program of China (Grant Nos. 2009CB119206 and 2006CB102005), the National Key Technology R&D Program of China (Grant No. 2008BADA5B02), the National Natural Science Foundation of China (Grant Nos. 31000880 and 30970473), the Zhejiang Provincial Natural Science Foundation of China (Grant No. Y3100176), the Hangzhou Science and Technology Development Program of China (Grant No. 20091133B08) and the Hangzhou normal university high-level talents start-up fund (Grant No.YS05203105).
References
- Wheeler hr DE, Martínez T. Storage proteins in ants (Hymenoptera: Formicidae) Comp Biochem Physiol. 1995;112:15–19. doi: 10.1016/0305-0491(95)00035-7. [DOI] [PubMed] [Google Scholar]
- Zhu YC, Muthukrishnan S, Kramer KJ. cDNA sequences and mRNA levels of two hexamerin storage proteins PinSP1 and PinSP2 from the Indianmeal moth, Plodia interpunctella. Insect Biochem Mol Biol. 2002;32:525–536. doi: 10.1016/S0965-1748(01)00131-X. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Kawooya JK, Ryan RD, Van Heusden MC, Ziegler R. Insect hemolymph proteins. Adv Insect Physiol. 1990;22:299–366. full_text. [Google Scholar]
- Telfer WH, Kunkel JG. The function and evolution of insect storage hexamers. Annu Rev Entomol. 1991;36:205–228. doi: 10.1146/annurev.en.36.010191.001225. [DOI] [PubMed] [Google Scholar]
- Bitondi MM, Nascimento AM, Cunha AD, Guidugli KR, Nunes FM, Simões ZL. Characterization and expression of the Hex 110 gene encoding a glutamine-rich hexamerin in the honey bee, Apis mellifera. Arch Insect Biochem Physiol. 2006;63(2):57–72. doi: 10.1002/arch.20142. [DOI] [PubMed] [Google Scholar]
- Mangum CP. Oxygen transport in invertebrates. Am J Physiol. 1985;248:505–514. doi: 10.1152/ajpregu.1985.248.5.R505. [DOI] [PubMed] [Google Scholar]
- Law JH, Wells MA. Insects as biochemical models. J Biol Chem. 1989;264(28):16335–16338. [PubMed] [Google Scholar]
- Willmer P, Stone G, Johnston I. Environmental Physiology of Animals. (Blackwell, Oxford); 2000. [Google Scholar]
- Martins JR, Nunes FM, Simões ZL, Bitondi MM. A honeybee storage protein gene, hex 70a, expressed in developing gonads and nutritionally regulated in adult fat body. J Insect Physiol. 2008;54(5):867–877. doi: 10.1016/j.jinsphys.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Hagner-Holler S, Pick C, Girgenrath S, Marden JH, Burmester T. Diversity of stonefly hexamerins and implication for the evolution of insect storage proteins. Insect Biochem Mol Biol. 2007;37(10):1064–1074. doi: 10.1016/j.ibmb.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Whitten JM. Comparative anatomy of the tracheal system. Annu Rev Entomol. 1972;17:373–402. doi: 10.1146/annurev.en.17.010172.002105. [DOI] [Google Scholar]
- Burmester T. Origin and evolution of arthropod hemocyanins and related proteins. J Comp Physiol B. 2002;172:95–117. doi: 10.1007/s00360-001-0247-7. [DOI] [PubMed] [Google Scholar]
- Beintema JJ, Stam WT, Hazes B, Smidt MP. Evolution of arthropod hemocyanins and insect storage proteins (hexamerins) Mol Biol Evol. 1994;11:493–503. doi: 10.1093/oxfordjournals.molbev.a040129. [DOI] [PubMed] [Google Scholar]
- Haunerland NH. Insect storage proteins: gene families and receptors. Insect Biochem Molec Biol. 1996;26:755–765. doi: 10.1016/S0965-1748(96)00035-5. [DOI] [PubMed] [Google Scholar]
- Burmester T, Massey HC, Zakharkin SO, Benes H. The evolution of hexamerins and the phylogeny of insects. J Mol Evol. 1998;47:93–108. doi: 10.1007/PL00006366. [DOI] [PubMed] [Google Scholar]
- Ryan RO, Kein PS, Wells MA, Law JH. Purification and properties of a predominantly female-specific protein from the haemolymph of the larva of tobacco hornworm, Manduca sexta. J Biol Chem. 1985;260:782–787. [PubMed] [Google Scholar]
- Markl J, Decker H. Molecular structure of the arthropod hemocyanins. Adv Comp Environ Physiol. 1992;13:325–376. [Google Scholar]
- Burmester T. Evolution and function of the insect hexamerins. Eur J Entomol. 1999;96:213–225. [Google Scholar]
- Wang YC. Insect Biochemistry. Beijing: China Agricultural Press; 2001. p. 144. (in Chinese) [Google Scholar]
- Gordadze AV, Korochkina SE, Zakharkin SO, Norton AL, Benes H. Molecular cloning and expression of two hexamerin cDNAs from the mosquito, Aedes aegypti. Insect Mol Biol. 1999;8(1):55–66. doi: 10.1046/j.1365-2583.1999.810055.x. [DOI] [PubMed] [Google Scholar]
- Moreira CK, Capurro ML, Walter M, Pavlova E, Biessmann H, James AA, deBianchi AG, Marinotti O. Primary characterization and basal promoter activity of two hexamerin gene of Musca domestica. J Insect Sci. 2004;4:1–10. doi: 10.1093/jis/4.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Yoon HJ, Park NS, Lee SM, Moon JY, Seo SJ, Jin BR, Sohn HD. Molecular cloning, expression, and characterization of a cDNA encoding the arylphorin-like hexameric storage protein from the mulberry longicorn beetle, Apriona germari. Arch Insect Biochem Physiol. 2003;53(2):49–65. doi: 10.1002/arch.10085. [DOI] [PubMed] [Google Scholar]
- Kim SR, Lee KS, Yoon HJ, Park NS, Lee SM, Je YH, Jin BR, Sohn HD. cDNA cloning, expression, and characterization of an arylphorin-like hexameric storage protein, AgeHex2, from the mulberry longicorn beetle, Apriona germari. Arch Insect Biochem Physiol. 2004;56(2):61–72. doi: 10.1002/arch.10144. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Rivera L, Hamada M, Soulages JL. Purification and characterization of recombinant lipid storage protein-2 from Drosophila melanogaster. Protein Pept Lett. 2008;15(9):1027–1032. doi: 10.2174/092986608785849191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Tarver MR, Bennett GW, Oi FM, Scharf ME. Two hexamerin genes from the termite Reticulitermes flavipes: Sequence, expression, and proposed functions in caste regulation. Gene. 2006;376(1):47–58. doi: 10.1016/j.gene.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Yoshiga T, Tojo S. cDNA cloning and deduced amino acid sequences of three storage proteins in the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae) Appl Entomol Zool. 2000;35:31–39. doi: 10.1303/aez.2000.31. [DOI] [Google Scholar]
- Nagamanju P, Hansen IA, Burmester T, Meyer SR, Scheller K, Dutta-Gupta A. Complete sequence, expression and evolution of two members of the hexamerin protein family during the larval development of the rice moth, Corcyra cephalonica. Insect Biochem Mol Biol. 2003;33(1):73–80. doi: 10.1016/S0965-1748(02)00178-9. [DOI] [PubMed] [Google Scholar]
- Telfer WH, Pan ML. Storage hexamer utilization in Manduca sexta. J Insect Science. 2003;3:1–6. doi: 10.1672/1536-2442(2003)003[0001:TMOAAE]2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar R. Ph.D Thesis. Bharathidasan University, Tiruchirappalli, India; 2006. Expression and sequestration of storage protein 1 (SP1) in differentiated fat body tissues in groundnut pest, Amsacta albistriga Walk. [Google Scholar]
- Chandrasekar R, Suganthi LM, Krishnan M. Intra-ovarian snthesis and tissue distribution of hexameric storage protein-1 in the ovary of red hairy caterpillar, Amsacta albistriga. J Asia-Pacific Entomol. 2007;10:1–10. doi: 10.1016/S1226-8615(08)60322-2. [DOI] [Google Scholar]
- Chandrasekar R, Jae SS, Krishnan M. Expression and localization of storage protein 1 (SP1) in differentiated fat body tissues of red hairy caterpillar, Amsacta albistriga Walker. Arch Insect Biochem Physiol. 2008;69(2):70–84. doi: 10.1002/arch.20266. [DOI] [PubMed] [Google Scholar]
- Ashfaq M, Sonoda S, Tsumuki H. Expression of two methionine-rich storage protein genes of Plutella xylostella (L.) in response to development, juvenile hormone-analog and pyrethroid. Comp Biochem Physiol. 2007;5:1756–1427. doi: 10.1016/j.cbpb.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Sum H, Haunerland NH. VHDL, a larval storage protein from the corn earworm, Helicoverpa zea, is a member of the vitellogenin gene family. Insect Biochem Mol Biol. 2007;37(10):1086–1093. doi: 10.1016/j.ibmb.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Tungjitwitayakul J, Singtripop T, Nettagul A, Oda Y, Tatun N, Sekimoto T, Sakurai S. Identification, characterization, and developmental regulation of two storage proteins in the bamboo borer Omphisa fuscidentalis. J Insect Physiol. 2008;54(1):62–76. doi: 10.1016/j.jinsphys.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Spyliotopoulos A, Gkouvitsas T, Fantinou A, Kourti A. Expression of a cDNA encoding a member of the hexamerin storage proteins from the moth Sesamia nonagrioides (Lef.) during diapause. Comp Biochem Physiol. 2007;148(1):44–54. doi: 10.1016/j.cbpb.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Gkouvitsas T, Kourti A. Juvenile hormone induces the expression of the SnoSP2 gene encoding a methionine-rich hexamerin in Sesamia nonagrioides (Lepidoptera) Comp Biochem Physiol B. 2009;153(2):206–215. doi: 10.1016/j.cbpb.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Ryan RO, Schmidt JO, Law JH. Arylphorin from the haemolymph of the larval honeybee. Insect Biochem. 1984;14:515–520. doi: 10.1016/0020-1790(84)90005-2. [DOI] [Google Scholar]
- Danty E, Arnold G, Burmester T, Huet JC, Huet D, Pernollet JC, Masson C. Identification and developmental profiles of hexamerins in antenna and hemolymph of the honeybee, Apis mellifera. Insect Biochem Mol Biol. 1998;28(5-6):387–397. doi: 10.1016/S0965-1748(98)00011-3. [DOI] [PubMed] [Google Scholar]
- Martínez T, Wheeler DE. Identification of two storage hexamers in the ant, Camponotus festinatus: accumulation in adult queenless workers. Insect Biochem Molec Biol. 1993;23:309–317. doi: 10.1016/0965-1748(93)90013-I. [DOI] [PubMed] [Google Scholar]
- Martínez T, Wheeler DE. Storage proteins in adult ants: Roles in colony founding by queens and in larval rearing by workers. J Insect Physiol. 1994;40:723–729. doi: 10.1016/0022-1910(94)90100-7. [DOI] [Google Scholar]
- Wheeler DE, Buck NA. Storage proteins in ants during development and colony founding. J Insect Physiol. 1995;41:885–894. doi: 10.1016/0022-1910(95)00028-S. [DOI] [Google Scholar]
- Tojo S, Nagata M, Kobayachi M. Storage proteins in the silkworm, Bombyx mori. Insect Biochem. 1980;10:289–303. doi: 10.1016/0020-1790(80)90024-4. [DOI] [Google Scholar]
- Tojo S, Yoshiga T. Purification and characterization of three storage proteins in the common cutworm, Spodoptera litura. Insect Biochem Mol Biol. 1994;24:729–738. doi: 10.1016/0965-1748(94)90061-2. [DOI] [Google Scholar]
- Kunkel J, Grossniklaus-Buergin C, Karpells S. Arylphorin of Trichoplusia ni: characterization and parasite-induced precocious increase in titer. Arch Insect Biochem Physiol. 1990;13:117–125. doi: 10.1002/arch.940130111. [DOI] [Google Scholar]
- Kramer KJ, Hopkins TL, Ahmed RF, Mueller D, Lookhart G. Tyrosine metabolism for cuticle tanning in the tobacco hornworm, Manduca sexta (L.) and other Lepidoptera: identification of beta-D-glucopyranosyl-O-L-tyrosine and other metabolites. Arch Biochem Biophys. 1980;205(1):146–155. doi: 10.1016/0003-9861(80)90093-4. [DOI] [PubMed] [Google Scholar]
- Willott E, Wang XY, Wells MA. cDNA and gene sequence of Manduca sexta arylphorin, an aromatic amino acid-rich larval serum protein. Homology to arthropod hemocyanins. J Biol Chem. 1989;264(32):19052–19059. [PubMed] [Google Scholar]
- Peter MG, Scheller K. In: The Physiology of Insect Epidermis. Retnakaran A, Binnington K, editor. CSIRO, Australia; 1991. Arylphorin and the integument; pp. 113–122. [Google Scholar]
- Brawn RP, Wyatt GW. Sequence of the hexamerin juvenile hormonebinding protein from the hemolymph of Locusta migratoria. J Biol Chem. 1996;6:31756–31762. doi: 10.1074/jbc.271.49.31756. [DOI] [PubMed] [Google Scholar]
- Gilbert LI, Granger NA, Roe RM. The juvenile hormones: historical facts and speculations on future research directions. Insect Biochem Mol Biol. 2000;30:617–644. doi: 10.1016/S0965-1748(00)00034-5. [DOI] [PubMed] [Google Scholar]
- Tawfik AI, Kellner R, Hoffman KH, Lorenz MW. Purification, characterization and titre of the haemolymph juvenile hormone binding proteins from Schistocerca gregaria and Gryllus bimaculatus. J Insect Physiol. 2006;52:255–268. doi: 10.1016/j.jinsphys.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Burmester T. Identification, molecular cloning and phylogenetic analysis of a non-respiratory pseudo-hemocyanin of Homarus americanus. J Biol Chem. 1999;274:13217–13222. doi: 10.1074/jbc.274.19.13217. [DOI] [PubMed] [Google Scholar]
- Terwilliger NB, Dangott LJ, Ryan MC. Cryptocyanin, a crustacean molting protein: evolutionary links to arthropod hemocyanin and insect hexamerins. Proc Natl Acad Sci. 1999;96:2013–2018. doi: 10.1073/pnas.96.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmester T. Molecular evolution of the arthropod hemocyanin superfamily. Mol Biol Evol. 2001;18(2):184–195. doi: 10.1093/oxfordjournals.molbev.a003792. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Kłudkiewicz B, Grzelak K, Beresewicz M, Cymborowski B. Hormonal regulation of the expression of two storage proteins in the larval fat body of the greater wax moth (Galleria mellonella) J Insect Physiol. 2003;49(6):551–559. doi: 10.1016/S0022-1910(03)00026-X. [DOI] [PubMed] [Google Scholar]
- Webb BA, Riddiford LM. Synthesis of two storage proteins during larval development of the tobacco hornworm, Manduca sexta. Dev Biol. 1988;130(2):671–681. doi: 10.1016/0012-1606(88)90359-4. [DOI] [PubMed] [Google Scholar]
- Hakim RS, Blackburn MB, Corti P, Gelman DB, Goodman C, Elsen K, Loeb MJ, Lynn D, Soin T, Smagghe G. Growth and mitogenic effects of arylphorin in vivo and in vitro. Arch Insect Biochem Physiol. 2007;64(2):63–73. doi: 10.1002/arch.20155. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(6):156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Aguila ED, Dutra MB, Silva JT, Paschoalin VM. Comparing protocols for preparation of DNA free total yeast RNA suitable for RT-PCR. BMC Molecular Biology. 2005;6:9. doi: 10.1186/1471-2199-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Chen XF, Liu Y, Tian HG, Liu J, Hu J, Xu WH, Zhang WQ. Characterization and expression patterns of a membrane-bound trehalase from Spodoptera exigua. BMC Molecular Biology. 2008;9:51. doi: 10.1186/1471-2199-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XF, Tian HG, Zou LZ, Tang B, Hu J, Zhang WQ. Disruption of Spodoptera exigua larval development by silencing chitin synthase gene A with RNA interference. Bull Entomol Res. 2008;98(6):613–619. doi: 10.1017/S0007485308005932. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. second. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Hoogerwerf WA, Hellmich HL, Micci MA, Winston JH, Zhou L, Pasricha PJ. Molecular cloning of the rat proteinase-activated receptor 4 (PAR4) BMC Molecular Biology. 2002;36:29. doi: 10.1186/1471-2199-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Alignment of the amino acid sequences deduced for (A) Hex and (B) SP1 genes in insects. The amino acid sequences deduced for insect Hex and SP1 genes were aligned using Vector NTI 9.0 software. Highly conserved regions are yellow and sky-blue.