Abstract
Background
The tree of life is significantly asymmetrical - a result of differential speciation and extinction - but general causes of such asymmetry are unclear. Differences in niche partitioning are thought to be one possible general explanation. Ecological specialization might lead to increases in diversification rate or, alternatively, specialization might limit the evolutionary potential of specialist lineages and increase their extinction risk. Here we compare the diversification rates of gall-inducing and non-galling insect lineages. Compared with other insect herbivores feeding on the same host plant, gall-inducing insects feed on plant tissue that is more nutritious and less defended, and they do so in a favorable microhabitat that may also provide some protection from natural enemies. We use sister-taxon comparisons to test whether gall-inducing lineages are more host-specific than non-galling lineages, and more or less diverse than non-gallers. We evaluate the significance of diversity bipartitions under Equal Rates Markov models, and use maximum likelihood model-fitting to test for shifts in diversification rates.
Results
We find that, although gall-inducing insect groups are more host-specific than their non-galling relatives, there is no general significant increase in diversification rate in gallers. However, gallers are found at both extremes - two gall-inducing lineages are exceptionally diverse (Euurina sawflies on Salicaceae and Apiomorpha scale insects on Eucalytpus), and one gall-inducing lineage is exceptionally species-poor (Maskellia armored scales on Eucalyptus).
Conclusions
The effect of ecological specialization on diversification rates is complex in the case of gall-inducing insects, but host range may be an important factor. When a gall-inducing lineage has a host range approximate to that of its non-galling sister, the gallers are more diverse. When the non-galler clade has a much wider host range than the galler, the non-galler is also much more diverse. There are also lineage-specific effects, with gallers on the same host group exhibiting very different diversities. No single general model explains the observed pattern.
Background
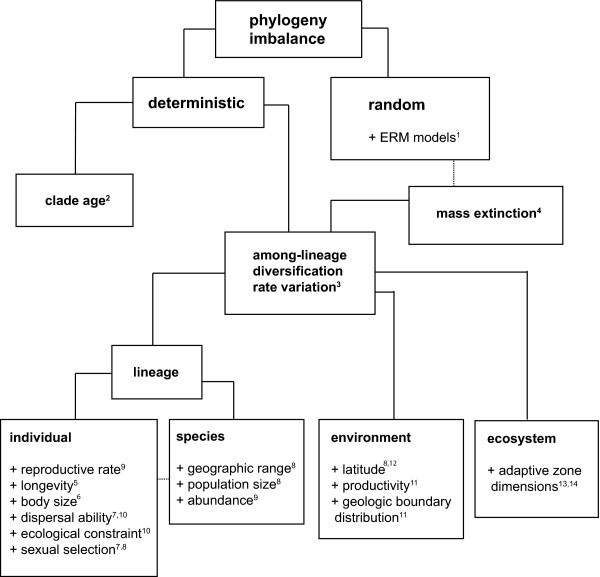
The tree of life is significantly less balanced than expected under a stochastic process of lineage divergence and extinction [1] - some lineages are diverse whereas others are species-poor. Deterministic explanations for the asymmetry include clade age [2], and among-lineage diversification rate variation [3] caused by mass extinction [4], lineage attributes [5-9], environmental attributes [10,11], and ecosystem attributes [12,13] (Figure 1). Lineage attributes affecting diversification rates can be divided into two classes: (1) phenotypic traits that are attributes of individuals, for example reproductive rate, dispersal ability, and the degree of ecological specialization; and (2) traits that are attributes of species, for example geographic range, population size, and local abundance. A key factor in the theory of diversification rate variation is resource availability and breadth, i.e. adaptive zone dimensions. Under an adaptive radiation model [14-16] it is argued that diversification is limited to the amount of free space in an adaptive zone, and that elevated rates of diversification are driven by ecological opportunities in geographic space (e.g. island colonization) or the evolution of an adaptive trait (key innovation). Well-studied examples of adaptive radiation include the Hawaiian silverswords [17], phytophagous beetles [12,18], and columbines [16]. Ecological specialization is thought to be an important process following expansion of a lineage's adaptive zone, and a major driving force generating species richness and diversity [15,19-21].
Figure 1.
Classification of causes of tree imbalance. Examples of attributes that are commonly discussed in the literature are preceded by '+,' with references in superscript. Mass extinction is shown as both random and deterministic; mass extinctions are stochastic events, but not the type captured by purely neutral ERM models.
Not all adaptive traits are expected to result in an increased rate of net diversification; on the contrary, some adaptive traits may result in a dramatic depression of the diversification rate e.g. in-bred sociality in theridiid spiders [22]. Reduced radiation rate as a result of failure to speciate is commonly interpreted to result in "evolutionary dead-ends" - lineages that have low adaptation potential and are thought likely to become extinct before they can diversify [23,24]. Thus, the expected effect of ecological specialization on diversification rate has been an area of debate [25]. On the one hand, diversification rate is likely to increase if specialists have smaller geographic ranges and population sizes [26]. On the other hand, tightly constrained niches are likely to be unstable over time, and it has been predicted that specialization should be associated with increased extinction rates due to a specialist's inability to adapt [23,27,28], a notion supported by some empirical analyses [29,30].
The evolution of gall-induction on plants is a major trophic shift that has occurred multiple times among insects, with over 13,000 described species with this habit [31]. Galls are believed to provide the inducer with enhanced nutrition, a favorable microclimate and, in some cases, protection from natural enemies [32]. Among nematine sawflies, gall-inducing species are targeted by fewer species of parasitoid and experience lower rates of mortality than external-feeding species [33], but in general natural enemies can inflict high mortalities on gallers, and top-down selection pressure is thought to be a critical force driving interspecific variation in gall form [32,34,35]. Insect galls are thus an extended phenotype of their inducers - with the gall exposed to selection pressures related to predation and host resistance [32].
Gall-inducing taxa appear to be exceptionally host- and tissue-specific. For example, less than 1% of the described species of gall wasps (Cynipidae) have been recorded from more than one host genus [36], and gall wasps on Quercus are the single most diverse lineage of herbivores associated with a single host genus (about 1000 described species [36]). All of the 640 described species of agaonid fig wasps induce galls within the flowers of species of Ficus [37]. Within-host-plant diversification has occurred in many groups of gall midges [38,39] with, for example, a monophyletic group of 14 species of Asphondylia (Diptera: Cecidomyiidae) inducing galls on the leaves, stems, buds and flowers of a single plant species (Larrea tridentate) [40]. Only three of the fourteen described species of the gall-inducing psyllid genus Calophya (Psylloidea: Calophyidae) associated with Schinus (Anacardiaceae) are known to develop on more than one host species - there are eight species of Schinus [41]. Gall-inducing species of aphids and scale insects also tend to be constrained to closely related hosts [42-45].
These observations of host-specificity, and the intimate relationship between gall-inducer and host, have led to the idea that gall-inducers are specialized relative to their externally-feeding progenitors [46,47]. Gall-inducing insects thus provide a good study system to examine the effects of ecological specialization on evolutionary diversification rates.
Here, we examine phylogenies of gall-inducing insects and their non-galling relatives, including a total of approximately 1,650 species (Additional files 1,2,3,4). We first use thirteen sister-taxon comparisons to test the assumption that gallers are more host-specific than non-gallers. We then evaluate the significance of any diversification asymmetry against an Equal Rates Markov (ERM) model. Finally, we use maximum likelihood to compare the fit of fixed (1 speciation rate) and flexible (2 or more speciation rates) birth-death models to phylogenies, to test whether inferred origins of gall induction correlate with a shift in diversification rate [48,49].
Results
Host-specificity
Gall-inducing insects are significantly more host-specific than their non-galling sister groups (exact one-tailed Wilcoxon signed-rank test p-value = 0.050). All five significant differences in host range favored gallers being more host specific than their non-galling sister group.
Diversification rates
Sister-taxon comparisons
The gall-inducing sister-clade was more diverse in seven of thirteen comparisons, and the non-galling sister was more diverse in the other six. We cannot reject the null model in which each sister has an equal chance of being more diverse with the binomial test (exact two-tailed test p-value = 1) or the signed-rank test (two-tailed p-value = 0.95).
One gall-inducing taxon was more diverse than expected under an Equal Rates Markov (ERM) null model: the scale insect genus Apiomorpha (p-value = 0.020). Two gall-inducing taxa were more diverse with marginal non-significance: Cerataphidini aphids (p-value of 0.053), sawflies in the subtribe Euurina (p-value = 0.059). Two gall-inducing taxa were markedly less diverse than expected: the armored scale insect genus Maskellia (p-value = 0.0023) and the sawfly genus Micronematus (p-value = 0.033).
ML (birth-death) modeling of shifts in radiation rate
Only two data sets were sufficiently sampled to satisfy criteria for tests of rate shifts along lineages. For the Acacia thrips, we were unable to reject the fixed-rate null model. In the LASER analysis, under no extinction (ϵ = 0), the likelihood ratio test (LRT) p-value was 0.096. Under high rates of extinction (ϵ = 0.95) the LRT p-value was 0.17. The MEDUSA analysis selected a one-rate model with a net diversification rate of 0.042 and a relative extinction rate of 0.13. All more complicated models had higher Akaike information criterion (AIC) scores.
For the nematine sawflies, the LASER analysis strongly favored the flexible-rate model (LRT p-value << 0.0001 under no extinction and high extinction) with an increase in diversification rate along the stem branch of the Salicaceae-galling Euurina. The MEDUSA analysis selected a model with 4 shift points. As in the LASER analysis, the single shift point resulting in the largest increase in likelihood was at the stem node of the Euurina. Background rates of net diversification (r) and relative extinction (ϵ) were estimated as 0.048 and 0.69 respectively. Within Euurina both the net diversification rate and relative extinction rate increased dramatically (r= 0.094, ϵ = 0.99).
Discussion
We confirm that, as expected and commonly stated, gall-inducing taxa are more host-specific than their non-galling relatives. The effect of ecological specialization in gall-inducing insects on diversification rates is complex, but host range appears to be a critical factor. When a gall-inducing lineage has a host range approximate to that of the non-galling sister, the gallers are more diverse. These include the Salicaceae-galling Euurina gall wasps that are sister to a group restricted to Salicaceae and Betulaceae, the Styrax-galling aphids Cerataphidini that are sister to a group restricted to Quercus, and the Eucalyptus-galling scale insect genus Apiomorpha that is sister to a group that occurs on Eucalyptus and Casuarinaceae. Conversely, when the non-galling clade has a much wider host range than the galler, the non-galler is much more diverse. In our analysis, the diversity bipartition that is most significantly weighted in favor of the non-galling taxon (Maskellia sister to a taxonomically heterogeneous group of armored scales) is also the comparison with the greatest disparity in host range, as measured by the estimated age of the most recent common ancestor of the hosts. The importance of host breadth as a determinant of diversification rate is consistent with the assumed importance of host-switching in speciation of phytophagous and parasitic lineages [46,50].
Thus adaptive zone dimensions may be of more fundamental importance to diversification rate variation than is the degree of ecological specialization. When clade host breadth was roughly equivalent between gall-inducing and non-galling sister groups, the gall-inducing group was more diverse. This could result from uneven diversification rates stemming from differences in species-level ecological specificity, or because, for a given set of hosts, the adaptive space available to a gall-inducing lineage is larger than that presented to a non-galling species.
Ecological traits of host lineages are also likely to affect diversification rates of gall-inducers. Two of the diverse gall-inducing groups in this study, Euurina and Apiomorpha, occur on host taxa (Salix and Eucalyptus respectively) that are persistent and locally abundant over vast geographic spaces, traits thought to reduce a lineage's probability of extinction. On the other hand, the most strikingly species-poor gall-inducing taxon included in this study, Maskellia, also occurs on Eucalyptus, and the gall-inducing thrips on Acacia are not as diverse as might be expected given the diversity of the host (more than 1000 described species of Acacia [51]), although it likely that much of the true species diversity of gall-inducing thrips is unrecognized [52].
Gall-inducer diversification rates could also be profoundly affected by top-down pressure from parasitoids and pathogens. Gall-inducing Euurina sawflies have been shown to have a depauperate fauna of natural enemies and suffer lower enemy-caused mortality than closely related external feeders. In contrast, species of Apiomorpha experience extremely high mortality rates (LGC personal observations).
Conclusions
Gall-inducing lineages tend to be more host-specific than their non-galling sister groups. A gall almost certainly represents an expansion of the ecological resource pool (e.g. new tissues and, at least initially, enemy-free spaces), but the gall-inducer's niche space may not be expanded. The effect of the evolution of gall-induction on net diversification rate appears to have been lineage-specific; no general trend was detected, but some gall-inducers were exceptionally diverse and others were exceptionally under-diverse. Although this study was not designed explicitly to test the effect of a lineage's host-breadth on its net diversification, our results suggest that host-breadth may be a general factor influencing the net diversification of phytophagous insect groups.
Methods
Data
We surveyed the literature for published phylogenies containing gall-inducing taxa and their non-galling relatives. We also used the PhyLoTA Browser rel. 1.01 [53] to survey the nucleotide sequence data deposited in GenBank for groups containing gall-inducers. Our dataset included thirteen galler and non-galler sister clades (Table 1). Unless otherwise noted, extant diversity estimates were derived from the literature (see Additional file 1). We estimated phylogenies when: (1) a sister relationship of interest had not been inferred with support in a published analysis and/or additional DNA sequence data had subsequently become available; (2) phylogenies and extant diversity were deemed sufficient (we would be able to allocate all extant species diversity among terminal nodes) for ML diversification rate model fitting. Details of the phylogenetic datasets and results for individual taxa are provided as Additional Material.
Table 1.
Comparisons between galling and non-galling insect sister groups
| Galling-sister | Species number | Host range | Gall host MRCA age (Ma) | ERM P galler | Non-galling sister | Species number | Host range | Non-gall host MRCA age (Ma) | ERM P non-galler |
|---|---|---|---|---|---|---|---|---|---|
| Euurina | 400 | Salix, Populus | 58 | 0.059 | Nematus melanaspis-group | 25 | Salicaceae, Betulaceae | 94 | 0.94 |
| Bacconematus | 1 | Ribes | 35 | 1.00 | Eitelius | 2 | Salix | 35 | 0.50 |
| Pristolina | 4 | Vaccinium | 0 | 0.50 | Pristicampus | 3 | Potentilla | 0 | 0.67 |
| Micronematus | 1 | Prunus | 70* | 0.97 | Pristiphora subgenus Sala | 30 | Fagaceae, Betulaceae, Salicaceae, Fabaceae, Rosaceae, Grossulariaceae, Malvaceae | 114 | 0.033 |
| Kladothrips | 24 | Acacia | 0 | 0.21 | Rhopalothripoides | 6 | Acacia | 0 | 0.83 |
| gall-inducing Cynipidae + Synergini | 1369 | Fagaceae, Rosaceae, Papaveraceae, Lamiaceae, Asteraceae, Anacardiaceae, Smilacaceae, Valerianaceae, Apiaceae, Sapindaceae | 144 | 0.24 | Figitidae (in part): Charpinae, Anacharitinae, Figitinae, Aspicerinae | 435 | Diptera, Neuroptera, Hymenoptera | 300 | 0.76 |
| Agaonidae + Otitesellinae | 690 | Ficus | 60 | 0.15 | Sycoryctinae | 121 | Agaonidae + Sycoryctinae | 60 | 0.85 |
| Eurostina | 41 | Asteraceae (Soldagininae: Solidago; Chrysothamnus; Gutierrezia)) | 0 | 0.54 | Euaresta | 46 | Asteraceae (Ambrosiinae: Ambrosia; Xanthium; Dicoria) | 0 | 0.48 |
| Oedapidina | 88 | Asteraceae | 42 | 0.71 | Tephrellini | 210 | Acanthaceae, Lamiaceae, Verbenaceae | 45 | 0.30 |
| Hexomyza | 16 | Liliopsida/Eudicotyledons | 144 | 0.95 | Ophiomyia + Tropicomyia | 282 | Liliopsida/Eudicotyledons | 144 | 0.054 |
| Apiomorpha | 150 | Eucalyptus | 65 | 0.020 | Ourococcus | 3 | Myrtaceae, Casuarinaceae | 104 | 0.99 |
| Maskellia | 2 | Eucalyptus | 65 | 1.00 | Aspidiotini; Pseudaonidina; Odonaspidini; non-pupillarial Parlatorini | 874 | Magnoliphyta/Coniferophyta | 366 | 0.0023 |
| Cerataphidini | 73 | Styrax | 0 | 0.053 | Thelaxes (Thelaxinae) | 4 | Quercus | 0 | 0.96 |
Phylogenetics
Unaligned FASTA files were downloaded from PhyLoTA, and aligned using MUSCLE v.3.6 [54]. Ribosomal alignments were filtered through the Gblocks server [55,56], with each of the options for less stringent selection chosen, to remove areas of high alignment ambiguity. Introns were excluded from nuclear protein-coding loci. If multiple loci were available, these were concatenated. Datasets were partitioned by genome, and by codon position for protein-coding loci. Maximum likelihood (ML) trees were inferred using RAxML v.7.0.3 [57], with the parameters of a general time reversible (GTR) nucleotide substitution model estimated independently for each data partition. Among-site rate variation was estimated under CAT approximation during 100 nonparametric bootstrap pseudoreplicates. Every fifth bootstrap tree was then used as a starting tree for more thorough ML optimization with gamma-distributed rate variation.
Host specificity analysis
In order to remove taxonomic bias from measures of host breadth, host range was measured as the age (Ma) of the most recent common ancestor (MRCA) of the hosts. This approach to quantifying host breadth is akin to phylogenetic diversity (PD) [58], the minimum total length of branches that span a given set of taxa on a phylogenetic tree. Our metric is distinct, however, in that branch lengths have been scaled to time rather than raw branch length. Because age is standard across analyses, it could be used to make comparisons across DNA sequence datasets with variable substitution rates. It is a measure of the evolutionary depth of host breadth, whereas PD also accounts for the phylogenetic density of host use. None of the sister taxa used in our comparisons was restricted to a single host species.
For each of 13 pairs of gall-inducing and non-galling insect sister pairs, we recorded the host range of each sister. In cases for which each sister in a comparison was restricted to a single host genus or family, and our knowledge of host phylogeny and/or insect host breadth was insufficient to identify an age for the MRCA, we assumed there was no difference in host range. We used the Wilcoxon Signed-Rank Test to assess if the observed disparity in host breadth departed significantly from that expected under a null model in which each sister has an equal chance of having a broader host range. The test was one-tailed, reflecting our prior expectation, derived from the literature, that gall-inducing taxa would be more host-specific.
Diversification rate analyses
Sister-taxon comparison
Some tree imbalance is expected under null models of stochastic diversification [59], and this needs to be taken into account in comparison of diversification rates. We compared the extant diversity of 13 monophyletic groups of gall-inducing insect species to the extant diversity of their sister taxa, against a null model in which the extant diversity of each sister has a 0.5 probability of being larger [7], and evaluated significance using the binomial and signed-rank tests, contrasting species richness with log-transformed ratios (as in [60], and recommended in [61]).
For each individual diversity bipartition we also calculated equal rates Markov (ERM) probabilities (using the equation 3 of Slowinski and Guyer [62]) for alternative hypotheses in which gall-inducing taxa were expected to more or less diverse, with a Bonferroni correction for multiple comparisons (α/2 = 0.025) to evaluate the significance of the departure from the null model. We did not follow the Slowinski-Guyer method of using Fisher's combined probability test to test the influence of a trait on diversification rates, because of the problems with that approach summarized by Vamosi and Vamosi [61].
ML tests for shifts in radiation rates
Optimal phylogenies were made ultrametric with nonparametric rate smoothing using r8 s v.1.70 [63]. As only the relative node heights were needed, an arbitrary root height of 100 was fixed for each tree. We used the modifications of the ML birth-death model fitting methods of Magallon and Sanderson [48] implemented in the R package LASER [64]. The likelihood was calculated by comparing the observed species diversity of a clade to an expected species diversity given a stem group age and a net diversification rate (speciation rate - extinction rate) estimated from the whole tree. To test for shifts in diversification rate, a fixed null model, in which a single diversification rate was estimated for all lineages, was compared to a flexible alternative model in which an ancestral diversification rate is permitted to shift to a descendent rate along some branch in the tree. The likelihood calculations were repeated for shifts along each branch of the tree. Significance of the model comparison was determined on the basis of likelihood ratio tests (LRT). To ensure that our inferences were robust over a range of extinction rates, analyses were repeated under two values for the ratio of the extinction rate to the speciation rate: 0 (no extinction) and 0.95 (very high extinction rates).
We also sought shifts in diversification rates using a stepwise birth-death model fitting approach based on the AIC implemented as MEDUSA in the R package GEIGER [65,66]. The estimateExtinction and cutAtStem parameters were set to True, and a cutoff of 4 units was selected for the improvement in AIC score required to accept a more complex model.
Automation
We provide a Python program, Systers (Additional file 5) to automate sister-taxon comparisons as outlined in Vamosi and Vamosi 2005 [62]. Species diversities of each sister clade are contrasted using raw differences [13], proportional differences [67], and log ratio differences [60]. Statistical significance is assessed in one of three ways, depending on the number of comparisons. For analyses with ≤ 10 contrasts, significance is assessed with a randomization test for matched pairs [68]. In brief, the sign of the contrast scores are permuted and the sum of the contrasts is found for each possible permutation. The fraction of possible sums more extreme than the observed sum is returned as a two-tailed p-value. For analyses with 11-19 contrasts, significance is assessed with a Wilcoxon signed-rank test, and for analyses with ≥ 20 contrasts significance is assessed by normal approximation of the Wilcoxon signed rank test.
List of abbreviations
AIC: Akaike information criterion; ERM: equal rates markov; GTR: general time reversible; LRT: likelihood ratio test; ML: maximum likelihood; MRCA: most recent common ancestor; PD: phylogenetic diversity.
Authors' contributions
NBH and LGC cooperated in the conception and design of the study as well as the drafting of the manuscript. NBH carried out the data-mining and analyses and wrote the Python program Systers. Each read and approved the final manuscript.
Supplementary Material
Phylogenetic datasets and results. Survey of DNA sequence-based phylogenetic studies including gall-inducing groups, and details of phylogenetic estimates performed here.
ML tree estimated from aphid DNA sequence data. Aphidoidea ML phylogeny estimated from analysis of EF1α, long-wavelength opsin, COI, COII, cytochrome b, NADH dehydrogenase 1, ATP synthase subunit 6, and mitochondrial ribosomal subunits 12 S and 16 S dataset partitioned by genome and codon position. Major lineages are labeled following the classification used by Blackman and Eastop.
ML tree estimated from cynipoid DNA sequence data. Cynipoidea ML phylogeny estimated from analysis of 28 S, 18 S, and COI dataset partitioned by genome and codon position. Deep relationships supported by >70% bootstrap proportions denoted by stars. Empty circle indicated clade of unpublished Cynipini sequences assumed to be misidentifications of synergine inquilines.
ML tree estimated form chalcidoid DNA sequence data. Chalcidoidea ML phylogeny estimated from analysis of 28 S sequences. Deep relationships supported by >70% bootstrap proportions denoted by stars. Group A composed of exemplars of the following families: Aphelinidae, Chalcididae, Encyrtidae, Eucharitidae, Leucospidae, Mymaridae, Pteromalidae, Perilampidae, Tetracampidae; Group B composed of exemplars of: Aphelinidae, Eurytomidae, Ormyridae, Perilampidae, Pteromalidae, Tanaostigmatidae, Tetracampidae, Torymidae.
Python script to automate sister taxon comparisons. A program that reads sister clade species diversities in a CSV file, calculates diversity contrasts with multiple metrics, and evaluates significance with either Siegel's randomization test for matched pairs, the Wilcoxon signed rank test, or normal approximation of the Wilcoxon signed rank test.
Contributor Information
Nate B Hardy, Email: nbhardy@gmail.com.
Lyn G Cook, Email: l.cook@uq.edu.au.
Acknowledgements
Mike Crisp and two anonymous reviewers provided helpful comments on the manuscript. We thank Dianne Cook for statistical advice. This study was partly funded by Australian Biological Resources Study grants to NBH and to LGC, an Australian Research Council Discovery Project grant to LGC, and another to Mike Crisp and LGC.
References
- Guyer C, Slowinsky JB. Adaptive radiation and the topology of large phylogenies. Evolution. 1993;47:253–263. doi: 10.2307/2410133. [DOI] [PubMed] [Google Scholar]
- McPeek AM, Brown JM. Clade age and not diversification rate explains species richness among animal taxa. The American Naturalist. 2007;169:E97–E106. doi: 10.1086/512135. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. Ecological limits on clade diversification in higher taxa. The American Naturalist. 2009. pp. 662–674. [DOI] [PubMed]
- Raup DM. Mass extinctions in the marine fossil record. Science. 1982;215:1501–1503. doi: 10.1126/science.215.4539.1501. [DOI] [PubMed] [Google Scholar]
- Gittleman JL, Purvis A. Body size and species-richness in carnivores and primates. Proceedings of the Royal Society of London B. 1998;265:113–119. doi: 10.1098/rspb.1998.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens IPF, Bennett PM, Harvey PH. Species richness among birds: body size, life history, sexual selection or ecology? Proceedings of the Royal Society of London B. 1999;266:933–939. doi: 10.1098/rspb.1999.0726. [DOI] [Google Scholar]
- Stuart-Fox D, Owens IPF. Species richness in agamid lizards: chance, body size, sexual selection or ecology? Journal of Evolutionary Biology. 2003;16:659–669. doi: 10.1046/j.1420-9101.2003.00573.x. [DOI] [PubMed] [Google Scholar]
- Isaac NJ, Jones KE, Gittleman JL, Purvis A. Correlates of species richness in mammals: body size, life history and ecology. The American Naturalist. 2005;165:600–607. doi: 10.1086/429148. [DOI] [PubMed] [Google Scholar]
- Phillimore AB, Freckleton RP, Orme CDL, Owens IPF. Ecology predicts large-scale patterns of phylogenetic diversification in birds. The American Naturalist. 2006;168:220–229. doi: 10.1086/505763. [DOI] [PubMed] [Google Scholar]
- Cracraft J. Biological diversification and its causes. Annals of the Missouri Botanical Gardens. 1985;72:794–822. doi: 10.2307/2399222. [DOI] [Google Scholar]
- Cardillo M, Orme CDL, Owens IPF. Testing for latitudinal bias in diversification rates: an example using new world birds. Ecology. 2005;86:2278–2287. doi: 10.1890/05-0112. [DOI] [Google Scholar]
- Mitter C, Farrell B, Weigman B. The phylogenetic study of adaptive zones. has phytophagy promoted insect diversification? The American Naturalist. 1988;132:107–128. doi: 10.1086/284840. [DOI] [Google Scholar]
- Wiegman BM, Mitter C, Farrell B. Diversification of carnivorous parasitic insects: extraordinary radiation or specialized dead end? The American Naturalist. 1993;142:737–754. doi: 10.1086/285570. [DOI] [Google Scholar]
- Rundell RJ, Price TD. Adaptive radiation, nonadaptive radiation, ecological specialization and nonecological specialization. Trends in Ecology and Evolution. 2009;7:394–399. doi: 10.1016/j.tree.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends in Ecology and Evolution. 2001;16:372–380. doi: 10.1016/S0169-5347(01)02198-X. [DOI] [PubMed] [Google Scholar]
- Hodges SA, Derieg NJ. Adaptive radiations: From field to genomic studies. Proceedings of the National Academy of Sciences USA. 2009;106:9947–9954. doi: 10.1073/pnas.0901594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin BG, Sanderson MJ. Age and rate of diversification of the Hawaiian silversword alliance (Compositae) Proceedings of the National Academy of Sciences. 1997;95:9402–9406. doi: 10.1073/pnas.95.16.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell BD, Sequeira AS. Evolutionary rates in the adaptive radiation of beetles on plants. Evolution. 2004;58:1984–2001. doi: 10.1111/j.0014-3820.2004.tb00484.x. [DOI] [PubMed] [Google Scholar]
- Mayr E. Systematics and the Origin of Species. New York: Columbia Biological Series; 1941. [Google Scholar]
- Funk DJ, Filchak KE, Feder JL. Herbivorous insects: model systems for the comparative study of speciation. Genetica. 2002;116:251–267. doi: 10.1023/A:1021236510453. [DOI] [PubMed] [Google Scholar]
- Via S, Hawthorne DJ. The genetic architecture of ecological specialization: correlated gene effects on host use and habitat choice in pea aphids. The American Naturalist. 2002;159:S76–S88. doi: 10.1086/338374. [DOI] [PubMed] [Google Scholar]
- Agnarsson I, Aviles L, Coddington JA, Maddison WP. Sociality in theridiid spiders: Repeated origins of an evolutionary dead end. Evolution. 2006;60:2342–2351. [PubMed] [Google Scholar]
- Simpson GG. The Major Features of Evolution. New York: Columbia University Press; 1953. [Google Scholar]
- Kelley ST, Farrell BD. Is specialization a dead end? The Phylogeny of Dendroctonus bark beetles (Scolytidae) Evolution. 1988;52:1731–1743. doi: 10.2307/2411346. [DOI] [PubMed] [Google Scholar]
- Futuyma DJ, Moreno G. The evolution of ecological specialization. Annual Review of Ecology and Systematics. 1988;19:207–233. doi: 10.1146/annurev.es.19.110188.001231. [DOI] [Google Scholar]
- Fryer G, Iles TD. Alternative routes to evolutionary success as exhibited by African cichlid fishes of the genus Tilapia and the species flocks of the Great Lakes. Evolution. 1969;23:359–369. doi: 10.2307/2406692. [DOI] [PubMed] [Google Scholar]
- Huxley JS. Evolution, the Modern Synthesis. London: Allen & Unwim; 1942. [Google Scholar]
- Stireman JO. The evolution of generalization? Parasitoid flies and the perils of inferring host range evolution from phylogenies. Journal of Evolutionary Biology. 2005;18:325–336. doi: 10.1111/j.1420-9101.2004.00850.x. [DOI] [PubMed] [Google Scholar]
- Jablonski D. Background and mass extinctions: The alteration of macroevolutionary regimes. Science. 1986;231:129–133. doi: 10.1126/science.231.4734.129. [DOI] [PubMed] [Google Scholar]
- Labandeira CC, Johnson KR, Wilf P. Impact of terminal Cretaceous event on plant-insect associations. Proceedings of the National Academy of Sciences USA. 2002;99:2061–2066. doi: 10.1073/pnas.042492999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman A, Schaefer CW, Withers TM. Biology, Ecology and Evolution of Gall-inducing Arthropods. Enfield: Science Publishers, Inc; 2005. [Google Scholar]
- Stone GN, Schönrogge K. The adaptive significance of insect gall morphology. Trends in Ecology ad Evolution. 2003;18:512–522. doi: 10.1016/S0169-5347(03)00247-7. [DOI] [Google Scholar]
- Price PW, Pschorn-Walcher H. Are galling insects better protected against parasitoids than exposed feeders?: a test using tenthredinid sawflies. Ecological Entomology. 1988;13:195–205. doi: 10.1111/j.1365-2311.1988.tb00347.x. [DOI] [Google Scholar]
- Hawkins BA, Cornell HV, Hochberg ME. Predators, parasitoids, and pathogens as mortality agents in phytophagous insect populations. Ecology. 1997;78:2145–2152. doi: 10.1890/0012-9658(1997)078[2145:PPAPAM]2.0.CO;2. [DOI] [Google Scholar]
- Bailey R, Schönrogge K, Cook JM, Melika G, Csóka G, Thuróczy C, Stone GN. Host niches and defensive extended phenotypes structure parasitoid wasp communities. PLoS Biology. p. e1000179. [DOI] [PMC free article] [PubMed]
- Ronquist F, Liljeblad J. Evolution of the gall wasp-host plant association. Evolution. 2001;55(12):2503–2522. doi: 10.1111/j.0014-3820.2001.tb00765.x. [DOI] [PubMed] [Google Scholar]
- Goulet H, Huber JT. Hymenoptera of the world: an identification guide to families. Ottawa: Agriculture Canada. 1993.
- Hawkins BA, Goeden RD, Gagne RJ. Ecology and taxonomy of the Asphondylia spp. (Diptera: Cecidomyiidae) forming galls on Atriplex spp. (Chenopodiaceae) in Southern California. Entomography. 1986;4:55–107. [Google Scholar]
- Gagné RJ. The Plant-feeding Gall Midges of North America. Ithaca: Cornell University Press; 1989. [Google Scholar]
- Joy JB, Crespi BJ. Adaptive radiation of gall-inducing insects within a single host-plant species. Evolution. 2007;61:784–795. doi: 10.1111/j.1558-5646.2007.00069.x. [DOI] [PubMed] [Google Scholar]
- Burckhardt D, Basset Y. The jumping plant-lice (Hemiptera, Psyloidea) associated with Schinus (Anacardiaceae): systematics, biogeography and host plant relationships. Journal of Natural History. 2000;34:57–155. doi: 10.1080/002229300299688. [DOI] [Google Scholar]
- Von Dohlen CD, Kurosu U, Aoki S. Phylogenetics and evolution of the eastern Asian--eastern North American disjunct tribe, Hormaphidini (Hemiptera: Aphididae) Molecular Phylogenetics and Evolution. 2002;23:257–267. doi: 10.1016/S1055-7903(02)00025-8. [DOI] [PubMed] [Google Scholar]
- Gullan PJ, Miller DR, Cook LG. In: Biology, Ecology, and Evolution of Gall-inducing Arthropods. Raman A, Schaefer CW, Withers TM, editor. Enfield: Science Publishers, Inc.; 2005. Gall-inducing scale insects (Hemiptera: Sternorrhyncha: Coccoidea) pp. 159–229. [Google Scholar]
- Wool D. In: Biology, Ecology, and Evolution of Gall-inducing Arthropods. Raman A, Schaefer CW, Withers TM, editor. Enfield: Science Publishers, Inc; Gall-inducing aphids: biology, ecology and evolution; pp. 73–132. [Google Scholar]
- Havill NP, Foottit RG, von Dohlen CD. Evolution of host specialization in the Adelgidea (Insecta: Hemiptera) inferred from molecular phylogenies. Molecular Phylogenetics and Evolution. 2007;44:357–370. doi: 10.1016/j.ympev.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Price PW. Evolutionary Biology of Parasites. Princeton: Princeton University Press; 1980. [Google Scholar]
- Gaston KJ, Reavey D, Valladares GR. Intimacy and fidelity: internal and external feeding by the British microlepidoptera. Ecological Entomology. 1992;17:86–88. doi: 10.1111/j.1365-2311.1992.tb01044.x. [DOI] [Google Scholar]
- Magallon S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. Likelihood methods for detecting temporal shifts in diversification rates. Evolution. 2006;60(6):1152–1164. [PubMed] [Google Scholar]
- Nyman T, Zinovjev AG, Vikberg V, Farrell BD. Molecular phylogeny of the sawfly subfamily Nematinae (Hymenoptera: Tentredinidae) Systematic Entomology. 2006;31:569–583. doi: 10.1111/j.1365-3113.2006.00336.x. [DOI] [Google Scholar]
- Orchard AE, Maslin BR. Proposal to conserve the name Acacia (Leguminosae: Mimosoideae) with a conserved type. Taxon. 2003;52(1584):362–363. doi: 10.2307/3647418. [DOI] [Google Scholar]
- McLeish MJ, Chapman TW, Schwartz MP. Host-driven diversification of gall-inducing Acacia thrips and the aridification of Australia. BMC Biology. 2007;5 doi: 10.1186/1741-7007-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ, Boss D, Chen D, Cranston KA, Wehe A. The PhyLoTA Browser: processing GenBank for molecular phylogenetic research. Systematic Biology. 2008;57:335–346. doi: 10.1080/10635150802158688. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Gblocks Server. http://molevol.cmima.csic.es/castresana/Gblocks_server.html
- Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Faith DP. Conservation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- Slowinsky JB, Guyer C. Testing the stochasticity of patterns of organismal diversity: an improved null model. The American Naturalist. 1989;134:907–921. doi: 10.1086/285021. [DOI] [Google Scholar]
- Barraclough TG, Harvey PH, Nee S. Rate of rbcL gene sequence evolution and species diversification in flowering plants (angiosperms) Proceedings of the Royal Society of London, B. 1996;263:589–591. doi: 10.1098/rspb.1996.0088. [DOI] [Google Scholar]
- Vamosi SM, Vamosi JC. Endless tests: guidelines for analyzing non-nested sister-group comparisons. Evolutionary Ecology Research. 2005;7:567–579. [Google Scholar]
- Slowinski JB, Guyer C. Testing whether certain traits have caused amplified diversification: an improved method based on a model of random speciation and extinction. The American Naturalist. 1993;142:1019–1024. doi: 10.1086/285586. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:310–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Rabosky DL. LASER: a maximum likelihood toolkit for inferring temporal shifts in diversification rates. Evol Bioinform Online. 2006;2:273–276. [PMC free article] [PubMed] [Google Scholar]
- Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Alfaro ME, Santini F, Brock C, Alamillo H, Dornburg A, Rabosky DL, Carnevale G, Harmon LJ. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proceedings of the National Academy of Sciences of the United States of America. 2009;32:13410–13414. doi: 10.1073/pnas.0811087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG, Harvey PH, Nee S. Sexual Selection and taxonomic diversity in passerine birds. Proceedings of the Royal Society of London B. 1995;259:211–215. doi: 10.1098/rspb.1995.0031. [DOI] [Google Scholar]
- Siegel S. Nonparametric Statistics for the Behavioral Sciences. New York: McGraw-Hill; 1956. [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin's abominable mystery: Insights from a supertree of the angiosperms. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic datasets and results. Survey of DNA sequence-based phylogenetic studies including gall-inducing groups, and details of phylogenetic estimates performed here.
ML tree estimated from aphid DNA sequence data. Aphidoidea ML phylogeny estimated from analysis of EF1α, long-wavelength opsin, COI, COII, cytochrome b, NADH dehydrogenase 1, ATP synthase subunit 6, and mitochondrial ribosomal subunits 12 S and 16 S dataset partitioned by genome and codon position. Major lineages are labeled following the classification used by Blackman and Eastop.
ML tree estimated from cynipoid DNA sequence data. Cynipoidea ML phylogeny estimated from analysis of 28 S, 18 S, and COI dataset partitioned by genome and codon position. Deep relationships supported by >70% bootstrap proportions denoted by stars. Empty circle indicated clade of unpublished Cynipini sequences assumed to be misidentifications of synergine inquilines.
ML tree estimated form chalcidoid DNA sequence data. Chalcidoidea ML phylogeny estimated from analysis of 28 S sequences. Deep relationships supported by >70% bootstrap proportions denoted by stars. Group A composed of exemplars of the following families: Aphelinidae, Chalcididae, Encyrtidae, Eucharitidae, Leucospidae, Mymaridae, Pteromalidae, Perilampidae, Tetracampidae; Group B composed of exemplars of: Aphelinidae, Eurytomidae, Ormyridae, Perilampidae, Pteromalidae, Tanaostigmatidae, Tetracampidae, Torymidae.
Python script to automate sister taxon comparisons. A program that reads sister clade species diversities in a CSV file, calculates diversity contrasts with multiple metrics, and evaluates significance with either Siegel's randomization test for matched pairs, the Wilcoxon signed rank test, or normal approximation of the Wilcoxon signed rank test.