Abstract
Background
Plasmodium falciparum apical membrane antigen-1 (AMA1) is a leading malaria vaccine candidate antigen that is expressed by sporozoite, liver and blood stage parasites. Since CD8+ T cell responses have been implicated in protection against pre-erythrocytic stage malaria, this study was designed to identify MHC class I-restricted epitopes within AMA1.
Methods
A recombinant adenovirus serotype 5 vector expressing P. falciparum AMA1 was highly immunogenic when administered to healthy, malaria-naive adult volunteers as determined by IFN-γ ELISpot responses to peptide pools containing overlapping 15-mer peptides spanning full-length AMA1. Computerized algorithms (NetMHC software) were used to predict minimal MHC-restricted 8-10-mer epitope sequences within AMA1 15-mer peptides active in ELISpot. A subset of epitopes was synthesized and tested for induction of CD8+ T cell IFN-γ responses by ELISpot depletion and ICS assays. A 3-dimensional model combining Domains I + II of P. falciparum AMA1 and Domain III of P. vivax AMA1 was used to map these epitopes.
Results
Fourteen 8-10-mer epitopes were predicted to bind to HLA supertypes A01 (3 epitopes), A02 (4 epitopes), B08 (2 epitopes) and B44 (5 epitopes). Nine of the 14 predicted epitopes were recognized in ELISpot or ELISpot and ICS assays by one or more volunteers. Depletion of T cell subsets confirmed that these epitopes were CD8+ T cell-dependent. A mixture of the 14 minimal epitopes was capable of recalling CD8+ T cell IFN-γ responses from PBMC of immunized volunteers. Thirteen of the 14 predicted epitopes were polymorphic and the majority localized to the more conserved front surface of the AMA1 model structure.
Conclusions
This study predicted 14 and confirmed nine MHC class I-restricted CD8+ T cell epitopes on AMA1 recognized in the context of seven HLA alleles. These HLA alleles belong to four HLA supertypes that have a phenotypic frequency between 23% - 100% in different human populations.
Background
The sterile protective immunity to malaria induced in humans by immunization with irradiated sporozoites is thought to be mediated by CD4+ and CD8+ T cells responding to malaria peptides expressed on the surface of hepatocytes or antigen presenting cells by secreting interferon-gamma (IFN-γ) and/or by cytotoxic responses, although anti-sporozoite antibodies may contribute [1-5]. Many sporozoite and liver stages antigens have been identified [6] that could play a role in sporozoite and liver stage immunity, including the circumsporozoite protein (CSP), the main antigenic component of the partially protective RTS, S vaccine currently undergoing Phase 3 testing in sub-Saharan Africa[7,8]. Although CSP contributes to the protection induced by irradiated sporozoites, it is not required, indicating the importance of other antigens[9,10]; additionally it has not been possible to consistently induce CD8+ T cell responses using recombinant CSP-based vaccines such as RTS, S[7,11,12]. Combining CSP with other pre-erythrocytic stage antigens and using a vaccine platform such as adenovirus vectors better able to induce class I restricted cell-mediated immunity might therefore more effectively target the hepatic stages of infection and reproduce the immunity induced by the irradiate sporozoite vaccine.
Apical Membrane Antigen-1 (AMA1) is a candidate antigen for inclusion with CSP in a multi-antigen malaria vaccine. AMA1 has previously been tested in several clinical trials as a recombinant protein and elicited both CD4+ and CD8+ T cell responses[13-18]. AMA1 is an integral membrane protein found in all species of Plasmodium and has traditionally been regarded as a blood stage antigen, since it is required for the invasion of red blood cells[19], monoclonal and polyclonal antibodies targeting AMA1 inhibit blood stage growth in vitro, naturally acquired anti-AMA1 antibodies correlate with protection against clinical malaria in endemic areas [20-24], and vaccines based on AMA1 elicit protection against blood stage infection [13,25] in animal models that appears to be antibody mediated [25,26]. However, AMA1 is also expressed in sporozoites and liver stage parasites[27], and thus may be a suitable target for CD8+ T cell responses directed toward liver stage parasites.
To test this hypothesis, two adenovirus-vectored vaccines encoding P. falciparum CSP and AMA1 were evaluated in a Phase 1 clinical trial. Volunteers were administered a single dose of the mixed CSP- and AMA1-encoding constructs (termed the NMRC-M3V-Ad-PfCA vaccine), either 2 × 1010 (1 × 1010 of each construct) or 1 × 1011 (5 × 1010 of each construct) particle units (pu). Robust CD4+ and CD8+ T cell responses were induced in both low dose and high dose groups against both antigens, as measured by ex vivo enzyme-linked immunospot (ELISpot) assay conducted using pools of 15-mer peptides spanning full length CSP or AMA1 as the stimulant. These responses were significantly higher in the low dose than the high dose group, and the vaccine consistently induced stronger CD8+ than CD4+ T cell responses in both groups (Sedegah M, Tamminga C, McGrath S, House B, Ganeshan H, Lejano J, Abot E, Banania GJ, Sayo R, Farooq F, Belmonte M, Manohar N, Richie NO, Wood C, Long CA, Regis D, Shi M, Chuang I, Spring M, Epstein JE, Mendoza-Silveiras J, Limbach K, Patterson NB, Bruder JT, Doolan DL, King CR, Soisson L, Diggs C, Carucci D, Dutta S, Hollingdale MR, Ockenhouse CF, Richie TL. Multi-stage adenovirus 5-vectored falciparum malaria vaccine elicits CD8+ and CD4+ T cell responses and limited antibodies in healthy, seronegative adults, submitted). The CD8+ T cell IFN-γ responses induced in this clinical trial provided the opportunity to identify the underlying minimal CD8+ T cell epitopes. Frozen peripheral blood mononuclear cells (PBMC) collected from five volunteers from the more immunogenic low dose group were therefore selected for this epitope mapping study.
The crystal structure of the AMA1 ectodomain shows a conserved central core and variable external loops formed by Domains I, II and III[28]. About 10% of amino acids are polymorphic and many of these cluster within the tertiary structure[28-32] on one external surface and are presumably accessible to antibodies[31,32]. The most polymorphic regions surround a hydrophobic groove containing cryptic and conserved epitopes[31,32]. A recent study in Mali identified 186 unique AMA1 haplotypes largely varying at these polymorphic sites[33], while other studies have shown minimal cross-reactivity among the various allelic variants[17]. This immune diversity would appear to represent a major barrier to developing antibody-based vaccines against AMA1 [26,31,34]. The inhibitory MAb 1F9 maps to variable epitopes[24] including one site that is also recognized by a putative receptor for AMA1 binding to red blood cells[35]. Less is known regarding the most important T cell epitopes of AMA1, and whether or not they fall in the regions of highest variability. Previously identified proliferative AMA1 epitopes in malaria-exposed individuals in Kenya were mapped to both variable and non-variable regions [36].
Approaches to mapping B and T epitopes have included using antibodies[37-42], peptides, cell arrayed polypeptides or phages[36,43-48], computer-based algorithms [49-53] such as NetMHC used in this study[54], and combinations of these approaches[55]. In this study, peptide-based mapping and NetMHC algorithms were combined to identify class I-restricted AMA1 epitopes. Peptide pools showing positive responses in ELISpot assays using PBMC from the five most responsive volunteers from the low dose group were deconvoluted by testing each individual 15-mer peptide. Next, computerized algorithms in NetMHC software[56] were used to predict the binding affinities of AMA1 8-10-mer sequences within the dominant peptides for defined HLA-A or HLA-B supertypes expressed by each immunized volunteer. A subset of the predicted epitopes was synthesized, and ELISpot depletion and intracellular cytokine staining (ICS) assays were performed to confirm recall responses by PBMC from the immunized volunteers and to show their CD8+ T cell-dependence. A pool of selected minimal epitopes provided a potentially suitable reagent to efficiently measure anti-AMA1 CD8+ T cell responses in genetically diverse populations. Finally, 10 of the 14 epitopes were localized to the tertiary structure of AMA1.
Methods
Ethics
This study was conducted according to the Declaration of Helsinki and the U.S. Code of Federal Regulations regarding the protection of human participants in research including The Nuremberg Code, The Belmont Report, 32 CFR 219 (The Common Rule) and all regulations pertinent to the Department of Defense, the Department of the Navy, the Department of the Army, the Bureau of Medicine and Surgery of the United States Navy and the internal policies for human subject protections and the standards for the responsible conduct of research of the Naval Medical Research Center (NMRC) and US Army Medical Research and Materiel Command (USAMRMC). NMRC holds a Department of Defense/Department of the Navy Federal Wide Assurance for human subject protections, and a Federal Wide Assurance (FWA 00000152) from the Office for Human Research Protections (OHRP) for cooperation with the Department of Health and Human Services. All NMRC key personnel are certified as having completed mandatory human research ethics education curricula and training under the direction of the NMRC Office of Research Administration (ORA) and Human Subjects Protections Program (HSPP). The trial was performed under US Food and Drug Investigational New Drug Application BB-IND-13003.
Vaccine and trial design
The NMRC-M3V-Ad-PfCA vaccine used in this study is a combination of two separate recombinant adenovirus 5 constructs, one expressing full length P. falciparum CSP (minus 16 repeats, and insertion of 23 amino acids derived from the 3'-noncoding bovine growth hormone polyadenylation sequence at the C-terminus) and the other expressing full length P. falciparum AMA1 (both strain 3D7). Epitope mapping was conducted using PBMC taken from volunteers 1 month after administration of one intramuscular injection of 2 × 1010 particle units (pu) of the combination vaccine, unless other intervals are indicated.
Volunteers and HLA typing
Five of the six immunized volunteers in the low dose group (v001, v002, v005, v008 and v012) were used in this study, as v006 gave poor post-immunization responses. Low to moderate resolution HLA molecular typing for HLA-A and HLA-B loci (Department of Defense Bone Marrow Donor Program using specific oligonucleotide probes to amplify HLA class I and II genes) provided a list of allelic codes from which it was possible to tentatively assign each volunteer to an HLA-A or HLA-B supertype using code lists http://bioinformatics.nmdp.org/HLA/Allele_Codes/Allele_Code_Lists/index.html (Table 1).
Table 1.
Volunteer HLA A and B supertypes
| Volunteer | A Supertype | B Supertype |
|---|---|---|
| 001 | A01/A02 | B44/B44 |
| 002 | A01/A02 | B08/B44 |
| 005 | A01/A02 | B08/B27 |
| 008 | A02/A03 | B27/B27 |
| 012 | A01/A03 | B44/B58 |
Low resolution molecular HLA typing permitted identification of the HLA supertype for each volunteer.
PBMC
PBMC cryopreserved in liquid nitrogen induced comparable ELISpot activity as fresh cells (data not shown) and were used in these experiments. Due to limitation of the samples collected at 1 month post immunization, PBMC collected at other time points were used in some cases (as indicated), as long as responses were still moderately strong.
Peptides and peptide pools
153 15-mer peptides overlapping by 11 amino acids and spanning the length of AMA1 were synthesized commercially (Mimotopes, VIC, Australia, >80% purity) and grouped into 12 peptide pools containing 10 to 13 peptides each (Table 2). Seven of these pools (Ap1, Ap3, Ap4, Ap7, Ap8, Ap10 and Ap11) containing 91 peptides elicited strong ELISpot responses among the volunteers. Proliferative T cell epitopes previously identified in Kenya[36,43] are shown for each peptide pool. All 91 peptides were assayed individually, identifying 16 individual 15-mers showing positive ELISpot responses. Fourteen predicted 8-10-mer epitopes identified within these 16 15-mers were synthesized (Alpha Diagnostics Intl. Inc., San Antonio, TX, USA >91% purity) and tested for recall responses. The positive control was commercially obtained Class I Peptide Pool Plus (Anaspec, USA). Negative control was media with all supplements but no antigen-specific stimulant.
Table 2.
AMA1 peptides used in ELISpot and ICS assays
| Pool | Amino acids | Number peptides | Class II T epitope | Residues | T epitope sequence |
|---|---|---|---|---|---|
| Ap1 | 1-63 | 13 | PL186 | 14-35 | EFTYMIFNGRGQNYWEHPYQKS |
| PL187 | 41-51 | INEHRPKEY | |||
| Ap2 | 53-115 | 13 | PL188 | 92-103 | NLFSSIEIVERS |
| Ap3 | 105-167 | 13 | |||
| Ap4 | 157-219 | 13 | PL189 | 188-204 | PLMSPMTLDEMRHFYKD |
| Ap5 | 209-271 | 13 | PL190 | 218-229 | SRHAGNMIPDND |
| PL191 | 259-271 | NGPRYCNKDE | |||
| Ap6 | 261-323 | 13 | PL192 | 279-288 | AKDISFQNYT |
| Ap7 | 313-375 | 13 | PL171 | 348-366 | DQPKQYEQHLTDYEKIKEG |
| Ap8 | 365-427 | 13 | PL193 | 390-402 | YKSHGKGYNWGNY |
| Ap9 | 417-479 | 13 | PL172 | 444-461 | SLYKNEIMKEIERESKRI |
| Ap10 | 469-531 | 13 | |||
| Ap11 | 521-583 | 13 | PL194 | 527-538 | EYKDEYADIPEH |
| PL173 | 571-588 | GNAEKYDKMDEPQHYGKS | |||
| Ap12 | 573-622 | 10 | PL173 | 571-588 | GNAEKYDKMDEPQHYGKS |
PfAMA1 peptide sequences and residue numbers were based on those of the P. falciparum clone FC27 Gene Bank ID 810891. Previously identified class II AMA1 epitopes (see text) were distributed among peptide pools, except pools 3 and 10. Seven immunodominant pools are indicated in bold type.
Ex vivo IFN-γ ELISpot assays
IFN-γ ELISpot assays were conducted as previously described[57]. Cryopreserved PBMC were suspended in 100 μL complete medium and stimulated with AMA1 peptides in 100 μL of complete medium at a final concentration of 10 μg/mL of each peptide tested[57]. Cultures were incubated for 36 hours at 37°C, 5% CO2. Depending on availability of cells, each PBMC sample was assayed in duplicate, triplicate, or quadruplicate and the number of IFN-γ-secreting spot forming cells (sfc) was counted using an automated ELISpot reader (AID, GmbH, Germany). In duplicate assays, all values were used in analysis. For triplicate or quadruplicate assays, outliers were rejected if any single value contributed more than 50% of the standard deviation of the replicates and if its value was three-fold greater or three-fold less than the average of the remaining two (or three) values. The mean number of sfc obtained in negative control wells was subtracted from the value of each test well from the same sample. Negative counts generated by this background subtraction were converted to zero. The mean number of spots of the test sample was then calculated and expressed as sfc/million (sfc/m). Based on testing five volunteers with 91 15-mer peptides, 40 sfc/m was used as a conservative cut-off for determining positive activity.
Characterization of ELISpot IFN-γ-producing cells by T cell subset depletions
PBMC were depleted of T cell subsets using anti-human CD4+ or anti-CD8+ coated Dynabeads M-450 (Dynal, Great Neck, NY) following the manufacturer's instructions. Mock depletion was done with Dynabeads coated with sheep anti-mouse IgG. Flow cytometry confirmed that T cell subset depletions were >99% in all experiments. Data are presented as the sfc/m and percent decrease or increase in activity after depletion.
Intracellular cytokine staining (ICS)
ICS was performed as published previously [58]. Cryopreserved PBMC were thawed, washed, and resuspended at 1 × 107 cells per mL in complete medium. Peptides were used at 10 μg/mL and costimulatory antibodies anti-CD28 and anti-CD4+9 d (BD Bioscience, San Jose, CA) were used at 1 μg/mL. Stimulants were added to cells and incubated at 37°C with 5% CO2 for 2 hours. Cells were stained with anti-CD3, anti-CD4+, anti-CD8+, anti-IFN-γ, anti-TNF-α, and anti-IL2 and the entire available sample was acquired on a BD LSRII using FACSDiVa (BD Bioscience) software. Data were analysed using FlowJo Software (Treestar, Inc.). The gating strategy involved progressively measuring total cells; viable cells; lymphocytes; T cells; CD4+ CD8+ populations; and finally a specific cell type expressing a specific cytokine. Results were transferred to Prism (GraphPad) for graphing and statistical analysis. Data for peptides were corrected for media responses. Results are expressed as total IFN- γ from all VD4+ or CD8+ T cells containing IFN- γ either alone or with other cytokines.
NetMHC-based epitope predictions and epitope down-selection
NetMHC[59] was used to predict HLA class I binding affinities of 15-mer peptides. NetMHC returns predicted binding affinity scores that approximate the half maximal inhibitory concentration (IC50) in nM. Thus, smaller IC50 values indicate stronger binding. Peptides with measured binding affinities less than 500 nM IC50 are considered binders.
Because lengths of typical peptides that bind to HLA class I molecules range from 8 to 10-mers, binding predictions were made for all possible 8-10-mers in a 15-mer. In order to discover a minimal epitope for each 15-mer that induced a CD8+ response, the peptide with the strongest predicted binding affinity for a given HLA allele was selected.
In practice, because of the low resolution of HLA typing data available for the volunteers, peptide binding predictions were made for all alleles that matched the provided data. For example, the HLA typing data indicates that volunteer v001 has an allele that belongs in the A02 supertype. The HLA typing data also provided a list of possible alleles that belong in the same supertype, which includes A*0201, A*0207 and A*0209. Peptide binding predictions were made for each allele, provided a predictor was available. Thus, for each 15-mer, the strongest predicted binder out of all 8-10-mer peptides across different possible alleles and its predicted binding affinity were recorded. This strongest predicted binder is referred to as the predicted minimal epitope. In the end, a single predicted minimal epitope was associated with each 15-mer after ranking candidates by binding affinity predictions.
Epitope mapping on the AMA1 3 D structure
The P. falciparum AMA1 Domains I and II model 1Z40 was fitted onto the P. vivax AMA1 Domain I, II and III model 1w81 (E chain) using Swiss Pdb-Viewer software [Swiss Institute of Bioinformatics (Basel, Switzerland)]. The final 3 D model was generated by combining Domains I and II of the P. falciparum AMA1 model with Domain III of the P. vivax AMA1 model, omitting Domains I and II of the P. vivax AMA1 model. Accessibility of the amino acid residues was determined by the same software.
Results
Volunteers
The five volunteers used in this study expressed a total of seven supertypes (Table 1), A01, A02, A03, B08, B27, B44 and B58, representing three of the six HLA-A and four of the six HLA-B supertypes[60]. Together these cover 100% of the Caucasian population, 27% or the African American population, and a variable per cent of sub-Saharan populations according to their genetic diversity[61]. There were no volunteers with other high frequency supertypes such as A24 or B07[61].
Identification of AMA1 15-mer peptides most active in ELISpot assay
The 91 15-mers contained within peptide pools Ap1, Ap3, Ap4, Ap7, Ap8, Ap10 and Ap11 were tested individually by ELISpot assay using PBMC from 1 month post immunization. Sixteen peptides were recognized by one or more of the five volunteers at levels > 40 sfc/m, and assigned numbers of 1-16 (Table 3). Some were recognized by more than one volunteer such as peptide 14 in pool 8 (Ap8-11) that was recognized by v001, v005 and v008.
Table 3.
ELISpot IFN-γ activity of AMA1 peptide pools and individual 15-mer peptides within these pools
| Pool | Vol. | sfc/m | 15-mer peptide | Amino acid number | Sequence | sfc/m | Peptide number |
|---|---|---|---|---|---|---|---|
| Ap1 | 001 | 119 | Ap1-3 | 9-23 | LLSAFEFTYMINFGR | 48 | 1 |
| Ap1 | 001 | 119 | Ap1-9 | 33-47 | QNSDVYRPINEHREH | 48 | 2 |
| Ap1 | 002 | 161 | Ap1-11 | 41-55 | INEHREHPKEYEYPL | 180 | 3 |
| Ap1 | 005 | 325 | Ap1-11 | INEHREHPKEYEYPL | 322 | ||
| Ap1 | 002 | 161 | Ap1-12 | 45-59 | REHPKEYEYPLHQEH | 66 | 4 |
| Ap1 | 005 | 325 | Ap1-12 | REHPKEYEYPLHQEH | 118 | ||
| Ap1 | 001 | 119 | Ap1-13 | 49-64 | KEYEYPLHQEHTYQQ | 42 | 5 |
| Ap4 | 005 | 208 | Ap4-5 | 173-187 | NQYLKDGGFAFPTE | 50 | 6 |
| Ap4 | 002 | 208 | Ap4-9 | 189-203 | LMSPMTLDEMRHFYK | 189 | 7 |
| Ap4 | 005 | 310 | Ap4-9 | LMSPMTLDEMRHFYK | 395 | ||
| Ap4 | 002 | 208 | Ap4-10 | 193-207 | MTLDEMRHFYKDNKY | 82 | 8 |
| Ap4 | 005 | 208 | Ap4-10 | MTLDEMRHFYKDNKY | 118 | ||
| Ap4 | 005 | 208 | Ap4-11 | 197-211 | EMRHFYKDNKYVKNL | 65 | 9 |
| Ap7 | 001 | 131 | Ap7-3 | 310-324 | EDIPHVNEFPAIDLF | 139 | 10 |
| Ap7 | 001 | 131 | Ap7-4 | 314-328 | HVNEFPAIDLFECNK | 96 | 11 |
| Ap7 | 001 | 131 | Ap7-7 | 336-350 | CNKLVFELSADQPK | 77 | 12 |
| Ap8 | 012 | 138 | Ap8-6 | 384-398 | FKADRYKSHGKGYNW | 143 | 13 |
| Ap8 | 001 | 78 | Ap8-11 | 405-419 | ETQKCEIFNVKPCL | 48 | 14 |
| Ap8 | 005 | 156 | Ap8-11 | ETQKCEIFNVKPCL | 218 | ||
| Ap8 | 008 | 78 | Ap8-11 | ETQKCEIFNVKPCL | 48 | ||
| Ap8 | 005 | 156 | Ap8-12 | 409-423 | CEIFNVKPTCLINNS | 127 | 15 |
| Ap10 | 001 | 172 | Ap10-13 | 517-531 | TSNNEVVVKEEYDE | 233 | 16 |
| Ap10 | 012 | 193 | Ap10-13 | TSNNEVVVKEEYDE | 250 |
A total of 16 individual peptides were identified from a total of 91 15-mer peptides from seven active pools. Peptides Ap1-11, Ap1-12, Ap4-9, Ap4-10, Ap8-11 and Ap10-13 stimulated strong responses in more than one volunteer.
ELISpot assays with 15-mer peptides after depletion of CD4+ and CD8+ T cells
In these depletion studies, the availability of PBMC from 1 month following immunization limited assays to five of the sixteen 15-mers and four of the volunteers listed in Table 3 (v008 excluded). CD8+ T cell depletion reduced ELISpot activity by 56-100% (Table 4) demonstrating the presence of at least one minimal CD8+-restricted epitope within each 15-mer tested for each of the four volunteers that was recognized in the context of supertypes A1, B44 or B08. As shown in Table 3, peptide 3 was recognized by v002 and v005, and peptide 16 by v001 and v012, and this is attributed to common HLA alleles between these volunteers (Table 1). In those cases where depletion of CD4+ T cells also reduced ELISpot responses, the effect was much smaller (0-57%) than for CD8+ T cells, suggesting that these peptides contain predominantly CD8+ T cell epitopes. Depletion of CD4+ T cells led to an increase in ELISpot activity with v001, suggesting the removal of suppressor mechanisms.
Table 4.
ELISpot IFN-γ activity of AMA1 15-mer peptides after depletion of CD4+ and CD8+ T cells
| Vol. | Pool | Peptide number* | Sequence | Control depletion sfc/m | CD4 depletion sfc/m (%) |
CD8 depletion sfc/m (%) |
|---|---|---|---|---|---|---|
| 001 | Ap1 | 155 | 222 (+43%) | 55 (-65%) | ||
| 1 | LLSAFEFTYMINFGR | 85 | 58 (-32%) | 37 (-56%) | ||
| 5 | KEYEYPLHQEHTYQQ | 72 | 168 (+133%) | 17 (-76%) | ||
| 16 | TSNNEVVVKEEYDE | 103 | 252 (+148%) | 13 (-88%) | ||
| 002 | Ap1 | 187 | 148 (-21%) | 5 (-97%) | ||
| 3 | INEHREHPKEYEYPL | 185 | 152 (-18%) | 11 (-94%) | ||
| 005 | Ap1 | 267 | 173 (-35%) | 11 (-96%) | ||
| 3 | INEHREHPKEYEYPL | 233 | 145 (-37%) | 0 (-100%) | ||
| Ap4 | 273 | 117 (-57%) | 0 (-100%) | |||
| 7 | LMSPMTLDEMRHFYK | 204 | 137 (-32%) | 1 (-100%) | ||
| 012 | Ap10 | 48 | 37 (-23%) | 0 (-100%) | ||
| 16 | TSNNEVVVKEEYDE | 79 | 90 (+14%) | 0 (-100%) |
The percent change in ELISpot activity is shown after depletion of CD4+ or CD8+ T cells. Depletion of CD8+ T cells reduced ELISpot activity by 56-100%, whereas CD4+ T cell depletion reduced activity by no more than 57%. Although this suggested that the primary epitopes within these peptide pools and 15-mers are CD8+ T cell epitopes, some peptides may also contain CD4+ T cell epitopes. v001 and v012 showed an increase in ELISpot activity after CD4+ T cell depletion that may indicate the removal of regulatory T cells that had been suppressing CD8+ T cell activity.
* See last column of Table 3.
NetMHC prediction of class I-restricted epitopes in AMA1 15-mer peptides
NetMHC predicted a series of HLA-A- and B-restricted 8-10-mer epitopes within the 16 15-mers active in ELISpot assays (Table 5). Each predicted epitope was restricted by a specific HLA allele but since the HLA typing of the volunteers was low to moderate resolution, this study selected the best binders for the several candidate alleles within each volunteer's HLA supertype. For example, peptide 1 contained a predicted epitope restricted by HLA B*1801 that matched the v001 HLA supertype B44 as shown in Table 5. Since overlapping peptides were used, in three cases predicted epitopes were contained in more than one peptide, for example epitope E3 was contained in two overlapping peptides 3 and 4 (Table 5). Furthermore, there was also one case where one 15-mer peptide, peptide 14, contained 2 predicted epitopes, E11 and E12, leaving a net of 14 unique epitopes.
Table 5.
Predicted CD8+ T cell-restricted epitopes within AMA1 15-mer peptides specific for each volunteer
| Peptide number | Predicted epitope | Amino acid number |
IC50 nM | HLA restriction | HLA supertype | Epitope number |
|---|---|---|---|---|---|---|
| 1 | LLSAFEFTYMINFGR | 13-21 | 5 | B*1801 | B44 | E1 |
| 2 | QNSDVYRPINEHREH | 35-44 | 14991 | B*4402 | B44 | E2 |
| 3 | INEHREHPKEYEYPL | 47-55 | 18 | B*0801 | B08 | E3 |
| 4 | REHPKEYEYPLHQEH | |||||
| 5 | KEYEYPLHQEHTYQQ | 51-59 | 39 | B*1801 | B44 | E4 |
| 6 | NQYLKDGGFAFPTE | 175-183 | 142 | B*0801 | B08 | E5 |
| 7 | LMSPMTLDEMRHFYK | 194-202 | 17 | A*0101 | A01 | E6 |
| 8 | MTLDEMRHFYKDNKY | |||||
| 9 | EMRHFYKDNKYVKNL | 198-207 | 4742 | A*0101 | A01 | E7 |
| 10 | EDIPHVNEFPAIDLF | 327-335 | 7 | B*1801 | B44 | E8 |
| 11 | HVNEFPAIDLFECNK | |||||
| 12 | CNKLVFELSADQPK | 339-346 | 5708 | A*0201 | A02 | E9 |
| 13 | FKADRYKSHGKGYNW | 389-397 | 201 | A*3002 | A01 | E10 |
| 14 | ETQKCEIFNVKPCL | 405-414 | 142 | A*6802 | A02 | E11 |
| ETQKCEIFNVKPCL | 406-414 | 919 | A*0201 | A02 | E12 | |
| 15 | CEIFNVKPTCLINNS | 410-419 | 617 | A*6802 | A02 | E13 |
| 16 | TSNNEVVVKEEYDE | 520-528 | 15 | B*1801 | B44 | E14 |
The 15-mer peptides that were recognized by the volunteers (Table 3) were analysed by NetMHC web-based software to predict potential high affinity HLA binding by minimal CD8+ T cell epitopes within each 15-mer. Each minimal epitope was specific for a known HLA allele within each supertype. Those minimal epitopes with the strongest binding affinities for the HLA supertype of each volunteer were selected. The Table shows the minimal epitopes in bold. Two different HLA-restricted epitopes were predicted with peptide 14.
Nine of the 14 resulting epitopes (64%) were classified as strong binders (<500 nM). Of these 14 minimal epitopes, two were predicted to be restricted by HLA-A*0101 and one by HLA-A*3002 (A01 supertype); two by HLA-A*0201 and two by HLA-A*6802 (A02 supertype); two by HLA-B*0801 (B08 supertype); four by HLA-B*1801 and one by HLA-B*4402 (B44 supertype). Epitopes predicted to bind to supertypes A03, B27 and B58 present in the volunteers were not identified.
ELISpot activity of predicted minimal epitopes compared to parent peptide pool
PBMC available from bleeds at 4 months had similar activity to those at 1 month for v001 and v002 and were, therefore, used for the evaluation of epitopes E1, E2, E3, E4, E6, E8, E9, E12 and E14. ELISpot activity of these predicted minimal epitopes compared favorably to the ELISpot activity of parent peptide pool activity for all nine (Table 6). Availability of PBMC from v005 and v008 was more restricted, and their ELISpot activity was too low to allow evaluation of E5, E7, E11 and E13. PBMC from v012 taken 7 months post-immunization confirmed positive responses for E8 and E14 but suggested that E10, where responses were ten-fold less than the parent peptide pool and below the cut-off of 40 sfc/m, did not constitute the minimal epitope for that volunteer. Therefore, nine of the predicted epitopes were recognized by one or more volunteers, four could not be adequately tested and one was not recognized. Of the nine confirmed epitopes, six had strong predicted binding affinities (< 500 nM) and three were >500 nm (Table 5).
Table 6.
ELISpot IFN-γ activity of the original peptide pool and the derived 8-10-mer epitopes
| Epitope number | Sequence | HLA supertype | sfc/m | Pool | sfc/m | Vol. |
|---|---|---|---|---|---|---|
| E1 | FEFTYMINF | B44 | 1214m | Ap1 | 1334m | 001 |
| E2 | SDVYRPINEH | B44 | 1144m | Ap1 | 1334m | 001 |
| E3 | HPKEYEYPL | B08 | 694m | Ap1 | 964m | 002 |
| E4 | YEYPLHQEH | B44 | 2224m | Ap1 | 1334m | 001 |
| E5 | YLKDGGFAF | B08 | 910d | Ap4 | 4610d | 005 |
| E6 | TLDEMRHFYK | A01 | 514m | Ap4 | 1144m | 002 |
| E7 | MRHFYKDNKY | A01 | 1510d | Ap4 | 4610d | 005 |
| E8 | NEFPAIDLF | B44 | 2244m | Ap7 | 2004m | 001 |
| - | - | - | 497m | Ap7 | 807m | 012 |
| E9 | KLVFELSA | A02 | 1694m | Ap7 | 2004m | 001 |
| E10 | RYKSHGKGY | A01 | 117m | Ap8 | 1087m | 012 |
| E11 | ETQKCEIFNV | A02 | 2110m | Ap8 | 5710m | 008 |
| E12 | TQKCEIFNV | A02 | 1724m | Ap8 | 1044m | 001 |
| E13 | EIFNVKPTCL | A02 | 3710m | Ap8 | 5710m | 008 |
| E14 | NEVVVKEEY | B44 | 3444m | Ap10 | 1554m | 001 |
| - | - | - | 1047m | Ap10 | 1517m | 012 |
Synthetic peptide 8-10-mers were tested in ELISpot assay using PBMC collected 10 days (v005), 4 months (v001, v002), 7 months (v012) or 10 months (v008) following immunization (PBMC collected at 1 month were no longer available). Activity was measured as sfc/m. Nine of the 14 epitopes (E1, E2, E3, E4, E6, E8, E9, E12 and E14) were confirmed as positive using the parent peptide pool as controls, in most cases recalling responses equivalent to those recalled by the parent peptide pool (epitope number and sequences in bold). Four of the epitopes (E5, E7, E11, E13) did not meet the 40 sfc/m cut-off but these results were not considered meaningful as the responses of the parent peptide pools were only marginally positive. The parent peptide pool for E10 recalled 108 sfc/m while the responses to the peptide were much lower (11 sfc/m), suggesting that E10 may not be correctly identified. Assays where the parent peptide pool did not exceed the cut-off of 40 sfc/m were not included in the table (see Methods).
ICS CD4+ and CD8+ T cell total IFN-γ responses to AMA1 peptide pools and epitopes
PBMC from the same time points were used in ICS assays to measure total IFN-γ from CD4+ and CD8+ T cell subsets in response to selected minimal epitopes and parent peptide pools (Figure 1). These assays showed that the frequency of CD8+ T cells producing total IFN-γ was far greater than that of CD4+ T cells. Four epitopes (E3, E6, E8 and E14) induced equal or more CD8+ IFN-γ than the parent peptide pool, whereas two epitopes (E10 and E11) induced weak CD8+ IFN-γ responses. PBMC were insufficient to perform assays for the other epitopes. These results supported the identification, as minimal MHC class I epitopes, of E3, E6, E8 and E14, while E10 and E11 could not be confirmed.
Figure 1.
AMA1 peptide pools and minimal epitopes elicit ICS CD4+ and CD8+ T cell IFN-γ responses. AMA1 peptide pools (P) and associated minimal CD8+ T epitopes (E) were tested in ICS assays using PBMC collected 10 days (v005), 4 months (v001, v002), 7 months (v012) or 10 months (v008) following immunization. Activity was measured as % cytokine producing CD4+ and CD8+ T cells producing IFN-γ.
ICS CD4+ and CD8+ T cell multifunctional responses to AMA1 peptide pools and minimal epitopes
The pattern of multifunctional CD8+ T cell responses (defined as cells expressing two or more cytokines) recalled by each minimal epitope (Figure 2) were generally similar to that of total CD8+ IFN-γ responses (Figure 1), although they were lower. Concordant with total IFN-γ responses, epitopes E10 and E11 did not appear to induce multifunctional responses with v008 and v012.
Figure 2.
AMA1 peptide pools and minimal epitopes elicit ICS CD4+ and CD8+ T cell multifunctional responses. AMA1 peptide pools (P) and associated minimal CD8+ T epitopes (E) were tested in ICS assays using PBMC collected 10 days (v005), 4 months (v001, v002), 7 months (v012) and 10 months (v008) following immunization. Multifunctional activity was measured as % cytokine producing CD4+ and CD8+ T cells producing at least two cytokines among IFN-γ, TNF-α and IL-2.
ELISpot assays with epitope mixture AMA1-14e, single AMA1 pool, individual pools and individual epitopes
The feasibility of using pooled epitopes to evaluate CD8+ T cell responses to AMA1 was investigated to establish that non-binding epitopes did not block specific HLA-restricted binding. Three volunteers who consistently gave high responses to peptide pools were tested with four different stimulants: (1) Ap1-Ap12: all 153 15-mer peptides at 1.25 μg/mL; (2) AMA1-14e: the 14 predicted epitopes mixed and tested at 10 μg/mL; (3) 15-mer peptide pools at 10 μg/ml that were strongly recognized by these volunteers; (4) The predicted epitope at 10 μg/ml within those selected 15-mer peptide pools previously shown to have ELISpot activity. As shown in Figure 3, AMA1-14e was as active or nearly as active in inducing ELISpot responses as a mixture of all 153 15-mers (Ap1-Ap12).
Figure 3.

AMA1-14e is as active in ELISpot as peptide pools and minimal epitopes. PBMC from each volunteer were tested in ELISpot assays with Ap1-12 (mixture of 153 15-mer overlapping peptides), AMA1-14e (mixture of 14 minimal epitopes), Ap1 or Ap10, and individual epitopes (consistent with that volunteer's HLA supertype - NEVVVKEEY is E14 and HPKEYEYPL is E3). AMA1-14e was as active as Ap1-12 with v002 and v005 and nearly as active with v001. Bars represent standard deviation of the mean response (duplicates).
Phenotype of cells involved in ELISpot and ICS assays with epitope mixture AMA1-14e
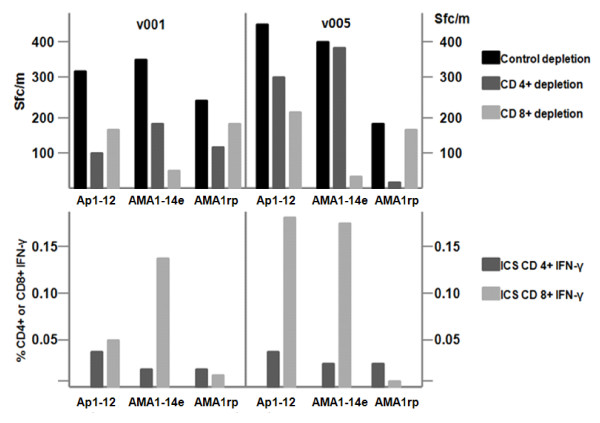
CD8+ T cell depletion reduced ELISpot activity to AMA1-14e by about 85% - 95% in the two volunteers tested, v001 and v005, and AMA1-14e much more strongly induced CD8+ than CD4+ T cell IFN-γ by ICS assay. In contrast, Ap1-Ap12 induced both CD4+ and CD8+ T cell responses, whereas AMA1 recombinant protein only recalled CD4+ T cell responses (Figure 4). Thus AMA1-14e may be a suitable reagent to demonstrate CD8+ T cell immunogenicity of AMA1-based vaccines in vaccine trials where volunteers have HLA alleles matching the epitopes in AMA1-14e.
Figure 4.
ELISpot activity against AMA1-14e after CD4+ or CD8+ T cell depletion. PBMC from v001 and v005 were tested in IFN-γ ELISpot assays after CD4+ or CD8+ T cell depletion (top panel), and in ICS assay for total % CD4+ or CD8+ T cell IFN-γ (lower panel), with Ap1-12 (all 153 15-mer peptides), AMA1-14e, and AMA1 recombinant protein (AMA rp). ELISpot depletion and ICS assays show that AMA1-14e and Ap1-12 are both strongly recognized by CD8+ T cells. However, recombinant AMA1 protein is preferentially recognized by CD4+ T cells.
Localization of the predicted minimal epitopes within AMA1
E1 lies within the signal domain, E2, E3, E4 within the prodomain, E5, E6 and E7 within Domain I, E8, E9, E10, E11, E12 and E13 within Domain II, and E14 within Domain III (Table 7). Three of the 14 predicted CD8+ minimal epitopes were fully or partially within proliferative epitopes identified in Kenya: E1 and PL186; E6 and PL189; E10 and PL193. The other 11 predicted epitopes were largely or wholly independent of these proliferative epitopes.
Table 7.
Summary of the properties of AMA1 epitopes
| Epitope | HLA-restriction | Amino acids | Sequence | Location | Crystal face |
Accessibility |
|---|---|---|---|---|---|---|
| E1 | B44 | 13-21 | FEFTYMINF | Signal | ||
| E2 | B44 | 35-44 | SDVYRPINEH | Prodomain | ||
| E3 | B08 | 47-55 | HPKEYEYPL | Prodomain | ||
| E4 | B44 | 51-59 | YEYPLHQEH | Prodomain | ||
| E5 | B08 | 175-183 | YLKDGGFAF | Domain I | F/B | YnaKD |
| E6 | A01 | 194-202 | TLDE+MRHFY | Domain I | B | D, E, H, F |
| E7 | A01 | 198-207 | MRHFYKDNKY | Domain I | F/B | H, F, D, K, Y |
| E8 | B44 | 327-335 | NEFPAIDLF | Domain II | F/B | E, P, I |
| E9 | A02 | 339-346 | KLVFELS*A | Domain II | F | E, L |
| E10 | A01 | 389-397 | RYKSHGKGY | Domain II | F | H, K, Y |
| E11 | A02 | 405-414 | ETQKCEIFNV | Domain II | B | E, T, Q, N |
| E12 | A02 | 406-414 | TQKCEIFNV | Domain II | F | T, Q, N |
| E13 | A02 | 410-419 | EIFNVK*PTCL | Domain II | F | NK |
| E14 | B44 | 520-528 | NEVVVKE*EY | Domain III | F | VEE |
Variable amino acids are highlighted in bold.
*Indicated that there is only one variant of <0.001% frequency
E+ is essential for binding of growth inhibitory MAb 1F9
F = front, B = back, F/B = wraps from front to back
Yna Accessibility not available for amino acid Y
Confirmed epitope numbers are indicated in bold
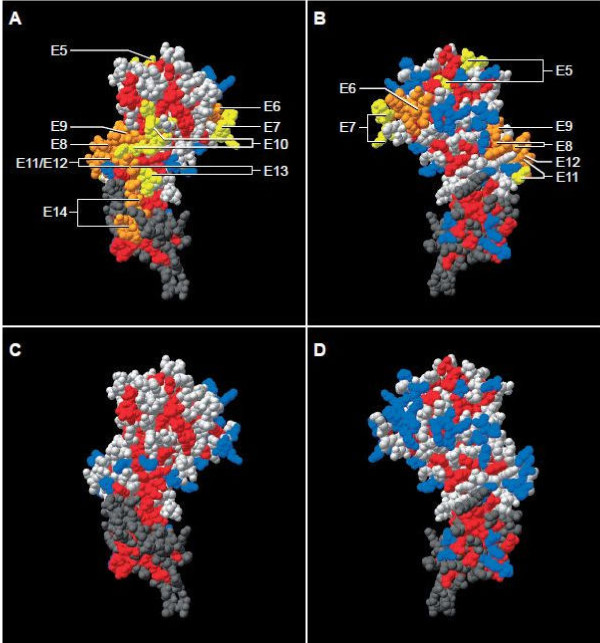
Based on the sequence of AMA1[26], epitopes E2, E3, E4, E5, E6, E7, E8, E10, E11 and E12 are variable, E9, E13, E14 have a low frequency (< 0.001%) of variable residues, whereas E1 is conserved. There are disulphide bonds between several cysteines (C) that form loops; epitopes E5, E6 and E7 are contained in a C-C loop in Domain I, and E8, E9, E10, E11, E12 and E13 are in a C-C loop in Domain II. Since all of these epitopes in C-C loops are variable, it is likely that they are exposed on the surface[32]. This was confirmed by mapping these epitopes to a model structure of AMA1, constructed from P. falciparum Domains I and II, and P. vivax Domain III[28,31,32]. The signal and prodomain containing E1-E4 were not contained in this model. The localization of epitopes E5-E14 is shown in Figure 5 and Table 7. Epitopes E8, E9, E10, E12, E13 and part of E14 form a continuous cluster on the front face, E5, E7, E8 and E9 wrap around from the front to back faces, and E6 (largely) and E11 localize to the back face. E5, though not confirmed as an epitope, is largely non-variant and immediately adjacent to the conserved hydrophobic cleft, whereas E6 is in a hypervariable region of Domain I also adjacent to the cleft. E7 is contiguous with E6 as might be expected from their overlapping sequences (Table 7). E9 is close to the binding site of inhibitory MAb 4G2 and contiguous with E10 that contains R389 that is essential for 4G2 binding[20]. E12 appears to be clustered with E8 and E9 although it is 60 amino acids C-terminal to E9, and close to E13. As expected from the linear sequences E11 appears to be adjacent to E12 but on the back face. Unexpectedly from the linear sequence, E14 is close to E13 and adjacent to the large cluster formed by E8, E9, E10, E12, E13 and part of E14.
Figure 5.
Location within model of AMA1 crystal structure of 10 predicted HLA-A and HLA-B epitopes. Spatial distribution of CD8 epitopes on the 3 D model structure of AMA1. The structure of PfAMA1 Domain I and II (1Z40) was fitted on the structure of PvAMA1 Domain I, II and III (1wk8), using Swiss Pdb-Viewer software http://www.expasy.org/spdbv/. Domain I and II of the PfAMA1 structure (light grey regions) and Domain III of PvAMA (dark grey region) are visible. Panels A & C show front view (largely conserved), B & D show back view (more polymorphic). Confirmed CD8 epitopes (E6, E8, E9, E12, E14) are orange in panels A & B, non-confirmed epitopes (E5, E7, E10, E11, E13) are yellow, while overlapping confirmed epitopes and non-confirmed epitopes are also in orange. Residues that are conserved among malaria species are shown in red and those that are polymorphic are shown in blue. Light grey (PfAMA) and dark grey (PvAMA1) are residues that differ between these species. Arrows point to individual epitopes. Overlapping epitopes may be indicated by multiple arrows. Confirmed epitopes E1 to E4 (see text) are not shown, as the part of the protein where these epitopes reside is not present in this crystal structure model.
Epitope polymorphism and surface accessibility
The polymorphic residues in all 14 epitopes are shown in Table 7. Only E1 is completely non-variant, although the frequencies of single amino acid polymorphisms in E9, E13 and E14 are rare (<0.001%). Overall there are 27 polymorphic residues in all 14 epitopes. For E5-E14 that could be mapped, there are 18 polymorphic residues of which 16 are surface accessible (Table 7). Thus, E6 contains variable amino acids at positions D196, E197, R199, H200 and F201, and all except R199 are accessible to the surface. Overall, R199 and S345 in these epitopes are polymorphic but not surface accessible. However, there are 10 amino acids that are surface accessible that are not polymorphic such as T406 and N413 in E12. Therefore polymorphic residues are mostly surface accessible, but there are other surface accessible residues that are not polymorphic.
Discussion
The aim of this study was to identify class I epitopes underlying the CD8+ T cell IFN-γ responses observed on ELISpot and ICS assays following administration of the NMRC-M3V-Ad-PfCA adenovirus-vectored P. falciparum malaria vaccine. Mapping class I epitopes was pursued to better understand the immune responses induced by AMA1 and to guide the development of a broadly protective malaria vaccine for a genetically diverse human population.
To map the epitopes, low to mid level HLA typing was performed to determine the HLA A and B supertype for each of five immunized volunteers. Subsequently, all possible 8-10 amino acid sequences from 15-mer AMA1 peptides inducing recall responses on ELISpot assay were evaluated in silico using NetMHC software, and ranked for binding affinity to the A and B alleles. Predicted top-binding epitopes were synthesized and ELISpot and ICS assays were used to confirm epitope identification by demonstrating that (1) the predicted epitope recalled IFN-γ responses as effectively as the parent peptide pool, (2) the responses of the parent 15-mer peptide were CD8+ T cell-dependent by ELISpot assay, and (3) the epitopes themselves stimulated IFN-γ production by CD8+ T cells on ICS assay.
Using this approach, sixteen 15-mer peptides were down-selected from the peptide pools on the basis of stimulating positive recall responses by ELISpot assay from the immunized volunteers. NetMHC predicted 14 class I 8-10-mer epitopes lying within these peptides for the five volunteers. Nine of these were shown by ELISpot assay to be active (E1, E2, E3, E4, E6, E8, E9, E12 and E14), eliciting IFN-γ responses comparable to those elicited by the parent peptide pools, while four could not be adequately assessed as the PBMC eliciting strong recall responses for these volunteers were no longer available (E5, E7, E11, E13), and one induced low recall responses relative to the parent peptide pool (E10).
To further evaluate the predicted epitopes, ELISpot depletion assays were conducted by stimulating PBMC from four volunteers with three parent peptide pools (Ap1, Ap4, Ap10) or with five individual 15-mer peptides from those pools (1, 3, 5, 7, 16). The 15-mers contained, respectively, the epitopes E1, E3, E4, E6 and E14. CD8+ T cell depletion reduced responses in five tests of the three peptide pools by 65%, 97%, 96%, 100% and 100%, respectively, and to the five 15-mers by 56-100% (mean of 88%). In contrast, depleting CD4+ T cells increased activity in Ap1 and two of the three peptides it contained as well as one peptide in Ap10, and reduced the ELISpot activity recalled by the remaining peptides by only 18- 37% (mean of 30%), strongly supporting the contention that the selected 15-mers contained minimal class I epitopes.
ICS assays performed using six minimal epitopes showed that four could recall IFN-γ responses from CD8+ T cells (E3, E6, E8 and E14), while two could not (E10 and E11). In contrast IFN-γ responses from CD4 cells were minimal or absent when stimulating with the same minimal epitopes.
In summary, of the nine epitopes active by ELISpot (E1, E2, E3, E4, E6, E8, E9, E12 and E14), subsequent ELISpot depletion assays support the identification of E1, E3, E4, E6 and E14 and ICS assays the identification of E3, E6, E8 and E14, while E2, E9 and E12 could not be adequately assessed due to PBMC supplies. Four additional epitopes (E5, E7, E11 and E13) have yet to be confirmed as class I-restricted by testing for reductions in ELISpot responses on CD8+ T cell depletion or by testing for CD8+ T cell responses on ICS assay. The remaining epitope, E10, recalled only 10% of the response recalled by the parent peptide pool on ELISpot assay and was not active on ICS assay, and thus may not be correctly identified.
While the focus of these experiments was induction of IFN-γ in ELISpot or ICS assays, four epitopes (E3, E6, E8 and E14) demonstrated recall of multifunctional responses (production of at least two cytokines of IFN-γ, TNF-α and IL-2) that might, due to the higher IFN-γ secretion levels in multifunctional T cells, contribute disproportionately to the overall IFN-γ response at 1 month following immunization, acting as more potent immune effectors.
Developing a reagent for class I responses to AMA1
The 14 putative minimal class I epitopes were combined into a reagent pool, denoted AMA1-14e, and this pool was shown to be an efficient stimulant for recalling T cell responses to AMA1. The AMA1-14e pool stimulated responses equivalent to parent 15-mer peptides and peptide pools, and these responses were CD8+ T cell-dependent by ELISpot depletion assay. Future trials with the NMRC-M3V-Ad-PfCA vaccine should reveal the usefulness of the AMA1-14e reagent. A previous study using DNA plasmids containing five malaria candidate vaccine genes relied on using HLA-matched peptides[62]; an HLA-diverse reagent similar to AMA1-14e would have greatly simplified this approach.
Population coverage associated with the identified epitopes
The 14 epitopes identified in this study were characterized with regard to the coverage provided for the HLA diverse human population. The epitopes were restricted by four of the seven HLA supertypes expressed by the volunteers: A01, A02, B08 and B44. MHC class 1 molecules from the other supertypes identified in the volunteers, A03, B27 and B58, may also have recognized AMA1 CD8+ T cell epitopes. Such epitopes binding to these allelic supertypes were predicted by NetMHC, but were not located within the 15-mers recalling the strongest responses by ELISpot assay, and thus were not synthesized or tested. A01, A02, B08 and B44 supertypes cover approximately 100% of Caucasian populations and 27% of US African Americans [61]. Therefore, to reach nearly 100% coverage, particularly of sub-Saharan Africans, other epitopes restricted by the prevalent supertypes A03, A24, B07 and B58 need to be identified. Previous observations identified 11 proliferative T cell epitopes in Kenya[36], one of which was associated with lower risk of parasitaemia[43]. Since these were long peptides (12-22 amino acids) they may contain CD4+ or CD8+ T cell epitopes.
Epitope variability and location within the AMA1 crystal structure
The structures of Domains I, II and III of P. falciparum AMA1 are relatively conserved among Plasmodium species [26,28], but there is nevertheless extensive polymorphism present as well [63-65], reflected by the strain-specificity of inhibitory antibodies [22,26,66]. Many of the previously defined proliferative epitopes (Table 2) map to variant regions of the protein [36,43]. The 14 putative class I epitopes predicted in this study were compared to available sequence data on AMA1 [26], and this analysis showed that 13 of the 14 epitopes likewise contained one or more polymorphic amino acids (all except E1), although three were variable at a low level (<0.001%) (E9, E13 and E14).
A model of the crystal structure of AMA1 was generated to allow localization of ten of the 14 epitopes within the tertiary structure (Figure 5). This is one of the first studies in which CD8+ T cell epitopes, rather than antibody epitopes, have been localized in this way. Many of the ten epitopes clustered together on the front face, whereas a few wrapped around from the front to back or were localized on the back face. Surface location was a surprising finding, since it is generally thought that class I-restricted epitopes occur independently of surface exposure. Surface location of T epitopes has been previously described in HIV where it was suggested that this resulted in increase accessibility to the antigen-processing pathway [67].
Epitope E6 localized to a hyper-variable region of AMA1 on the back of the molecule, adjacent to the putative hydrophobic cleft[32] and E197, one of the variable residues in E6 (see E197 in Table 7), is crucial to the binding of the growth inhibitory MAb 1F9 and also forms part of a putative AMA1 receptor[35]. E197 is also contained within the variable epitope 189 identified in Kenya[36].
Amino acid polymorphism in AMA1 is associated with surface accessibility, whether on the front or back face. Immune pressure may have driven polymorphism in residues surrounding the putative binding site in the hydrophobic cleft[31]. Other studies have attributed polymorphism within T cell epitope regions to host evasion of T cell recognition irrespective of molecular function [26,34,68] in which parasite strains are favored through reduced affinity of molecular sequences for binding to HLA A or B alleles[31]. An association between immune pressure and T cell epitope variability has been proposed for other malaria antigens such as CSP and will need to be overcome for successful vaccine development[14,34]. In one study, a diversity-covering approach that took into account genetic linkages among polymorphic sequences improved allelic recognition by ELISA and growth inhibitory antibodies for AMA1[69], and may also be applicable to T cell-dependent AMA1 vaccines.
Four HLA-B8/B44-restricted epitopes are close to the N-terminal in the signal sequence and prodomain (E1-E4), while seven of the 10 remaining epitopes are HLA-A1/A2-restricted. The signal sequence and prodomain are cleaved off first during merozoite release and red cell invasion[70] during which AMA1 re-localizes around the merozoite surface, and then Domains I and II and part of Domain III are shed as the merozoite invades[26]. It is therefore possible that this sequence of events influences AMA1 uptake by antigen-presenting cells, such that epitopes in the signal sequence and prodomain may be presented to HLA-B-restricted T cells, while the later shedding of the domains favors presentation to HLA-A-restricted T cells. It is also interesting that the single non-polymorphic epitope E14 is located in the part of Domain III which remains on the merozoite during invasion.
Conclusions
This study identified fourteen putative minimal CD8+ T cell-dependent epitopes within AMA1 that are restricted by four HLA supertypes that together are expressed by 100% of Caucasians, but only 27% of African Americans. Nine epitopes were confirmed using ELISpot or ELISpot and ICS assays. Some of these clustered on the front face of AMA1, while others were more randomly distributed but still mostly surface accessible. When the 14 epitopes were mixed together, they recalled CD8+ T cell responses in volunteers with different HLA supertypes providing a reagent useful for measuring CD8+ T cell responses in genetically different populations.
Further testing of the NMRC-M3V-Ad-PfCA vaccine in volunteers representing different HLA A and B supertypes would provide the opportunity to identify additional class I epitopes. This could support the design of a chimeric AMA1 molecule able to elicit broadly protective responses among an HLA diverse population. Determining the potential contribution of these epitopes for inducing protective responses awaits future testing of the NMRC-M3V-Ad-PfCA vaccine in challenge studies.
Competing interests
JTB, CRK and DLD are inventors listed on U.S. Patent No., U.S. Patent No. 2009-0148477 A1, and international patent application PCT/US06/33982, titled "Adenoviral Vector-based Malaria Vaccines"; JTB, CRK, TLR and DLD are inventors listed on U.S. Patent Application 12/522,335, and international patent application PCT/US08/50565 titled "Adenoviral Vector-based Malaria Vaccines". JTB, GenVec, Inc. contributed to the design of the study, interpretation of data, editing of drafts and final manuscript approval but he did not contribute to protocol development or study execution.
Authors' contributions
MS and TLR designed the research; TLR, DLD, JB and CRK designed the vaccine; HG, JL, EA, GB, MB, and RS performed ELISpot assays; SM and FF performed the ICS assays; YK, BP and AS used NetMHC to predict epitopes; CO, CT, DR, and IC were investigators in the clinical trial; BF and ER performed the crystal localization studies; MS, YK, DLD, TLR and MRH wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Martha Sedegah, Email: martha.sedegah@med.navy.mil.
Yohan Kim, Email: ykim.biology@gmail.com.
Bjoern Peters, Email: bpeters@liai.org.
Shannon McGrath, Email: smcgrath18@gmail.com.
Harini Ganeshan, Email: Harini.Ganeshan@med.navy.mil.
Jennylynn Lejano, Email: jnlejano@gmail.com.
Esteban Abot, Email: Steve.Abot@med.navy.mil.
Glenna Banania, Email: glenna.banania@med.navy.mil.
Maria Belmonte, Email: maria.belmonte@med.navy.mil.
Renato Sayo, Email: renato.sayo@med.navy.mil.
Fouzia Farooq, Email: fouzia.farooq@med.navy.mil.
Denise L Doolan, Email: Denise.Doolan@qimr.edu.au.
David Regis, Email: David.Regis@med.navy.mil.
Cindy Tamminga, Email: cindy.tamminga1@med.navy.mil.
Ilin Chuang, Email: Ilin.Chuang@med.navy.mil.
Joseph T Bruder, Email: jbruder@genvec.com.
C Richter King, Email: RKing@iavi.org.
Christian F Ockenhouse, Email: christian.f.ockenhouse@us.army.mil.
Bart Faber, Email: Faber@bprc.nl.
Edmond Remarque, Email: Remarque@bprc.nl.
Michael R Hollingdale, Email: mikedc110@gmail.com.
Thomas L Richie, Email: thomas.richie@med.navy.mil.
Alessandro Sette, Email: alex@liai.org.
Acknowledgements
The authors thankfully acknowledge Henk van Westbroek who created Figure 5. DR, CT, IC, CFO and TLR are active duty military personnel at the time they contributed to this work; MS is a US Government employee. The work of these individuals was prepared as part of official government duties. Title 17 U.S.C. §105 provides that 'Copyright protection under this title is not available for any work of the United States Government.' Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties. The work of authors affiliated with the Naval Medical Research Center was supported by work unit number 6000.RAD1.F.A0309. Major funding for this work was provided by USAID, the Military Infectious Diseases Research Program, and the Congressionally Directed Medical Research Program. DLD was supported by a Pfizer Australia Senior Research Fellowship. The study protocol for the clinical trial presented in this manuscript was approved by the National Naval Medical Center, Naval Medical Research Center and Walter Reed Army Institute of Research Institutional Review Boards, in compliance with all applicable Federal regulations governing protection of human subjects. All study subjects gave written informed consent. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of the Army, the Department of Defense, or the U.S. Government.
References
- Doolan DL, Martinez-Alier N. Immune response to pre-erythrocytic stages of malaria parasites. Curr Mol Med. 2006;6(2):169–185. doi: 10.2174/156652406776055249. [DOI] [PubMed] [Google Scholar]
- Beeson JG, Osier FH, Engwerda CR. Recent insights into humoral and cellular immune responses against malaria. Trends Parasitol. 2008;24(12):578–584. doi: 10.1016/j.pt.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Frevert U, Nardin E. Cellular effector mechanisms against Plasmodium liver stages. Cell Microbiol. 2008;10(10):1956–1967. doi: 10.1111/j.1462-5822.2008.01211.x. [DOI] [PubMed] [Google Scholar]
- Bongfen SE, Torgler R, Romero JF, Renia L, Corradin G. Plasmodium berghei-infected primary hepatocytes process and present the circumsporozoite protein to specific CD8+ T cells in vitro. J Immunol. 2007;178(11):7054–7063. doi: 10.4049/jimmunol.178.11.7054. [DOI] [PubMed] [Google Scholar]
- Trimnell A, Takagi A, Gupta M, Richie TL, Kappe SH, Wang R. Genetically attenuated parasite vaccines induce contact-dependent CD8+ T cell killing of Plasmodium yoelii liver stage-infected hepatocytes. J Immunol. 2009;183(9):5870–5878. doi: 10.4049/jimmunol.0900302. [DOI] [PubMed] [Google Scholar]
- Williams CT, Azad AF. Transcriptional analysis of the pre-erythrocytic stages of the rodent malaria parasite, Plasmodium yoelii. PLoS One. 2010;5(4):e10267. doi: 10.1371/journal.pone.0010267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy VS, Ballou WR. Immunological mechanisms underlying protection mediated by RTS,S: a review of the available data. Malar J. 2009;8(1):312. doi: 10.1186/1475-2875-8-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vekemans J, Leach A, Cohen J. Development of the RTS, S/AS malaria candidate vaccine. Vaccine. 2009;27(Suppl 6):G67–71. doi: 10.1016/j.vaccine.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Mauduit M, Tewari R, Depinay N, Kayibanda M, Lallemand E, Chavatte JM, Snounou G, Renia L, Gruner AC. Minimal role for the circumsporozoite protein in the induction of sterile immunity by vaccination with live rodent malaria sporozoites. Infect Immun. 2010;78(5):2182–2188. doi: 10.1128/IAI.01415-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KA, Sano G, Boscardin S, Nussenzweig RS, Nussenzweig MC, Zavala F, Nussenzweig V. The circumsporozoite protein is an immunodominant protective antigen in irradiated sporozoites. Nature. 2006;444(7121):937–940. doi: 10.1038/nature05361. [DOI] [PubMed] [Google Scholar]
- Kester KE, Cummings JF, Ofori-Anyinam O, Ockenhouse CF, Krzych U, Moris P, Schwenk R, Nielsen RA, Debebe Z, Pinelis E. et al. Randomized, Double-Blind, Phase 2a Trial of Falciparum Malaria Vaccines RTS, S/AS01B and RTS, S/AS02A in Malaria-Naive Adults: Safety, Efficacy, and Immunologic Associates of Protection. J Infect Dis. 2009;200(3):337–346. doi: 10.1086/600120. [DOI] [PubMed] [Google Scholar]
- Barbosa A, Naniche D, Aponte JJ, Manaca MN, Mandomando I, Aide P, Sacarlal J, Renom M, Lafuente S, Ballou WR. et al. Plasmodium falciparum-specific cellular immune responses after immunization with the RTS, S/AS02 D candidate malaria vaccine in infants living in an area of high endemicity in Mozambique. Infect Immun. 2009;77(10):4502–4509. doi: 10.1128/IAI.00442-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul A, Lawrence G, Allworth A, Elliott S, Anderson K, Rzepczyk C, Martin LB, Taylor D, Eisen DP, Irving DO. et al. A human phase 1 vaccine clinical trial of the Plasmodium falciparum malaria vaccine candidate apical membrane antigen 1 in Montanide ISA720 adjuvant. Vaccine. 2005;23(23):3076–3083. doi: 10.1016/j.vaccine.2004.09.040. [DOI] [PubMed] [Google Scholar]
- Thera MA, Doumbo OK, Coulibaly D, Diallo DA, Kone AK, Guindo AB, Traore K, Dicko A, Sagara I, Sissoko MS. et al. Safety and Immunogenicity of an AMA-1 Malaria Vaccine in Malian Adults: Results of a Phase 1 Randomized Controlled Trial. PLoS ONE. 2008;3(1):e1465. doi: 10.1371/journal.pone.0001465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicko A, Sagara I, Ellis RD, Miura K, Guindo O, Kamate B, Sogoba M, Niambele MB, Sissoko M, Baby M. et al. Phase 1 study of a combination AMA1 blood stage malaria vaccine in Malian children. PLoS ONE. 2008;3(2):e1563. doi: 10.1371/journal.pone.0001563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huaman MC, Mullen GE, Long CA, Mahanty S. Plasmodium falciparum apical membrane antigen 1 vaccine elicits multifunctional CD4 cytokine-producing and memory T cells. Vaccine. 2009;27:5239–5246. doi: 10.1016/j.vaccine.2009.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring MD, Cummings JF, Ockenhouse CF, Dutta S, Reidler R, Angov E, Bergmann-Leitner E, Stewart VA, Bittner S, Juompan L. et al. Phase 1/2a study of the malaria vaccine candidate apical membrane antigen-1 (AMA-1) administered in adjuvant system AS01B or AS02A. PLoS One. 2009;4(4):e5254. doi: 10.1371/journal.pone.0005254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin EM, Diemert DJ, McArthur JH, Perreault JR, Miles AP, Giersing BK, Mullen GE, Orcutt A, Muratova O, Awkal M. et al. Phase 1 clinical trial of apical membrane antigen 1: an asexual blood-stage vaccine for Plasmodium falciparum malaria. Infect Immun. 2005;73(6):3677–3685. doi: 10.1128/IAI.73.6.3677-3685.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, Crabb BS, Cowman AF. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol Microbiol. 2000;38(4):706–718. doi: 10.1046/j.1365-2958.2000.02175.x. [DOI] [PubMed] [Google Scholar]
- Collins CR, Withers-Martinez C, Bentley GA, Batchelor AH, Thomas AW, Blackman MJ. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J Biol Chem. 2007;282(10):7431–7441. doi: 10.1074/jbc.M610562200. [DOI] [PubMed] [Google Scholar]
- Hodder AN, Crewther PE, Anders RF. Specificity of the protective antibody response to apical membrane antigen 1. Infect Immun. 2001;69(5):3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Mwangi T, Kocken CH, Thomas AW, Dutta S, Lanar DE, Remarque E, Ross A, Williams TN, Mwambingu G. et al. Human antibodies to recombinant protein constructs of Plasmodium falciparum Apical Membrane Antigen 1 (AMA1) and their associations with protection from malaria. Vaccine. 2004;23(5):718–728. doi: 10.1016/j.vaccine.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Coley AM, Campanale NV, Casey JL, Hodder AN, Crewther PE, Anders RF, Tilley LM, Foley M. Rapid and precise epitope mapping of monoclonal antibodies against Plasmodium falciparum AMA1 by combined phage display of fragments and random peptides. Protein Eng. 2001;14(9):691–698. doi: 10.1093/protein/14.9.691. [DOI] [PubMed] [Google Scholar]
- Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, Harris KS, Batchelor AH, Anders RF, Foley M. The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody. Infect Immun. 2006;74(5):2628–2636. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers AW, Kennedy MC, Keegan BP, Saul A, Long CA, Miller LH. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect Immun. 2002;70(12):6961–6967. doi: 10.1128/IAI.70.12.6961-6967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remarque EJ, Faber BW, Kocken CH, Thomas AW. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2008;24(2):74–84. doi: 10.1016/j.pt.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Silvie O, Franetich JF, Charrin S, Mueller MS, Siau A, Bodescot M, Rubinstein E, Hannoun L, Charoenvit Y, Kocken CH. et al. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J Biol Chem. 2004;279(10):9490–9496. doi: 10.1074/jbc.M311331200. [DOI] [PubMed] [Google Scholar]
- Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, Collins CR, Withers-Martinez C, Hackett F, Blackman MJ, Faber BW, Remarque EJ, Kocken CH. et al. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science. 2005;308(5720):408–411. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- Chesne-Seck ML, Pizarro JC, Vulliez-Le Normand B, Collins CR, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. Structural comparison of apical membrane antigen 1 orthologues and paralogues in apicomplexan parasites. Mol Biochem Parasitol. 2005;144(1):55–67. doi: 10.1016/j.molbiopara.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Dutta S, Lee SY, Batchelor AH, Lanar DE. Structural basis of antigenic escape of a malaria vaccine candidate. Proc Natl Acad Sci USA. 2007;104(30):12488–12493. doi: 10.1073/pnas.0701464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruder JT, Angov E, Limbach KJ, Richie TL. Molecular vaccines for malaria. Hum Vaccin. 2010;6(1):54–77. doi: 10.4161/hv.6.1.10463. [DOI] [PubMed] [Google Scholar]
- Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci USA. 2005;102(36):12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouattara A, Mu J, Takala-Harrison S, Saye R, Sagara I, Dicko A, Niangaly A, Duan J, Ellis RD, Miller LH. et al. Lack of allele-specific efficacy of a bivalent AMA1 malaria vaccine. Malar J. 2010;9(1):175. doi: 10.1186/1475-2875-9-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala SL, Plowe CV. Genetic diversity and malaria vaccine design, testing and efficacy: preventing and overcoming 'vaccine resistant malaria'. Parasite Immunol. 2009;31(9):560–573. doi: 10.1111/j.1365-3024.2009.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Casey JL, Coley AM, Karas JA, Sabo JK, Tan YY, Dolezal O, Norton RS, Hughes AB, Scanlon D. et al. Rapid optimization of a peptide inhibitor of malaria parasite invasion by comprehensive N-methyl scanning. J Biol Chem. 2009;284(14):9361–9371. doi: 10.1074/jbc.M808762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal AA, Hughes MA, Oliveira DA, Nelson C, Bloland PB, Oloo AJ, Hawley WE, Hightower AW, Nahlen BL, Udhayakumar V. Identification of T-cell determinants in natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in an adult population exposed to malaria. Infect Immun. 1996;64(3):1054–1059. doi: 10.1128/iai.64.3.1054-1059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abboud N, De Jesus M, Nakouzi A, Cordero RJ, Pujato M, Fiser A, Rivera J, Casadevall A. Identification of linear epitopes in Bacillus anthracis protective antigen bound by neutralizing antibodies. J Biol Chem. 2009;284(37):25077–25086. doi: 10.1074/jbc.M109.022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Suguitan AL, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ. et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120(5):1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang R, Wu Y, Zhang D, He Z, Pan W. Epitope mapping of PfCP-2.9, an asexual blood-stage vaccine candidate of Plasmodium falciparum. Malar J. 2010;9(1):94. doi: 10.1186/1475-2875-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger T, Becker MR, Schadlich L, Waterboer T, Gissmann L. Identification of B-cell epitopes on virus-like particles of cutaneous alpha-human papillomaviruses. J Virol. 2009;83(24):12692–12701. doi: 10.1128/JVI.01582-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H, Zhou Z, Feng K, Tian X, Li X, Li H, Xing K, Chen J, Li C, Zhou R. Epitope mapping and cross-reactivity analysis of the monoclonal antibodies against hexon protein of human adenovirus type 3. Virus Res. 2009;146(1-2):58–65. doi: 10.1016/j.virusres.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Hall RA, Tan SE, Selisko B, Slade R, Hobson-Peters J, Canard B, Hughes M, Leung JY, Balmori-Melian E, Hall-Mendelin S. et al. Monoclonal antibodies to the West Nile virus NS5 protein map to linear and conformational epitopes in the methyltransferase and polymerase domains. J Gen Virol. 2009;90(Pt 12):2912–2922. doi: 10.1099/vir.0.013805-0. [DOI] [PubMed] [Google Scholar]
- Udhayakumar V, Kariuki S, Kolczack M, Girma M, Roberts JM, Oloo AJ, Nahlen BL, Lal AA. Longitudinal study of natural immune responses to the Plasmodium falciparum apical membrane antigen (AMA-1) in a holoendemic region of malaria in western Kenya: Asembo Bay Cohort Project VIII. Am J Trop Med Hyg. 2001;65(2):100–107. doi: 10.4269/ajtmh.2001.65.100. [DOI] [PubMed] [Google Scholar]
- Maier RH, Maier CJ, Rid R, Hintner H, Bauer JW, Onder K. Epitope mapping of antibodies using a cell array-based polypeptide library. J Biomol Screen. 2010;15(4):418–426. doi: 10.1177/1087057110363821. [DOI] [PubMed] [Google Scholar]
- Singh R, Singh S, Sharma PK, Singh UP, Briles DE, Hollingshead SK, Lillard JW Jr. Helper T cell epitope-mapping reveals MHC-peptide binding affinities that correlate with T helper cell responses to pneumococcal surface protein A. PLoS One. 2010;5(2):e9432. doi: 10.1371/journal.pone.0009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, Cai X, Jia W, Liu B, Zhang S, Wang P, Yan H, Luo X. Mapping of Taenia solium TSOL18 antigenic epitopes by phage display library. Parasitol Res. 2010;106(5):1151–1157. doi: 10.1007/s00436-010-1786-1. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pan Y, Guo Y, Qiu L, Ding X, Che X. Comprehensive mapping of immunodominant and conserved serotype- and group-specific B-cell epitopes of nonstructural protein 1 from dengue virus type 1. Virology. 2010;398(2):290–298. doi: 10.1016/j.virol.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Freund NT, Enshell-Seijffers D, Gershoni JM. Phage display selection, analysis, and prediction of B cell epitopes. Curr Protoc Immunol. 2009;Chapter 9(Unit 9 8) doi: 10.1002/0471142735.im0908s86. [DOI] [PubMed] [Google Scholar]
- Chung JL, Sun J, Sidney J, Sette A, Peters B. IMMUNOCAT-a data management system for epitope mapping studies. J Biomed Biotechnol. 2010;2010:856842. doi: 10.1155/2010/856842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ru B, Li S, Lin H, Guo FB. SAROTUP: scanner and reporter of target-unrelated peptides. J Biomed Biotechnol. 2010;2010:101932. doi: 10.1155/2010/101932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS. In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine. 2009;27(46):6471–6479. doi: 10.1016/j.vaccine.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Wu D, Xu T, Wang X, Xu X, Tao L, Li YX, Cao ZW. SEPPA: a computational server for spatial epitope prediction of protein antigens. Nucleic Acids Res. 2009. pp. W612–616. [DOI] [PMC free article] [PubMed]
- Wulf M, Hoehn P, Trinder P. Identification of human MHC class I binding peptides using the iTOPIA- epitope discovery system. Methods Mol Biol. 2009;524:361–367. doi: 10.1007/978-1-59745-450-6_26. full_text. [DOI] [PubMed] [Google Scholar]
- Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni JM, Roitburd-Berman A, Siman-Tov DD, Tarnovitski Freund N, Weiss Y. Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs. 2007;21(3):145–156. doi: 10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12(5):1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Epstein J, Baraceros FM, Gorak EJ, Charoenvit Y, Carucci DJ, Hedstrom RC, Rahardjo N, Gay T, Hobart P. et al. Induction of CD4(+) T cell-dependent CD8(+) type 1 responses in humans by a malaria DNA vaccine. Proc Natl Acad Sci USA. 2001;98(19):10817–10822. doi: 10.1073/pnas.181123498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart VA, McGrath SM, Dubois PM, Pau MG, Mettens P, Shott J, Cobb M, Burge JR, Larson D, Ware LA. et al. Priming with an adenovirus 35-circumsporozoite protein (CS) vaccine followed by RTS, S/AS01B boosting significantly improves immunogenicity to Plasmodium falciparum CS compared to that with either malaria vaccine alone. Infect Immun. 2007;75(5):2283–2290. doi: 10.1128/IAI.01879-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, Buus S, Lund O. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008;4(7):e1000107. doi: 10.1371/journal.pcbi.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50(3-4):201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- Wang R, Richie TL, Baraceros MF, Rahardjo N, Gay T, Banania JG, Charoenvit Y, Epstein JE, Luke T, Freilich DA. et al. Boosting of DNA vaccine-elicited gamma interferon responses in humans by exposure to malaria parasites. Infect Immun. 2005;73(5):2863–2872. doi: 10.1128/IAI.73.5.2863-2872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Chokejindachai W, Conway DJ. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165(2):555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Grebert HM, Isea R, Goldman IF, Basco L, Magris M, Biswas S, Kariuki S, Lal AA. A study of genetic diversity in the gene encoding the circumsporozoite protein (CSP) of Plasmodium falciparum from different transmission areas--XVI. Asembo Bay Cohort Project. Mol Biochem Parasitol. 2002;125(1-2):83–90. doi: 10.1016/S0166-6851(02)00216-5. [DOI] [PubMed] [Google Scholar]
- Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect Immun. 2003;71(3):1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MC, Wang J, Zhang Y, Miles AP, Chitsaz F, Saul A, Long CA, Miller LH, Stowers AW. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect Immun. 2002;70(12):6948–6960. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surman S, Lockey TD, Slobod KS, Jones B, Riberdy JM, White SW, Doherty PC, Hurwitz JL. Localization of CD4+ T cell epitope hotspots to exposed strands of HIV envelope glycoprotein suggests structural influences on antigen processing. Proc Natl Acad Sci USA. 2001;98(8):4587–4592. doi: 10.1073/pnas.071063898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zevering Y, Khamboonruang C, Good MF. Human and murine T-cell responses to allelic forms of a malaria circumsporozoite protein epitope support a polyvalent vaccine strategy. Immunology. 1998;94(3):445–454. doi: 10.1046/j.1365-2567.1998.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remarque EJ, Faber BW, Kocken CH, Thomas AW. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect Immun. 2008;76(6):2660–2670. doi: 10.1128/IAI.00170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AW, Trape JF, Rogier C, Goncalves A, Rosario VE, Narum DL. High prevalence of natural antibodies against Plasmodium falciparum 83-kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture-enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am J Trop Med Hyg. 1994;51(6):730–740. doi: 10.4269/ajtmh.1994.51.730. [DOI] [PubMed] [Google Scholar]