Abstract
ATP-gated purinergic P2X4 receptors (P2X4Rs) are expressed in the central nervous system and are sensitive to ethanol at intoxicating concentrations. P2XRs are trimeric; each subunit consists of two transmembrane (TM) α-helical segments, a large extracellular domain, and intracellular amino and carboxyl terminals. Recent work indicates that position 336 (Met336) in the TM2 segment is critical for ethanol modulation of P2X4Rs. The anthelmintic medication ivermectin (IVM) positively modulates P2X4Rs and is believed to act in the same region as ethanol. The present study tested the hypothesis that IVM can antagonize ethanol action. We investigated IVM and ethanol effects in wild-type and mutant P2X4Rs expressed in Xenopus oocytes by using a two-electrode voltage clamp. IVM antagonized ethanol-induced inhibition of P2X4Rs in a concentration-dependent manner. The size and charge of substitutions at position 336 affected P2X4R sensitivity to both ethanol and IVM. The first molecular model of the rat P2X4R, built onto the X-ray crystal structure of zebrafish P2X4R, revealed a pocket formed by Asp331, Met336, Trp46, and Trp50 that may play a role in the actions of ethanol and IVM. These findings provide the first evidence for IVM antagonism of ethanol effects in P2X4Rs and suggest that the antagonism results from the ability of IVM to interfere with ethanol action on the putative pocket at or near position 336. Taken with the building evidence supporting a role for P2X4Rs in ethanol intake, the present findings suggest that the newly identified alcohol pocket is a potential site for development of medication for alcohol use disorders.
Introduction
ATP-gated purinergic P2X receptors (P2XRs) are a superfamily of ligand-gated ion channels (Khakh et al., 2001; North, 2002) that are becoming a focus of investigation in alcohol studies. Recent evidence suggests that P2XRs may play a role in ethanol consumption (Tabakoff et al., 2009).
P2XRs are broadly distributed in the central and peripheral nervous systems (Rubio and Soto, 2001; North, 2002). Currently, seven subunits of the P2XRs have been identified (P2X1–P2X7) that form functional ATP-activated homomeric channels (e.g., P2X2, P2X4) and heteromeric receptors (e.g., P2X2/3, P2X4/6) in mammals (Aschrafi et al., 2004). P2XRs are trimeric; each subunit consists of two α-helical transmembrane (TM) segments, a large extracellular domain (ectodomain), and intracellular amino and carboxyl terminals (North, 2002). The TM1 and TM2 membrane-spanning segments are involved in ion channel gating and ion pore formation (Burnstock, 2004; Li et al., 2008). The ectodomain contains an ATP-binding site and is a site for channel regulation (Chizh and Illes, 2001; North, 2002). Recent crystallographic investigations (Kawate et al., 2009) confirmed the previous predictions that functional P2XR channels result from the assembly of three subunits (Jiang et al., 2003; Aschrafi et al., 2004) with TM2 segments lining the pore.
P2X2, P2X3, and P2X4Rs expressed in Xenopus oocytes are sensitive to ethanol at intoxicating and anesthetic concentrations (Xiong et al., 2000; Davies et al., 2002, 2005). Previous studies found that residues contained within the ectodomain–TM segment interfaces are important for ethanol action in P2X3Rs (Asatryan et al., 2008). Extending the investigation to P2X4Rs resulted in the identification of two key residues in the TM2 segment near the ectodomain interface (Asp331 and Met336) that, when substituted to alanine, caused a significant reduction in ethanol (10–200 mM) inhibition of ATP-gated currents (Popova et al., 2010). Preliminary investigations also identified position 46 (Trp46) in the TM1 segment near the ectodomain as a potential target for ethanol action. Another study found that substitution of a histidine residue for alanine at position 241 in the ectodomain region switched the mechanism of ethanol inhibition in P2X4Rs from competitive to noncompletive (Xiong et al., 2005). These findings led Xiong et al. to suggest that ethanol seems to inhibit receptor channels in H241A P2X4Rs by interacting with a yet unknown allosteric site. Taken together, these findings identified sites in the ectodomain region that can alter the mechanism of ethanol action and suggest that the actions of ethanol are initiated by interaction with positions 46, 331, and/or 336 in the TM regions.
Ivermectin (IVM), a macrocyclic lactone, is a member of a class of lipophilic compounds (avermectins) and is widely used in animals and humans as a broad spectrum anthelmintic medication (Richard-Lenoble et al., 2003; Geary, 2005). The therapeutic effect (antiparasitic properties) of IVM is attributed to its action on a nonmammalian, glutamate-gated inhibitory chloride channel (Dent et al., 1997). IVM has been shown to act as an anticonvulsant in mice (Dawson et al., 2000) and was originally thought to act on mammalians via potentiation of GABAA and glycine receptors [for review see Dawson et al. (2000) and Shan et al. (2001))]. However, more recent work suggests that IVM also acts on targets other than GABAA or glycine receptors (Sung et al., 2009). Within the P2XR superfamily, IVM is a selective P2X4R allosteric modulator that is routinely used as a pharmacological tool to identify the contribution of P2X4Rs in ATP-mediated processes (Khakh et al., 1999). Recent reports suggest that IVM binds at the lipid–protein interface, acting on sites located in the TM segments at the ectodomain–TM interface of the P2X4R. Individual mutations of residues at these interfaces indicated that positions Val47, Trp50, Val60, and Asn338 may play an important role in IVM regulation of the channel function (Jelínková et al., 2006, 2008; Silberberg et al., 2007).
Collectively, these findings suggest that the ectodomain–TM interface of P2X4Rs is a site of action and/or modulation for both ethanol and IVM. This notion led to the prediction that IVM would interfere with and, thus, reduce the sensitivity of P2X4Rs to ethanol. To test this hypothesis, we investigated the effects of IVM on ethanol inhibition of ATP-induced currents in wild-type (WT) and mutant P2X4Rs expressed in Xenopus oocytes. The findings support the hypothesis and led to a new molecular model of the sites of action for ethanol and IVM in P2X4Rs based on the recent X-ray crystal structure of zebrafish P2X4R (Kawate et al., 2009).
Materials and Methods
Materials.
ATP, ethanol (190 proof, USP), IVM (ivermectin B1A), and collagenase were purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions of IVM (10 mM) were made in dimethyl sulfoxide and kept at −20°C. The highest dimethyl sulfoxide concentration in final solutions was 0.1%. All other chemicals were of reagent grade.
Isolation of Xenopus laevis Oocytes and cRNA Injections.
Xenopus oocytes were isolated and maintained as described previously (Davies et al., 2002, 2005). All procedures for the maintenance of Xenopus laevis frogs and oocyte isolation were approved by The Institutional Animal Care and Use Committee of the University of Southern California. Stage V and VI oocytes were used for cRNA (20 ng) injections by using the Nanoject II Nanoliter injection system (Drummond Scientific, Broomall, PA) 1 day after isolation. Injected oocytes were stored at 16°C in incubation medium containing 96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5 mM HEPES, 0.6 mM theophylline, and 2.5 mM pyruvic acid with 1% horse serum and 0.05 mg/ml gentamycin. The oocytes were used in electrophysiological recordings for 3 to 7 days after cRNA injections.
Site-Directed Mutagenesis and cRNA Synthesis.
The cDNA of rat P2X4R (GenBank accession no. X87763) was subcloned into pcDNA3 vector (Invitrogen, Carlsbad, CA). Mutagenesis was performed to introduce single point mutations by using the QuikChange IIXL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). The sequences of mutant constructs were verified by automated DNA sequencing (Norris DNA Core Facility, University of Southern California).
The DNA of WT or mutant receptors was linearized and transcribed with the mMESSAGE mMACHINE kit (Ambion, Austin, TX) to result in cRNA, which was stored at −70°C until injection.
Whole-Cell Voltage-Clamp Recordings.
Two-electrode voltage-clamp recordings of oocytes were performed with an oocyte clamp amplifier (model OC-725C; Warner Instruments, Hamden, CT) using procedures published previously (Asatryan et al., 2008; Popova et al., 2010).
Experimental Procedures.
To induce currents and test the effects of alcohols and IVM in our experiments, we used maximal (100 μM) or submaximal ATP concentrations (EC5–10, hereafter referred to as EC10). We have shown previously that the use of EC10 can maximize the effects of ethanol and minimize receptor desensitization (Davies et al., 2002, 2005). The washout time between agonist applications was at least 5 to 10 min to allow complete resensitization of the receptors (Davies et al., 2002, 2005).
Effects of the drugs were tested after a stable response to ATP EC10 was obtained. After a sufficient washout time (5–15min) the cells were challenged with the same ATP EC10 to ensure an accurate response to the drugs and assess any change in the baseline level.
IVM and Alcohol Applications.
Alcohols (ethanol, butanol, and hexanol) were coapplied with ATP EC10 for 20 s. To allow comparison of the effects of IVM with those of ethanol, we used the same coapplication protocol for IVM. Alcohols and IVM did not affect the resting membrane currents in uninjected oocytes, and neither drug induced any currents when applied without ATP. To study the simultaneous effects of IVM and alcohols (either ethanol or hexanol), we coapplied both drugs with ATP EC10. UV spectra of IVM (OD244 nm) were measured in the presence of differing concentrations of ethanol. No change in the UV characteristics confirmed that IVM and ethanol did not interact in the solution.
Surface Biotinylation and Western Blotting.
Oocytes expressing P2X4R cRNA were treated with 3 μM IVM in the presence of 1 μM ATP and incubated with 1.0 mg/ml membrane-impermeable sulfo-NHS-SS-biotin (Pierce Chemical, Rockford, IL) for 1 h on ice. After lysis, the biotinylated proteins were precipitated by overnight incubation with streptavidin beads (Pierce Chemical) and extracted by using SDS sample buffer. Protein lysates of total and biotinylated fractions were then run on 10% SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and blotted with polyclonal P2X4R antibody (Millipore Bioscience Research Reagents, Temecula, CA). After incubation with the secondary antibody, the P2X4R bands were visualized by enhanced chemiluminescence (Pierce Chemical). Densitometry analysis of the protein bands was performed with Scion Image software (Scion Corporation, Frederick, MD). Data are presented as a ratio of biotinylated over total fractions.
Molecular Modeling.
We used the 3.1-Å resolution X-ray crystal structure of the zebrafish P2X4-A (Protein Data Bank ID 3I5D) (Kawate et al., 2009) as a template for the homology models of the rat P2X4R. Protein Data Bank file 3I5D has unresolved electron density at the N-terminal and C-terminal ends. As a result, the primary sequence of rat P2X4R was edited to fit the length of the zebrafish P2X4R structure before sequence alignment. The homology was high, and it was possible to align the sequences without gaps. We threaded the rat sequence onto the backbone atoms of the template by using the Modeler module of Discovery Studio 2.5.5 (Accelrys, San Diego, CA), essentially as described previously for GABAAR and GlyR models (Perkins et al., 2009). Then we used the autorotamer module of Discovery Studio to optimize the positions of the side chains while the backbone atoms were tethered. We relaxed the resulting model by tethering the backbone atoms with a harmonic restraint of 10 kcal/A2, then optimizing to a gradient of 0.001 kcal/A and running molecular dynamics for 10,000 1-fs steps at 300 K. We placed the resulting model in a periodic water box with dimensions assigned automatically with the solvation module of Discovery Studio 2.5.5 with the CHARMm force field. We optimized the assembly as above, while imposing the same harmonic restraints on the backbone atoms, and then performed molecular dynamics at 300 K for 100,000 1-fs steps. Then we built a molecule of ethanol at the same scale and manually positioned it into the final frame of the molecular dynamics trajectory, near the obvious site between Asp331, Trp46, Trp50, Met336, and the ectodomain. To compare what IVM would look like in this site, we built a model of IVM by reducing the double bond of avermectin as described by Sattelle et al. (2009). We then manually positioned this IVM model into the same site where we positioned ethanol; we tried to follow the orientation of IVM in the α-helical bundle as shown by Sattelle et al. (2009). That is, we positioned the spiroketal and benzofuran rings near Trp46 and Trp50 while pointing the disaccharide along the long axis of the α-helices. We should note that the ethanol or the IVM models were not positioned with a “docking” program. They are meant to give a sense of the relative scale of the two ligands.
Data Analysis.
Data were obtained from several batches of oocytes of at least three different frogs and are expressed as mean ± S.E.M. Effects of alcohols and IVM are presented as percentage change of peak currents evoked by ATP EC10 alone. Data were fitted to a concentration–response curve by using the following logistic equation: I = Imax × [drug]/([drug] + (EC50)), where I is the percentage of the maximum obtainable response (Imax) and EC50 is the concentration producing a half-maximal response. Prism software (GraphPad Software, Inc., San Diego, CA) was used for data analysis and curve fitting. Statistical analysis was performed by using unpaired t test with significance set at P < 0.05.
Results
Ethanol and IVM Modulate P2X4Rs.
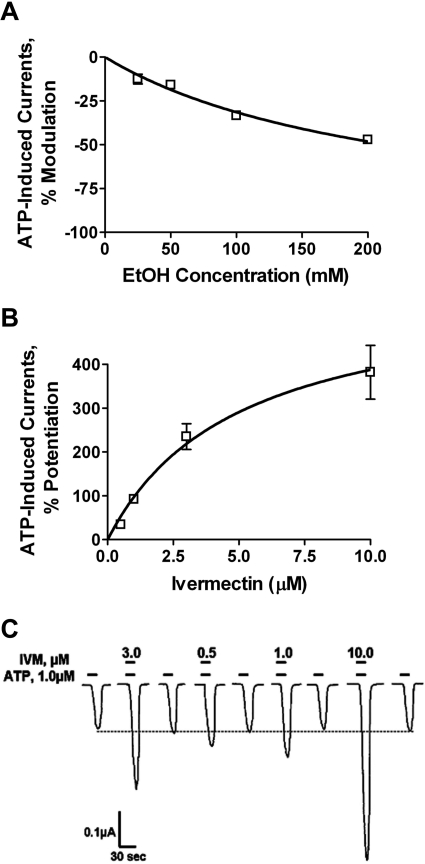
In agreement with previous work (Davies et al., 2002; Popova et al., 2010), ethanol (25–200 mM) coapplication with ATP inhibited ATP EC10 -induced currents in P2X4Rs in a concentration-dependent manner (Fig. 1A). Using the same protocol, IVM (0.5–10 μM) significantly potentiated EC10 ATP-gated currents in P2X4Rs in a concentration-dependent and reversible manner (Fig. 1, B and C). The direction and magnitude of these findings with IVM are consistent with prior studies (Khakh et al., 1999; Priel and Silberberg, 2004), particularly when taking into account our short 20-s incubation.
Fig. 1.
Ethanol and IVM differentially modulate P2X4Rs. A and B, ethanol inhibits (A) and IVM potentiates (B) ATP-gated currents in P2X4Rs in a concentration-dependent manner. Data in A and B are presented as mean ± S.E.M. of three to eight oocytes per data point. C, representative tracings of IVM modulation of ATP-induced currents in P2X4Rs.
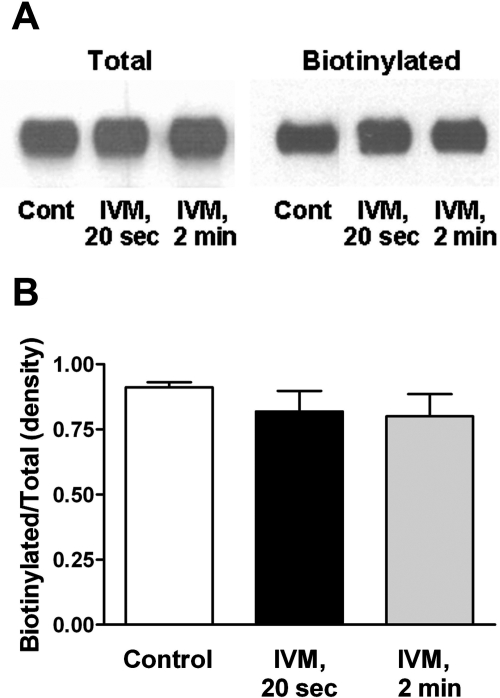
Exposure to IVM and ATP for either 20 s or 2 min did not significantly change the surface expression levels of P2X4Rs in oocytes (Fig. 2). This lack of change in cell surface expression during the time frame of our experiments is consistent with prior studies indicating that IVM potentiates ATP-gated currents in P2X4Rs by direct action on the receptor (Khakh et al., 1999; Priel and Silberberg, 2004) rather than by increasing surface expression (Toulmé et al., 2006).
Fig. 2.
IVM does not affect surface expression of P2X4Rs. A, a representative blot showing that there is no change in the surface expression (biotinylated fraction) of P2X4Rs after treatment with IVM. Control (Cont), 1 μM ATP; IVM 20 sec, 3 μM IVM + 1 μM ATP; IVM 2 min, oocytes treated with 3 μM IVM for 2 min before the addition of 1 μM ATP. B, analysis of the data from five separate experiments. Data are presented as ratio of biotinylated and total fractions after densitometry analysis of the protein bands.
IVM Antagonizes Ethanol Inhibition of P2X4Rs.
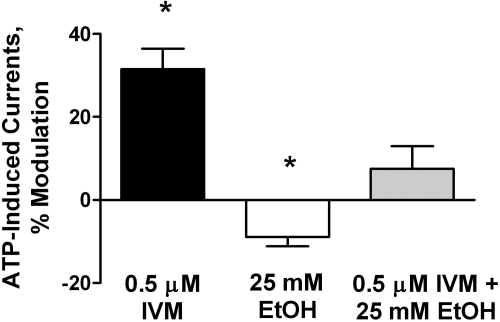
To begin testing the hypothesis that IVM will interfere with the sensitivity of P2X4Rs to ethanol, we investigated the effect of 0.5 μM IVM alone and in combination with a behaviorally relevant concentration of ethanol (25 mM) on ATP-activated currents in P2X4Rs. The results are shown in Fig. 3. In agreement with previous studies, ethanol significantly inhibited ATP EC10-induced currents in P2X4Rs (Davies et al., 2005; Popova et al., 2010), and IVM significantly potentiated ATP EC10-induced currents in P2X4Rs (Khakh et al., 1999; Priel and Silberberg, 2004). As predicted by the hypothesis, IVM antagonized the inhibitory effect of ethanol (Fig. 3). In this single-dose study, the antagonism was complete.
Fig. 3.
IVM antagonizes the effects of a behaviorally relevant concentration of ethanol in P2X4Rs. Exposure to 0.5 μM IVM eliminated the significant inhibitory effect of 25 mM ethanol on EC10 ATP-gated currents in WT P2X4Rs. Data are presented as mean ± S.E.M. of 8 to 10 oocytes per data point. *, P < 0.05 compared with ATP control.
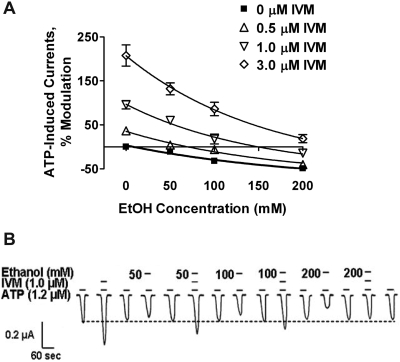
Follow-up concentration–response studies found that IVM caused a right shift of the ethanol inhibition curve (Fig. 4A). The degree of shift depended on the concentration of IVM and ethanol. IVM at 0.5 μM completely blocked the inhibitory effects of 50 and 100 mM ethanol, but did not significantly alter the effect of 200 mM ethanol. Higher concentrations of IVM reduced the inhibitory effects of ethanol at all ethanol concentrations (50–200 mM; Fig. 4A). These results clearly indicate that IVM antagonizes the effects of ethanol.
Fig. 4.
IVM right-shifts the ethanol concentration–response curve. A, the effects of ethanol (50–200 mM) and IVM (0.5–3 μM) alone and in combination on ATP-gated currents in P2X4Rs. Data are presented as mean ± S.E.M. of 5 to 10 oocytes per data point. B, representative tracings showing modulation of ATP-induced currents by individual applications of IVM (1 μM) and ethanol (50, 100, and 200 mM) and the antagonistic effect of IVM on ethanol inhibition during their combined action. Horizontal short bars above the currents depict ATP or drug applications. Numbers next to the short bars represent ethanol concentrations. Dotted line represents the baseline responses for ATP.
To test whether the current findings could be explained by additive effects of IVM potentiation offsetting ethanol inhibition, we compared the experimentally obtained results (presented in Figs. 3 and 4) to the estimated algebraic additive values of the individual effects of the two drugs. As shown in Table 1, the experimentally obtained data were significantly lower than the estimated additive values of the effects of the two drugs obtained individually. These findings indicate that IVM antagonism of ethanol in P2X4Rs cannot be explained by independent additive interactions.
TABLE 1.
The effects of IVM and ethanol on ATP-gated currents in P2X4Rs are nonadditive
Comparison of the data expressed as percent potentiation of ATP-induced currents either by different combinations of ethanol and IVM (experimental) or mathematical addition of the individual effects of the two drugs (estimated). Significant differences between the two groups were found: *, P < 0.05; **, P < 0.01. Data are presented as mean ± S.E.M. of 5 to10 oocytes per data point.
| EtOH | 0.5 μM IVM |
1.0 μM IVM |
3.0 μM IVM |
|||
|---|---|---|---|---|---|---|
| Experimental | Estimated | Experimental | Estimated | Experimental | Estimated | |
| mM | ||||||
| 25 | 9.9 ± 6.1* | 27.4 ± 4.6 | ||||
| 50 | −4.4 ± 3.4** | 26.1 ± 6.0 | 60 ± 7.3** | 95 ± 8.0 | 132 ± 13.7* | 195.8 ± 24.1 |
| 100 | −12.5 ± 4.5* | 6.9 ± 3.9 | 35.0 ± 15.0** | 70 ± 8.5 | 94.3 ± 15.4** | 177.6 ± 24.0 |
| 200 | −5.5 ± 9.5** | 56.2 ± 7.0 | 11.1 ± 6.2** | 160.6 ± 24.0 | ||
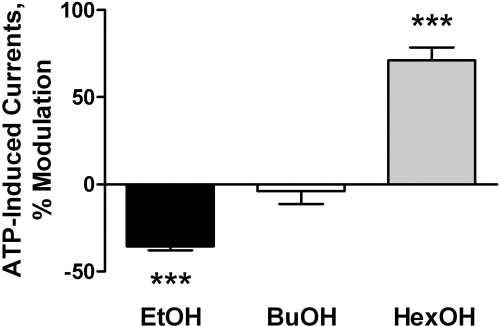
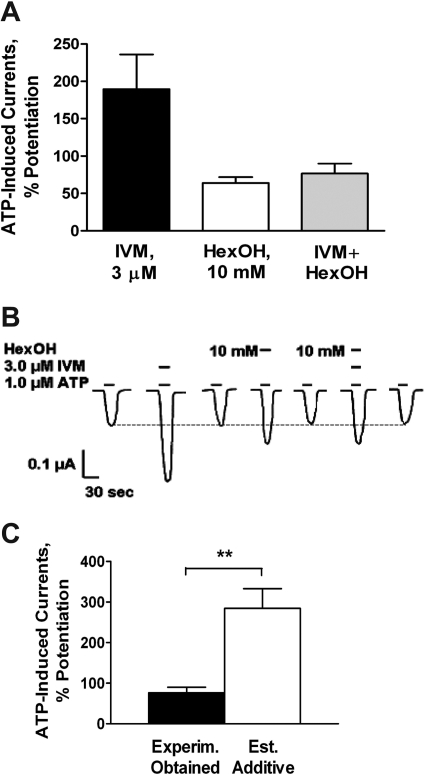
Further studies were conducted with hexanol. The use of hexanol was based on a study that showed a crossover from inhibition to potentiation of ATP-gated currents in P2X4Rs as the chain length of the n-chain alcohol increased from ethanol to hexanol (Fig. 5). Hexanol and IVM each potentiated P2X4R function when applied individually (Fig. 6). Their combined effects were significantly less than would be predicted based on independent interactions at different sites (Fig. 6). These findings support the contention that the actions of IVM on ethanol are not based on an additive interaction.
Fig. 5.
The effect of alcohols on ATP-gated currents in P2X4Rs crosses over from inhibition to potentiation as the n-chain increases from ethanol to hexanol. Ethanol (EtOH; C2, 200 mM) inhibited, and butanol (BuOH; C4, 50 mM) produced minimal effect, whereas hexanol (HexOH; C6, 10 mM) significantly potentiated ATP-induced currents in P2X4R. ***, P < 0.001 compared with ATP only. Data are presented as mean ± S.E.M. of 12 to 16 oocytes per data point.
Fig. 6.
The effects of IVM and hexanol on ATP-gated currents in P2X4Rs are nonadditive. A, IVM (3 μM) and hexanol (10 mM) significantly potentiated ATP-induced currents when applied individually or in combination. P < 0.05 compared with ATP only. B, representative tracings showing modulation of ATP-induced currents by individual applications of IVM (3 μM), hexanol (10 mM), and coapplication of IVM and hexanol. Horizontal short bars above the currents depict ATP or drug applications. Numbers next to the short bars represent hexanol concentration. Dotted line represents the baseline responses for ATP. C, comparison of experimentally obtained data of the combined effects of IVM and hexanol (filled bar) and estimated additive value of the effects of individual drugs (empty bar) shows a significant difference. **, P < 0.01. Data in A and C are presented as mean ± S.E.M. of three to five oocytes per data point.
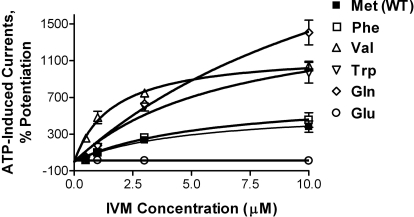
Position 336 Is Important for the Action of IVM.
Further studies created mutations at position 336 to directly test the possibility that IVM antagonizes ethanol by interfering with its action on a site in the TM2 segment of P2X4Rs. These studies tested the prediction that mutations at position 336 known to alter the sensitivity of P2X4Rs to ethanol (Popova et al., 2010) would also alter sensitivity of the receptor to IVM, if position 336 plays a role in the actions of both ethanol and IVM. As shown in Fig. 7, concentration–response studies found that mutations at position 336 caused marked changes in the sensitivity of P2X4Rs to IVM. It is noteworthy that Met336 substitutions (Ala, Val) that affected IVM and ethanol sensitivity of P2X4Rs also significantly reduced the effects of hexanol (p < 0.05; n = 5–14). Collectively, these findings suggest that position 336 plays a role in the actions of IVM, ethanol, and hexanol. It is noteworthy that taken with prior ethanol studies (Popova et al., 2010) these findings support the notion that IVM antagonizes ethanol by interfering with its action at or near position 336 in the TM2 segment.
Fig. 7.
Mutations at position 336 change the sensitivity of P2X4Rs to modulation by IVM. IVM (0.5–10 μM) concentration–response curves for WT and mutant P2X4Rs at position 336 are shown. Data were produced by using ATP EC10-induced currents and are presented as mean ± S.E.M. of 3 to 12 oocytes per data point. The ATP EC10 values for WT and mutant receptors are presented in Table 2.
The effects of these mutations provide insight into the physical–chemical properties at position 336 that affect sensitivity to IVM and ethanol. Table 2 shows the results for the effects of 3 μM IVM and 50 mM ethanol. Substituting Met336 with the small nonpolar Ala or Val residues markedly increased the degree of IVM potentiation and significantly decreased ethanol inhibition. Substituting the bulky nonpolar Phe or Trp residues increased IVM potentiation, but less so than with the small nonpolar amino acids (i.e., Ala, Val). Phe substitutions did not cause a significant change in ethanol sensitivity, whereas Trp substitution resulted in a decreased response to ethanol. Substituting Met with the polar bulky Gln residue had approximately the same effect on IVM sensitivity as substituting the nonpolar bulky residues and reduced sensitivity to ethanol. Substituting charged positive (Arg) or negative (Glu) amino acids for the bulky nonpolar Met eliminated sensitivity to IVM and ethanol.
TABLE 2.
Met336 substitutions with amino acids with different physical–chemical properties significantly alter the sensitivity to ethanol and IVM
Data of 50 mM ethanol are adopted from Popova et al. (2010). Data are presented as mean ± S.E.M. of 3 to 10 oocytes per data point. Significant differences were found for all the mutants except for the ethanol effect in Phe mutant (P < 0.05 compared with the WT P2X4R).
| IVM, 3 μM | EtOH, 50 mM | ATP EC10 Range | |
|---|---|---|---|
| % Potentiation | % Inhibition | μM | |
| Met (WT) | 208 ± 24 | 22.0 ± 1.6 | 1.0–1.2 |
| Ala | 1154 ± 51 | 9.3 ± 4.6 | 0.5–0.8 |
| Val | 737 ± 42 | 6.5 ± 3.7 | 0.8–1.0 |
| Phe | 324 ± 25 | 19.0 ± 3.5 | 0.8–1.0 |
| Trp | 531 ± 96 | 5.9 ± 2.5 | 0.25–0.3 |
| Gln | 508 ± 77 | 4.0 ± 5.5 | 0.6–0.8 |
| Glu | 15.5 ± 5.9 | −1.0 ± 3.6 | 0.6–0.8 |
| Arg | −4.8 ± 1.8 | 0.8 ± 1.4 | 0.1–0.2 |
Collectively, it seems that the size and the charge of the substitutions at position 336 play key roles in determining sensitivity to both IVM and ethanol, whereas polarity is important only for the sensitivity of the receptor to ethanol. Together, these mutational studies (Popova et al., 2010) add further evidence that position 336 plays a role in the actions of both ethanol and IVM on P2X4Rs.
Molecular Modeling of the Rat P2X4R Reveals Met336 Is at the Interdomain Interface.
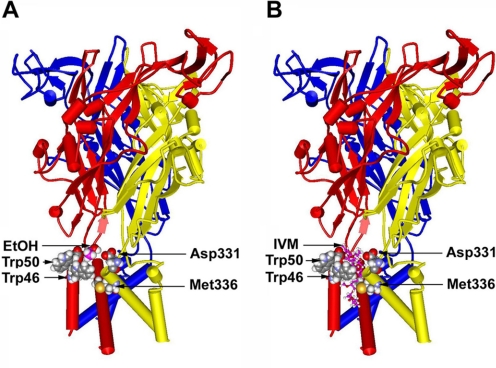
To visualize the potential molecular interactions of Met336 and other key residues such as Trp46 (our preliminary results) and Trp50 (Jelínková et al., 2006), Asp331 (Popova et al., 2010), and their potential role in IVM antagonism of ethanol action, we generated a homology model of the rat P2X4R. The model is based on the crystal structure of zebrafish P2X4R (Kawate et al., 2009). Based on the current and prior studies reviewed in the Introduction, Fig. 8 highlights residues Trp46 and Trp50 in the first α-helix of one subunit and Asp331 and Met336 in the final α-helix of the adjacent subunit. The location of the four residues (Trp46, Trp50, D331, and Met336) is at the interface of the ectodomain and TM segments. During the short molecular dynamics simulation, the C-α to C-β bonds of Trp46 and Trp50 rotated to be nearly perpendicular to each other and to form a site in which the two Trp side chains and the sulfur of Met336 interact (Fig. 8). The Trp46 and Trp50 side chains of the first helix faced toward Met336 in the final helix of the adjacent subunit, creating a putative pocket in which we manually positioned an ethanol molecule (Fig. 8A) or an IVM (Fig. 8B) molecule.
Fig. 8.
Molecular model of the rat P2X4R built by threading the edited primary sequence onto the X-ray crystal structure of zebrafish P2X4R. A, a side view of the rat P2X4R showing the ectodomain and the six α-helices of TM1 and TM2 segments of three different P2X4R subunits with a single ethanol molecule manually inserted at the interface. All residues of interest were rendered as space-filling surfaces; the ethanol molecule is colored pink to distinguish it. Residues Trp46 and Trp50 in the first α-helix of one subunit and Asp331 and Met336 in the final α-helix of the adjacent subunit form a pocket that demonstrates a good fit for a molecule of ethanol at the same scale. B, a similar view of the rat P2X4R, but with a model of IVM (rendered in balls and sticks) inserted into a putative binding site in a position between the α-helices such as that described in nicotinic acetylcholine receptors (Sattelle et al., 2009).
Discussion
The present study tested the hypotheses that IVM can affect ethanol sensitivity of P2X4Rs because of its action on sites located close to those for ethanol. Our findings support the hypotheses by demonstrating that IVM antagonizes the effects of ethanol in P2X4Rs. In addition, these studies provide evidence that IVM and ethanol both act on position 336 in the TM2 segment near the ectodomain. The roles of position 336 and other key residues in ethanol and IVM action are described in the context of a new molecular model of the rat P2X4R.
IVM alone, as previously reported (Khakh et al., 1999; Priel and Silberberg, 2004), produced a concentration-dependent increase in ATP-activated currents in P2X4Rs. The majority of previous studies indicate that IVM potentiation of agonist currents in P2X4Rs reflects allosteric modulation of the receptor (Khakh et al., 1999; Priel and Silberberg, 2004; Silberberg et al., 2007). However, one study suggested that the increase may result primarily from IVM-induced up-regulation of cell surface expression of the receptor (Toulmé et al., 2006). This contention was not supported by a subsequent study (Silberberg et al., 2007). In the present investigation, exposure to IVM and ATP for either 20 s or 2 min did not significantly change the surface expression levels of P2X4Rs in oocytes (Fig. 2). This lack of change indicates that effects of IVM on P2X4Rs in the present study reflect direct action of IVM on the receptor and cannot be explained by IVM-induced increases in cell surface expression.
The present findings are the first to show that IVM antagonizes the effects of ethanol. The degree of antagonism by IVM was inversely related to the ethanol concentration. The addition of 0.5 μM IVM completely eliminated the inhibitory effect of 25, 50, and 100 mM ethanol on ATP-activated currents without evidence of causing potentiation in P2X4Rs. An ethanol concentration of 25 mM is approximately 1.5 times the blood ethanol concentration (0.08%) that is considered to be a legally intoxicating dose in the United States. Therefore, these findings suggest that IVM has a potential application for antagonizing the effects of ethanol in humans.
The mechanism by which IVM antagonized ethanol in P2X4Rs is not known. The current findings provide some insight into this. First, IVM seemed to produce a nonparallel right shift in the ethanol concentration–response curve. The ability of higher concentrations of IVM to completely antagonize ethanol is consistent with a competitive mechanism of antagonism. However, the apparent absence of a parallel shift suggests that the interaction may be more complicated.
Second, it is notable that IVM alone potentiates ATP-induced currents. This introduces the possibility that ethanol antagonism by IVM results in the algebraic addition of the independent actions of IVM (potentiation) and ethanol (inhibition). This possibility is supported by the net potentiation observed when high concentrations of IVM were coapplied with ethanol. This possibility is also supported by prior work that found Zn2+, another positive allosteric modulator of P2X4Rs thought to act in the extracellular domain (Coddou et al., 2007), can eliminate the inhibitory effects of ethanol and that this interaction is additive (Davies et al., 2005). However, the experimentally obtained data with ethanol did not fit an additive model. Nonetheless, interpretation of these findings with ethanol is complicated by the opposing effects of ethanol and IVM on agonist action. Therefore, further studies were conducted with hexanol, an n-chain alcohol that was shown to potentiate ATP currents in P2X4Rs. This crossover in the effects of n-chain alcohols from inhibition to potentiation on ligand-gated ion channel function has been shown previously for nicotinic acetylcholine receptors (Borghese et al., 2002). In agreement with the ethanol findings, the combined effects of IVM and hexanol were significantly less than would be predicted based on independent interactions at different sites. These findings with IVM and hexanol eliminate the complications from opposing effects of ethanol and IVM and support the notion that IVM interferes with the actions of n-chain alcohols at a target in the TM segment of P2X4Rs.
Third, further studies tested the possibility that IVM antagonizes ethanol by interfering with its action on position 336 in the TM2 segment of P2X4Rs. This work demonstrated that mutations at position 336, which are known to alter the sensitivity of P2X4Rs to ethanol (Popova et al., 2010), also altered the receptor's sensitivity to IVM. These mutational studies support the notion that position 336 plays a role in the actions of both ethanol and IVM on P2X4Rs.
Moreover, the effects of these mutations provide insight into the physical–chemical requirements of the residue at position 336 for sensitivity to IVM and ethanol. In particular, substituting a positively charged bulky Arg or a negatively charged Glu at position 336 for the WT nonpolar, bulky Met made the receptor insensitive to IVM and ethanol. These findings suggest that a charge at position 336, regardless of its nature, interferes with the initiation or transduction of the actions of IVM and ethanol. This effect suggests that a hydrophilic side chain or moiety could be critical for entrance/docking to a site at or near position 336. In this scenario, highly polar, charged substitutions at position 336 (i.e., Arg or Glu) might offer the greatest degree of repulsion, thus accounting for the abolishment of the ethanol effect. It is also possible that charged residues interact with other neighboring residues, thus causing structural changes in the above indicated region that decrease the interaction of IVM and ethanol with the receptor.
The size of the substitutions at position 336 also seems to play a key role in determining sensitivity to both IVM and ethanol. Substituting small nonpolar amino acids (i.e., Ala, Val) for the bulky nonpolar Met markedly increased the receptor's sensitivity to IVM, but reduced its sensitivity to ethanol. If ethanol and/or IVM act on the receptor via complete or partial entry into a putative pocket that Met336 is a part of, then these small residues may induce changes in the size of this hypothetical pocket. This, in turn, may increase the accessibility and subsequent docking of the larger IVM molecule and thus explain the increased potentiation. On the other hand, the change in the size of the pocket could reduce the interaction with ethanol, leading to a decrease of ATP-gated currents. These findings add evidence for the importance of position 336 in the actions of both agents and demonstrate that varying the size of the residue at this position produces opposite effects on the magnitude of the receptor's response to these agents. Collectively, these finding support the notion that position 336 plays an important role in initiating and/or transducing the actions of IVM and ethanol on P2X4Rs.
To examine possible interactions between Met336 and other key residues thought to play a role in ethanol and/or IVM action (Jelínková et al., 2006, 2008; Popova et al., 2010), we built a homology model of the rat P2X4R by using the recently published 3.1-Å X-ray crystal structure of zebrafish P2X4R (Kawate et al., 2009). This model demonstrates that the Trp46 and Trp50 side chains of the first helix face Asp331 and Met336 in the final α-helix of the adjacent subunit. There is substantial literature about interactions of the antibonding orbitals of sulfur atoms with aromatic rings (Viguera and Serrano, 1995). In addition, interactions of the sulfur atom of methionine residues with multiple aromatic groups have been referred to as “hot spots” in protein–protein interactions (Ma and Nussinov, 2007). In the latter study, there are examples of multiple tryptophan or tyrosine rings oriented at right angles to each other that is “edge on.” In these conformations the positive partial atomic charges of the aromatic ring hydrogens can interact with the face of a neighboring tryptophan ring in a cation–π interaction (Pless et al., 2008). Manual rotations of the C-α to C-β bonds of Trp46 and Trp50 were sufficient to form a site in which the two Trp side chains and the sulfur of Met336 interact.
We then considered the possibility that an alcohol molecule could “bind” in this site by a combination of cation–π interactions (Pless et al., 2008) between the hydrogen of ethanol and the tryptophans and additional interactions between the methionine sulfur atom and the oxygen atom of ethanol. To visualize this possibility, we manually positioned an ethanol molecule in this site formed by Trp46, Trp50, Asp331, and Met336. This manipulation showed that ethanol fits nicely into this putative pocket. It is noteworthy that manual positioning indicated that IVM can also fit into the same pocket.
It is noteworthy that the location of the four residues forming the pocket is at the interface of the ectodomain and TM segments. All four of these residues are in the region in P2X4R that Kawate et al. (2009) suggested mediated transduction of binding energy between the ATP binding site and the constriction in the ion pore. Given previous studies indicating that this region of the P2X4R may play a role in ethanol action (Popova et al., 2010), these models illustrate how IVM binding in this region could antagonize ethanol by interfering with its ability to interact at a site within the putative pocket. Testing of these suggestions must await future more elaborate docking of ethanol and IVM molecules and long time scale molecular dynamics studies. Nonetheless, this model of P2X4Rs revealed that several amino acid residues known to be important for the effects of alcohol on these receptors, although seemingly widely scattered in the primary sequence of P2X4R, were actually clustered around a small cavity in the three-dimensional structure.
The present findings on rat P2X4Rs may have relevance to human P2X4Rs. This notion is supported by prior studies that found that the effects of IVM on human P2X4Rs are similar to those seen in the present study (Priel and Silberberg, 2004). Moreover, our preliminary findings suggest that the effects of ethanol on human P2X4Rs expressed in human embryonic kidney 293 cells parallel the current findings. Furthermore, the sequence of the TM2 segment of rat P2X4Rs is highly homologous with its human counterpart (Townsend-Nicholson et al., 1999). Collectively, these findings suggest that IVM will antagonize the effects of ethanol on P2X4Rs in humans.
In conclusion, the current studies suggest that IVM is a competitive antagonist of ethanol on P2X4Rs. Moreover, the findings and the new molecular model presented suggest that IVM acts by interfering with the actions of ethanol on a pocket at or near position 336 in the TM2 segment. Taken in the context with recent genomic evidence that P2X4Rs may play a role in modulating alcohol consumption in rats (Tabakoff et al., 2009), our studies suggest that the pocket comprised of position 336 and other key residues may represent a target for medication development for alcohol abuse. More than 18 million humans are treated with IVM each year via oral, topical, or subcutaneous injections (Burkhart, 2000). The wide use of IVM can be attributed to its therapeutic index and ease of administration (Davis et al., 1999; Burkhart, 2000). Central nervous system side effects have been reported in some patients over the years, but those incidences are very rare and seem to be linked to alterations of P-glycoprotein function at the blood-brain barrier (Edwards, 2003; Geyer et al., 2009). The findings of the present study and our recent preliminary evidence showing a significant reduction of alcohol consumption in IVM-treated mice suggest that IVM may be rapidly developed as a novel indication or improved on to help reduce alcohol consumption and abuse.
Acknowledgments
We thank Miriam Fine for conducting the molecular biology work and Amelie Nguyen and Mark Ambroso for providing technical assistance.
This work was supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism [Grants KO1-AA017243] (to L.A.), RO1-AA013890 (to D.L.D.), RO1-AA03972 (to R.L.A.), RO1-AA013378 (to J.R.T.)]; the National Institutes of Health Ruth L. Kirschstein National Research Service Awards [F31-AA017029 (to M.P.), F31-AA017569 (to D.I.P.)]; the Integrative Neurosciences Initiative on Alcoholism [Pilot Project AA013517] (to D.L.D.); and the University of Southern California School of Pharmacy.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.167908.
- P2XR
- purinergic P2X receptor
- IVM
- ivermectin
- TM
- transmembrane
- EtOH
- ethanol
- HexOH
- hexanol
- WT
- wild type
- GABAA
- γ-aminobutyric acid A.
References
- Asatryan L, Popova M, Woodward JJ, King BF, Alkana RL, Davies DL. (2008) Roles of ectodomain and transmembrane regions in ethanol and agonist action in purinergic P2X2 and P2X3 receptors. Neuropharmacology 55:835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Sadtler S, Niculescu C, Rettinger J, Schmalzing G. (2004) Trimeric architecture of homomeric P2X2 and heteromeric P2X1+2 receptor subtypes. J Mol Biol 342:333–343 [DOI] [PubMed] [Google Scholar]
- Borghese CM, Ali DN, Bleck V, Harris RA. (2002) Acetylcholine and alcohol sensitivity of neuronal nicotinic acetylcholine receptors: mutations in transmembrane domains. Alcohol Clin Exp Res 26:1764–1772 [DOI] [PubMed] [Google Scholar]
- Burkhart CN. (2000) Ivermectin: an assessment of its pharmacology, microbiology, and safety. Vet Hum Toxicol 42:30–35 [PubMed] [Google Scholar]
- Burnstock G. (2004) Introduction: P2 receptors. Curr Top Med Chem 4:793–803 [DOI] [PubMed] [Google Scholar]
- Chizh BA, Illes P. (2001) P2X receptors and nociception. Pharmacol Rev 53:553–568 [PubMed] [Google Scholar]
- Coddou C, Acuña-Castillo C, Bull P, Huidobro-Toro JP. (2007) Dissecting the facilitator and inhibitor allosteric metal sites of the P2X4 receptor channel: critical roles of CYS132 for zinc potentiation and ASP138 for copper inhibition. J Biol Chem 282:36879–36886 [DOI] [PubMed] [Google Scholar]
- Davies DL, Kochegarov AA, Kuo ST, Kulkarni AA, Woodward JJ, King BF, Alkana RL. (2005) Ethanol differentially affects ATP-gated P2X(3) and P2X(4) receptor subtypes expressed in Xenopus oocytes. Neuropharmacology 49:243–253 [DOI] [PubMed] [Google Scholar]
- Davies DL, Machu TK, Guo Y, Alkana RL. (2002) Ethanol sensitivity in ATP-gated P2X receptors is subunit dependent. Alcohol Clin Exp Res 26:773–778 [PubMed] [Google Scholar]
- Davis JA, Paylor R, McDonald MP, Libbey M, Ligler A, Bryant K, Crawley JN. (1999) Behavioral effects of ivermectin in mice. Lab Anim Sci 49:288–296 [PubMed] [Google Scholar]
- Dawson GR, Wafford KA, Smith A, Marshall GR, Bayley PJ, Schaeffer JM, Meinke PT, McKernan RM. (2000) Anticonvulsant and adverse effects of avermectin analogs in mice are mediated through the γ-aminobutyric acid A receptor. J Pharmacol Exp Ther 295:1051–1060 [PubMed] [Google Scholar]
- Dent JA, Davis MW, Avery L. (1997) avr-15 encodes a chloride channel subunit that mediates inhibitory glutamatergic neurotransmission and ivermectin sensitivity in Caenorhabditis elegans. EMBO J 16:5867–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards G. (2003) Ivermectin: does P-glycoprotein play a role in neurotoxicity? Filaria J 24(Suppl 1):S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary TG. (2005) Ivermectin 20 years on: maturation of a wonder drug. Trends Parasitol 21:530–532 [DOI] [PubMed] [Google Scholar]
- Geyer J, Gavrilova O, Petzinger E. (2009) Brain penetration of ivermectin and selamectin in mdr1a,b P-glycoprotein- and bcrp-deficient knockout mice. J Vet Pharmacol Ther 32:87–96 [DOI] [PubMed] [Google Scholar]
- Jelínkova I, Vávra V, Jindrichova M, Obsil T, Zemkova HW, Zemkova H, Stojilkovic SS. (2008) Identification of P2X(4) receptor transmembrane residues contributing to channel gating and interaction with ivermectin. Pflugers Arch 456:939–950 [DOI] [PubMed] [Google Scholar]
- Jelínková I, Yan Z, Liang Z, Moonat S, Teisinger J, Stojilkovic SS, Zemková H. (2006) Identification of P2X4 receptor-specific residues contributing to the ivermectin effects on channel deactivation. Biochem Biophys Res Commun 349:619–625 [DOI] [PubMed] [Google Scholar]
- Jiang LH, Kim M, Spelta V, Bo X, Surprenant A, North RA. (2003) Subunit arrangement in P2X receptors. J Neurosci 23:8903–8910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T, Michel JC, Birdsong WT, Gouaux E. (2009) Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature 460:592–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, Burnstock G, Kennedy C, King BF, North RA, Séguéla P, Voigt M, Humphrey PP. (2001) International union of pharmacology. XXIV. Current status of the nomenclature and properties of P2X receptors and their subunits. Pharmacol Rev 53:107–118 [PubMed] [Google Scholar]
- Khakh BS, Proctor WR, Dunwiddie TV, Labarca C, Lester HA. (1999) Allosteric control of gating and kinetics at P2X4 receptor channels. J Neurosci 19:7289–7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chang TH, Silberberg SD, Swartz KJ. (2008) Gating the pore of P2X receptor channels. Nat Neurosci 11:883–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Nussinov R. (2007) Trp/Met/Phe Hot spots in protein–protein interactions: potential targets in drug design. Curr Top Med Chem 7:999–1005 [DOI] [PubMed] [Google Scholar]
- North RA. (2002) Molecular physiology of P2X receptors. Physiol Rev 82:1013–1067 [DOI] [PubMed] [Google Scholar]
- Perkins DI, Trudell JR, Crawford DK, Asatryan L, Alkana RL, Davies DL. (2009) Loop 2 structure in glycine and GABA(A) receptors plays a key role in determining ethanol sensitivity. J Biol Chem 284:27304–27314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pless SA, Millen KS, Hanek AP, Lynch JW, Lester HA, Lummis SC, Dougherty DA. (2008) A cation–π interaction in the binding site of the glycine receptor is mediated by a phenylalanine residue. J Neurosci 28:10937–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova M, Asatryan L, Ostrovskaya O, Wyatt LR, Li K, Alkana RL, Davies DL. (2010) A point mutation in the ectodomain-transmembrane 2 interface eliminates the inhibitory effects of ethanol in P2X4 receptors. J Neurochem 112:307–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A, Silberberg SD. (2004) Mechanism of ivermectin facilitation of human P2X4 receptor channels. J Gen Physiol 123:281–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Lenoble D, Chandenier J, Gaxotte P. (2003) Ivermectin and filariasis. Fundam Clin Pharmacol 17:199–203 [DOI] [PubMed] [Google Scholar]
- Rubio ME, Soto F. (2001) Distinct localization of P2X receptors at excitatory postsynaptic specializations. J Neurosci 21:641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattelle DB, Buckingham SD, Akamatsu M, Matsuda K, Pienaar IS, Pienaar I, Jones AK, Sattelle BM, Almond A, Blundell CD. (2009) Comparative pharmacology and computational modeling yield insights into allosteric modulation of human α7 nicotinic acetylcholine receptors. Biochem Pharmacol 78:836–843 [DOI] [PubMed] [Google Scholar]
- Shan Q, Haddrill JL, Lynch JW. (2001) Ivermectin, an unconventional agonist of the glycine receptor chloride channel. J Biol Chem 276:12556–12564 [DOI] [PubMed] [Google Scholar]
- Silberberg SD, Li M, Swartz KJ. (2007) Ivermectin interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron 54:263–274 [DOI] [PubMed] [Google Scholar]
- Sung YF, Huang CT, Fan CK, Lin CH, Lin SP. (2009) Avermectin intoxication with coma, myoclonus, and polyneuropathy. Clin Toxicol (Phila) 47:686–688 [DOI] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, et al. (2009) Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol 7:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmé E, Soto F, Garret M, Boué-Grabot E. (2006) Functional properties of internalization-deficient P2X4 receptors reveal a novel mechanism of ligand-gated channel facilitation by ivermectin. Mol Pharmacol 69:576–587 [DOI] [PubMed] [Google Scholar]
- Townsend-Nicholson A, King BF, Wildman SS, Burnstock G. (1999) Molecular cloning, functional characterization, and possible cooperativity between the murine P2X4 and P2X4a receptors. Mol Brain Res 64:246–254 [DOI] [PubMed] [Google Scholar]
- Viguera AR, Serrano L. (1995) Side-chain interactions between sulfur-containing amino acids and phenylalanine in α-helices. Biochemistry 34:8771–8779 [DOI] [PubMed] [Google Scholar]
- Xiong K, Hu XQ, Stewart RR, Weight FF, Li C. (2005) The mechanism by which ethanol inhibits rat P2X4 receptors is altered by mutation of histidine 241. Br J Pharmacol 145:576–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong K, Li C, Weight FF. (2000) Inhibition by ethanol of rat P2X4 receptors expressed in Xenopus oocytes. Br J Pharmacol 130:1394–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]