Abstract
Allosteric modulation of neuronal nicotinic acetylcholine receptors (nAChRs) is considered to be one of the most promising approaches for therapeutics. We have previously reported on the pharmacological activity of several compounds that act as negative allosteric modulators (NAMs) of nAChRs. In the following studies, the effects of 30 NAMs from our small chemical library on both human α4β2 (Hα4β2) and human α3β4 (Hα3β4) nAChRs expressed in human embryonic kidney ts201 cells were investigated. During calcium accumulation assays, these NAMs inhibited nAChR activation with IC50 values ranging from 2.4 μM to more than 100 μM. Several NAMs showed relative selectivity for Hα4β2 nAChRs with IC50 values in the low micromolar range. A lead molecule, KAB-18, was identified that shows relative selectivity for Hα4β2 nAChRs. This molecule contains three phenyl rings, one piperidine ring, and one ester bond linkage. Structure–activity relationship (SAR) analyses of our data revealed three regions of KAB-18 that contribute to its relative selectivity. Predictive three-dimensional quantitative SAR (comparative molecular field analysis and comparative molecular similarity indices analysis) models were generated from these data, and a pharmacophore model was constructed to determine the chemical features that are important for biological activity. Using docking approaches and molecular dynamics on a Hα4β2 nAChR homology model, a binding mode for KAB-18 at the α/β subunit interface that corresponds to the predicted pharmacophore is described. This binding mode was supported by mutagenesis studies. In summary, these studies highlight the importance of SAR, computational, and molecular biology approaches for the design and synthesis of potent and selective antagonists targeting specific nAChR subtypes.
Introduction
Neuronal nicotinic acetylcholine receptors (nAChRs) are ligand-gated ion channels composed of five protein subunits encoded by a family of related, but distinct, genes (Anand et al., 1991; Cooper et al., 1991). Multiple nAChR subtypes have been described based on subunit (α2–α10 and β2–β4) composition; the three most prominent native neuronal nAChR subtypes are α4β2, α3β4, and α7 (Lukas et al., 1999). nAChRs mediate fast synaptic transmission and modulate the release of several neurotransmitters (e.g., norepinephrine, serotonin, GABA, dopamine) and neurohormones (e.g., epinephrine) in many parts of the vertebrate central and peripheral nervous systems. nAChRs are associated with many physiologically important mechanisms (e.g., cognition, arousal, pain sensation, addiction) and a number of neurological diseases (e.g., depression, schizophrenia, Alzheimer's disease, Tourette's syndrome, and autism) (Lloyd and Williams, 2000; Gotti et al., 2006). However, the specific nAChR subtypes involved with most of these physiological processes or diseases are not known.
The discovery of small molecules that target specific subtypes of nAChRs would contribute significantly to our understanding of the role specific nAChR subtypes play in normal and pathophysiological states and hold clinical promise for the treatment of several neuropathologies. Despite their obvious importance, the discovery of new drugs targeting specific nAChR subtypes has been slow. The focus of most nAChR drug discovery programs is on drugs that target orthosteric sites. One difficulty with this approach is the high degree of amino acid sequence homology among nAChR α and β subunits in the ligand binding domain for acetylcholine, making it difficult to develop drugs that specifically target nAChR subtypes. Thus, the selectivity of most nAChR drugs directed at orthosteric sites is modest at best. Looking for drugs targeting “nonorthosteric” sites of nAChRs (e.g., allosteric, noncompetitive sites) may be a more promising approach.
Allosteric binding sites are distinct from the orthosteric sites, allowing allosteric and orthosteric agents to bind simultaneously. Allosteric sites typically are defined by drugs that bind to the receptor (at nonorthosteric sites) and modulate function. Moaddel et al. (2007) described several noncompetitive, negative allosteric sites on nAChRs: central luminal sites, the ethidium binding site, and the quinacrine binding site. Several drugs bind the internal lumen of the nAChR-associated ion channel. These drugs include histrionicotoxin, phencyclidine, and mecamylamine and are often referred to as channel blockers (Changeux et al., 1986; Gallagher et al., 2001; Moaddel et al., 2007). The ethidium binding site, which also binds chlorpromazine and clozapine, was characterized through photoaffinity labeling and localized at the extracellular portion of the receptor, 46 Å above the transmembrane region (Pratt et al., 2000). The quinacrine binding site was identified within the transmembrane domain, 7 to 12 Å below the extracellular transmembrane domain interface (Arias, 1998).
We have discovered a class of nAChR NAMs that target a novel negative allosteric site on nAChRs (McKay et al., 2007; González-Cestari et al., 2009). Using our small chemical library of structurally similar NAMs, the present study describes structure–activity relationship (SAR) analyses on recombinant human α4β2 (Hα4β2) and recombinant human α3β4 (Hα3β4) nAChRs. 3D-QSAR [comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA)] models were generated from these data, and a pharmacophore was constructed to determine the chemical features that are important for biological activity. Using docking approaches and molecular dynamics (MD) on a homology model for the Hα4β2 nAChR, a binding mode for our lead molecule, KAB-18, at the α/β interface is described. Site-directed mutagenesis studies were designed to validate the existence of this site on Hα4β2 nAChRs. Overall, these studies present a novel class of NAMs that show relative selectivity for Hα4β2 nAChRs. In addition, these data present the novel site of interaction for this class of NAMs on Hα4β2 nAChRs.
Materials and Methods
Materials.
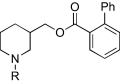
Fluo-4-acetoxymethyl ester (Fluo-4-AM), probenecid, and Pluronic F-127 were obtained from Molecular Probes (Eugene, OR). Dulbecco's modified Eagle medium, penicillin, streptomycin, and l-glutamine were obtained from Invitrogen (Carlsbad, CA). nAChR agonists and antagonists (Table 1) were purchased from Sigma-Aldrich (St. Louis, MO) with the exception of KAB-18. All other reagents were purchased from Thermo Fisher Scientific (Pittsburgh, PA). In general, molecules (Tables 2, 3, 4, and 5) were prepared by reaction of hydroxymethyl piperidine with the appropriate alkyl halide to provide the N-alkyl hydroxymethyl piperidine as reported previously (Bergmeier et al., 1999, 2004; Bryant et al., 2002; Huang et al., 2008). This molecule was then coupled to the appropriate carboxylic acid to provide the target molecule (Bergmeier et al., 2004). All molecules were >98% pure as shown by 1H NMR, 13C NMR, and high-resolution mass spectrometry. All compounds were converted to their hydrochloride or oxalate salts. For pharmacological evaluation, because of solubility problems all compounds were initially dissolved in 100% DMSO (0.01 M stocks). Further dilutions of compounds were made in double distilled H2O (≤100 μM).
TABLE 1.
Agonist and antagonist study on human nAChRs
| Hα4β2 nAChRs |
Hα3β4 nAChRs |
|||
|---|---|---|---|---|
| EC50 or IC50 Valuea | nH | EC50 or IC50 Valuea | nH | |
| Epibatidine (EC50) | 18.1 (6.5–50.6) nM | 0.6 | 37.4 (10.7–71.4) nM | 0.9 |
| Nicotine (EC50) | 8.9 (5.7–13.9) μM | 1.2 | 9.1 (4.5–18.6) μM | 0.9 |
| d-tubocurarine (IC50) | 2.3 (1.9–3.9) μM | −0.7 | 1.6 (0.4–3.5) μM | −0.9 |
| Mecamylamine (IC50) | 0.5 (0.3–0.8) μM | −0.9 | 0.2 (0.1–0.3) μM | −0.8 |
| KAB-18 (IC50) | 13.5 (9.7–18.5) μM | −0.7 | NA | |
nH, Hill coefficient; NA, no activity at concentrations up to 100 μM.
Values represent geometric means (confidence limits), n = 4–8.
TABLE 2.
Region 1 SAR studies
 |
R | Hα4β2 nAChRs |
Hα3β4 nAChRs |
||
|---|---|---|---|---|---|
| IC50 Value (μM)a | nH | IC50 Value (μM)a | nH | ||
| KAB-18 | 13.5 (9.7–18.5) | −0.7 | NA | ||
| COB-1 | −H | 7.1 (5.4–9.4) | −1.1 | 8.1 (4.2–15.7) | −0.7 |
| COB-3 | −(CH3)2 | 2.4 (1.6–3.8) | −1.0 | 6.4 (4.4–9.3) | −0.7 |
| COB-2 | −CH3 | 6.9 (4.2–11.3) | −1.0 | 4.8 (2.8–8.3) | −1.2 |
| DDR-15 | 4.3 (2.7–7.0) | −0.7 | 1.8 (0.7–5.1) | −1.1 | |
| DDR-13 | 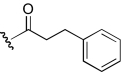 |
5.7 (3.2–10.3) | −0.4 | NA | |
| DDR-18 | 6.4 (3.7–11.1) | −1.0 | NA | ||
| APB-8 | 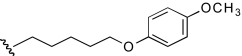 |
7.6 (2.0–29.0) | −0.8 | 22.4 (11.8–42.4) | −0.7 |
nH, Hill coefficient; NA, no activity at concentrations up to 100 μM.
Values represent geometric means (confidence limits), n = 4–8.
TABLE 3.
Region 2 SAR studies
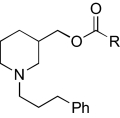 |
R | Hα4β2 nAChRs |
Hα3β4 nAChRs |
||
|---|---|---|---|---|---|
| IC50 Value (μM)a | nH | IC50 Value (μM)a | nH | ||
| KAB-18 |  |
13.5 (9.7–18.5) | −0.7 | NA | |
| DDR-1 | 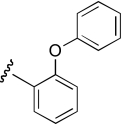 |
25.5 (17.2–37.7) | −1.1 | 13.2 (6.4–27.3) | −0.6 |
| DDR-2 | 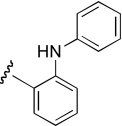 |
10.6 (3.5–32.1) | −0.7 | 19.5 (6.7–56.9) | −0.6 |
| KAB-21 | 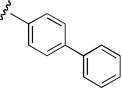 |
6.3 (1.9–20.9) | −0.9 | 18.5 (7.3–46.2) | −0.7 |
| DDR-19 | 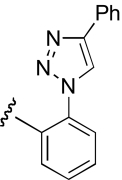 |
2.8 (1.3–6.1) | −0.9 | 4.3 (1.6–11.4) | −0.9 |
| KAB-24 | 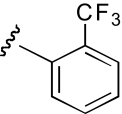 |
8.0 (4.2–15.3) | −1.2 | 5.5 (1.7–17.4) | −0.9 |
| KAB-22 | 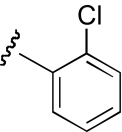 |
9.4 (5.5–16.1) | −1.3 | 23.1 (11.1–48.2) | −1.2 |
| KAB-23 | 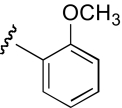 |
7.9 (4.9–12.7) | −0.7 | 12.1 (8.1–17.9) | −0.9 |
| KAB-19 | 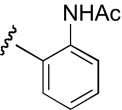 |
5.5 (2.0–15.2) | −1.1 | 4.3 (1.0–17.7) | −0.9 |
| KAB-20 | 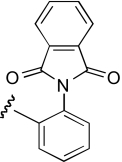 |
6.0 (4.4–8.2) | −1.5 | 5.9 (4.1–8.4) | −0.8 |
| KAB-30 | 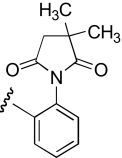 |
4.9 (2.5–9.5) | −1.5 | 5.0 (3.0–8.3) | −1.3 |
| IB-2 | 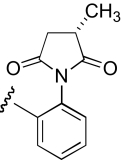 |
10.5 (7.4–14.8) | −0.8 | 10.9 (5.1–23.3) | −0.9 |
nH, Hill coefficient; NA, no activity at concentrations up to 100 μM.
Values represent geometric means (confidence limits), n = 4–8.
TABLE 4.
Region 3 SAR studies
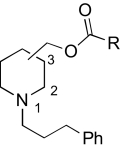 |
Piperidine Substitution | R | Hα4β2 nAChRs |
Hα3β4 nAChRs |
||
|---|---|---|---|---|---|---|
| IC50 Value (μM)a | nH | IC50 Value (μM)a | nH | |||
| KAB-18 | 3-position, racemic | 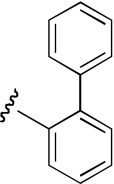 |
13.5 (9.7–18.5) | −0.7 | NA | |
| PPB-12 | 3-position, (S)-enantiomer | 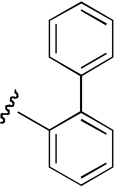 |
17.5 (10.9–28.3) | −1.0 | NA | |
| PPB-13 | 3-position, (R)-enantiomer | 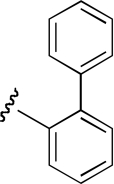 |
9.8 (4.4–21.6) | −0.9 | NA | |
| IB-2 | 3-position, (S)-enantiomer | 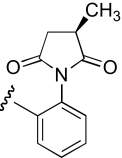 |
10.5 (7.2–14.8) | −0.8 | 10.9 (5.1–23.3) | −0.9 |
| IB-4 | 3-position, (R)-enantiomer | 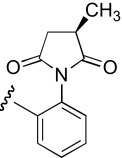 |
9.0 (4.3–14.0) | −0.9 | 9.5 (5.0–17.7) | −0.9 |
| COB-4 | 4-position, racemic |  |
8.1 (2.1–30.7) | −0.8 | 10.5 (7.6–14.4) | −0.9 |
nH, Hill coefficient; NA, no activity at concentrations up to 100 μM.
Values represent geometric means (confidence limits), n = 4–8.
TABLE 5.
Region 4 SAR studies
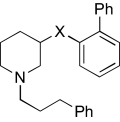 |
X | Hα4β2 nAChRs |
Hα3β4 nAChRs |
||
|---|---|---|---|---|---|
| IC50 Value (μM)a | nH | IC50 Value (μM)a | nH | ||
| KAB-18 |  |
13.5 (9.7–18.5) | −0.7 | NAc | |
| DDR-5 | 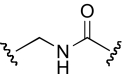 |
19.8 (8.3–47.1) | −0.8 | NAc | |
| DDR-3 | 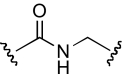 |
16.6 (9.7–28.6) | −0.9 | 14.5 (4.9–43.1) | −0.7 |
| JHB-9 | 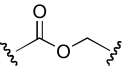 |
17.6 (11.6–26.8) | −0.9 | 10.2 (3.4–30.5) | −0.9 |
| APB-21 | 12.6 (3.0–53.8) | −0.8 | 15.5 (8.4–29.0) | −0.9 | |
| DDR-14 | 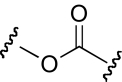 |
6.4 (4.1–10.4) | −1.1 | 2.7 (0.4–17.9) | −1.2 |
nH, Hill coefficient; NA, no activity at concentrations up to 100 μM.
Values represent geometric means (confidence limits), n = 4–8.
Calcium Accumulation Assays.
A previously reported procedure that has been used for rat α3β4 nAChRs stably expressed in HEK ts201 cells has been adapted for this work (McKay et al., 2007; González-Cestari et al., 2009). For the calcium accumulation assays, HEK ts201 cells stably expressing either Hα4β2 or Hα3β4 nAChRs (obtained from Professor Jon Lindstrom, University of Pennsylvania, Philadelphia, PA) were used. These Hα4β2 stably transfected HEK cells express predominantly low-sensitivity (α4)3(β2)2 nAChRs (Nelson et al., 2003). Cells were plated at a density of 2.0 to 2.3 × 105 cells per well in clear 96-well culture plates coated with poly-l-ornithine. Plated cells were incubated at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 10 mM l-glutamine, 0.7 mg/ml G418 (Geneticin), 100 units/ml penicillin, 100 μg/ml streptomycin, and 100 μg/ml zeocin. Cells were used for experiments at ∼100% confluence, typically 24 to 48 h after plating. On the day of the experiment, cells were washed with 100 μl of HEPES-buffered Krebs (HBK) solution (155 mM NaCl, 4.6 mM KCl, 1.2 mM MgSO4, 1.8 mM CaCl2, 6 mM glucose, and 20 mM HEPES, pH 7.4) and incubated (protected from light) for 30 min at 37°C followed by 30 min at 24°C with 40 μl of HBK containing 2 μM fluo-4-AM solution, 2.5 mM probenecid, and 0.05% Pluronic F127. Fluo-4-AM and Pluronic F127 were dissolved in DMSO (100 and 20% w/v, respectively), resulting in a final DMSO concentration of <0.1%. After loading the cells with fluo-4-AM, the cells were washed (1×), and 80 μl of HBK was added to each well. The plates were then placed into a fluid handling integrated fluorescence plate reader (FlexStation; Molecular Devices, Sunnyvale, CA). Fluo-4 fluorescence was read at an excitation of 494 nm and an emission of 520 nm from the bottom of the plate, and changes in fluorescence were monitored at ∼1.5-s intervals. Probenecid (2.5 mM) was included in all of the solutions once the cells were loaded with fluo-4 to prevent its leakage from the cells. The experimental design involved three treatment periods (20, 40, and 60 s) and three treatment groups (control-sham treated, control-agonist treated, and antagonist treated). All groups were initially incubated in HBK, and fluorescence was monitored for 20 s. For the control-agonist treated groups, HBK (40 μl) was then added, and the fluorescence was monitored for 40 s, followed by the addition of epibatidine (40 μl of a 12 μM solution) to achieve a final concentration of 3 μM. The effects after agonist addition were monitored for 60 s. For the antagonist-treated groups, the antagonist (40 μl of a 3× solution) was added, and fluorescence was monitored for 40 s, followed by the addition of epibatidine with the antagonist (at 60 s). The control-sham groups were treated with HBK during each of the treatment periods with fluorescence being monitored continuously to establish resting (background) levels of cell fluorescence. Functional responses were quantified by first calculating the net fluorescence changes (the difference between control sham-treated and control agonist-treated groups). Net peak (maximum) fluorescence values during the third treatment period for both the control-agonist treatment group and the antagonist (with agonist) treatment group were determined. Results were expressed as a percentage of control-agonist groups. Because of solubility problems, compound concentrations more than 100 μM were not used in our concentration–response studies. The DMSO concentration at this compound concentration was 1% and had no effect on basal or agonist-induced increases in fluorescence intensity.
GASP.
GASP (Genetic Algorithm Similarity Program) is a toolset of the molecular modeling program SYBYL designed for the construction of pharmacophore models. GASP uses an alignment of molecules to develop pharmacophores by using similarity constraints to optimize the orientation of molecule sets. For these alignments, the population size was set to 125, and the allele mutate weight was set to 96. The fitness increment was set to 0.02. Alignments for each set of molecules were repeated for 10 runs, and the optimal model was selected based on visual inspection. For generation of the pharmacophore model, four molecules (KAB-18, DDR-13, DDR-18, and DDR-5) were selected based on their relative selectivity for α4β2 nAChRs.
CoMFA.
CoMFA is a toolset of SYBYL used to evaluate 3D-QSAR. In this approach, evaluation of electrostatic and steric interactions is correlated to biological activity for the determination of important structural features within sets of molecules. For these studies, a cutoff value of 30 kcal/mol was used. Interaction fields were built on a 2.0-Å spaced lattice and continued beyond each structure by 4.0 Å in all directions. Of the molecules used, six were selected to construct models and be used as a training set to correlate predicted and experimentally derived IC50 values (see Table 7). For the test set, 11 molecules were randomly selected (see Table 8) outside of the training set. Model statistics were generated as described previously (McKay et al., 2007) where the dependent variables (biological descriptors) were IC50 values generated by using the calcium accumulation assay with Hα4β2 nAChRs.
TABLE 7.
Comparison of predicted and experimentally derived IC50 values of training set compounds
| CoMFA Log IC50 Values | CoMSIA Log IC50 Values | Hα4β2 nAChRs Log IC50 Valuesa | |
|---|---|---|---|
| M | |||
| KAB-18 | −4.94 | −4.94 | −4.87 |
| DDR-13 | −5.24 | −5.24 | −5.24 |
| DDR-18 | −5.20 | −5.20 | −5.19 |
| DDR-5 | −4.70 | −4.70 | −4.70 |
| KAB-18(R) | −4.94 | −4.94 | −5.00 |
| KAB-18(S) | −4.76 | −4.76 | −4.76 |
TABLE 8.
Comparison of predicted and experimentally derived IC50 values of analogs for test set compounds
| CoMFA Log IC50 Values | CoMSIA Log IC50 Values | Hα4β2 nAChRs Log IC50 Valuesa | |
|---|---|---|---|
| M | |||
| COB-1 | −5.03 | −5.03 | −5.14 |
| COB-2 | −5.02 | −5.02 | −5.15 |
| APB-8 | −5.01 | −5.00 | −5.10 |
| IB-2 | −4.99 | −5.00 | −4.98 |
| KAB-23 | −5.04 | −5.02 | −5.10 |
| DDR-3 | −4.55 | −4.72 | −4.78 |
| APB-21 | −4.97 | −4.90 | −4.90 |
| KAB-24 | −5.00 | −5.08 | −5.10 |
| DDR-2 | −5.02 | −4.93 | −4.98 |
| JHB-9 | −4.90 | −4.98 | −4.99 |
| KAB-22 | −5.04 | −5.01 | −5.03 |
CoMSIA.
CoMSIA is a toolset of SYBYL similar to CoMFA. CoMSIA uses Gaussian functions as opposed to Lennard-Jones potential and Coulombic potentials. The same training set and test set molecules used in the CoMFA model were selected for the CoMSIA model.
Docking and Molecular Dynamics.
KAB-18 was docked to a homology model of the Hα4β2 LBD to determine a precise binding mode. The model was initially built with the MODELLER software in the unbound state as reported earlier (González-Cestari et al., 2009). Before docking KAB-18, epibatidine was docked to the agonist binding site so that the noncompetitive binding mode of KAB-18 could be determined. Dockings were performed with the Lamarckian genetic algorithm as implemented in AutoDock 4.0 (Morris et al., 1998). Each docking job consisted of 100 individual runs in which each run used a population size of 150 with a cutoff of 100,000,000 energy evaluations. The final docking poses were clustered with an all-atom root mean square deviation of 2 Å. MD simulations of some docking modes were evaluated to determine the stability of the ligand in the docked position. MD simulations were performed with the Amber suite of programs (Case et al., 2005). Simulation protocol involved first solvating the entire extracellular domain complex and a TIP3P water model octahedron with a 15-Å buffer around all edges, adding counterions, then energy-minimizing the system. After energy minimization, the system was simulated for 200 ps during which all protein and ligand atoms were restrained with a 50 kcal/mol harmonic potential to allow the water molecules to equilibrate around the surface of the receptor. During this equilibration stage, the temperature was increased from 0 to 300 K. After equilibration, the system was simulated at a constant 300 K with a heat bath coupling constant of 2.0 ps. All simulations were performed at a constant 1 atm with a pressure relaxation time of 2.0 ps. Nonbonded interaction calculations were cut off at 8 Å, and the electrostatic energy was computed by using the Particle Mesh Ewald method.
Site-Directed Mutagenesis and Transient Expression of α4β2 nAChRs.
Human nAChR α4 and β2 full-length cDNAs in the vector pSP64 (poly A) were obtained from Prof. Jon Lindstrom and used as the templates for mutagenesis. A single mutation was made in both the α4 subunit and the β2 subunit by using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Stratagene) following the manufacturer's instructions. Primers were designed by using the QuikChange Primer Design Program (Stratagene) and Oligo 4.0 (National Biosciences, Plymouth, MN). Primers were synthesized by Invitrogen. Primers were designed to replace the α4 amino acid with one in a similar position in the human α3 subunit, whereas the β2 sequence was changed to an amino acid in a similar position in the human β4 subunit. The following complementary primers were designed to change the arginine (AGG) at position 187 in the α4 subunit to isoleucine (ATA): α4 mutant (+), 5′-GGGCACCTACAACACCATAAAGTACGAGTGCTGTGC-3′ and α4 mutant (−), 5′-GCACAGCACTCGTACTTTATGGTGTTGTAGGTGCCC-3′. The following primer was designed to change the threonine (ACC) at position 58 in the β2 subunit to lysine (AAG): β2 mutant, 5′-CCACCAATGTCTGGCTGAAGCAGGAGTGGGAAGATTATCG-3′. The underlined nucleotides defined the mutations. The mutant α4 cDNA was then subcloned into the eukaryotic expression vector pcDNA 3.1+ (Invitrogen), whereas the mutant β2 cDNA was subcloned into pcDNA 3.1+Zeo (Invitrogen). The wild-type human α4 and β2 cDNAs were also subcloned into the pcDNA 3.1+ and pcDNA 3.1+ Zeo vectors, respectively. All cDNA clones were completely sequenced with a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) at the Ohio State University Plant-Microbe Genomics Facility. DNAs used for transfection were purified by using PureLink High Pure Mini or Midi Kits (Invitrogen). HEK ts201 cells (kind gift of Dr. Rene Anand, Ohio State University, Columbus, OH) were transiently transfected with mutant Hα4β2 and wild-type Hα4β2 cDNAs by using Lipofectamine 2000 (Invitrogen) in 60-mm dishes. After 8 h, the cells were plated into clear 96-well plates at a density of 2.6 × 105 cells per well for the intracellular calcium accumulation assays. The net peak (maximum) fluorescence values obtained with these transiently transfected cell lines (wild-type and mutant) were identical and ∼77% of the net peak fluorescence values of the stably transfected cell line.
Calculations and Statistics.
Functional data were calculated from the number of observations performed in triplicate. Curve fitting was performed with Prism software (GraphPad Software, Inc., San Diego, CA) using the equation for a single-site sigmoidal dose–response curve with a variable slope. Linear regression analyses were performed with Prism with the level of significance set at p < 0.05. EC50 and IC50 values are expressed as geometric means (95% confidence limits). In Table 9, when statistical analyses were performed, the t test was used.
TABLE 9.
Effects of agonists and antagonists on wild-type and mutant human α4β2 nAChRs
Values represent geometric means (confidence limits), n = 5–7.
| Wild-Type Hα4β2 nAChRs |
Mutant Hα4β2 nAChRs |
|||
|---|---|---|---|---|
| EC50 or IC50 Values | nH | EC50 or IC50 Values | nH | |
| Epibatidine (EC50) | 36.8 (25.4–53.4) nM | 0.9 | 34.7 (21.4–56.2) nM | 1.1 |
| d-tubocurarine (IC50) | 5.5 (3.4–9.0) μM | −1.0 | 7.6 (5.6–10.5) μM | −0.8 |
| Mecamylamine (IC50) | 0.2 (0.1–0.3) μM | −1.4 | 0.2 (0.1–0.4) μM | −1.1 |
| KAB-18 (IC50) | 10.0 (5.5–18.0) μM | −1.2 | 110 (80.4–150) μMa | −0.6 |
| KAB-24 (IC50) | 5.9 (3.8–9.2) μM | −1.1 | 9.7 (5.4–17.5) μM | −1.2 |
nH, Hill coefficient.
Significant difference, P < 0.0001.
Results
Lead Molecule.
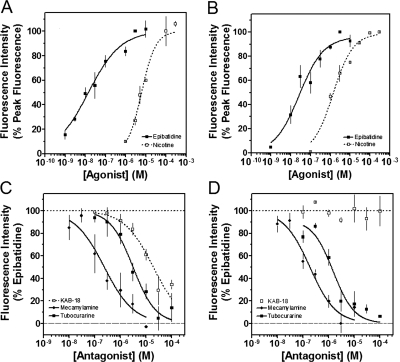
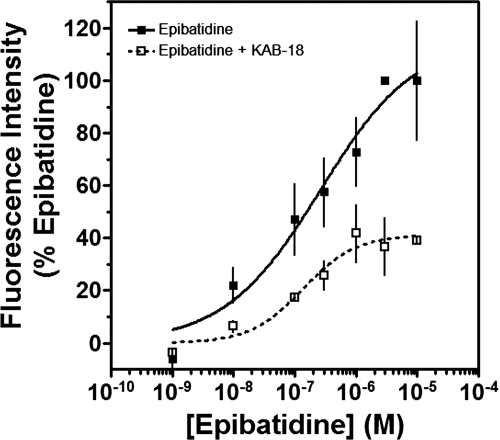
We had previously investigated the effects of novel NAMs on native bovine α3β4* nAChRs [the asterisk indicating the possible presence of additional subunits (Lukas et al., 1999)] and recombinant rat α3β4 nAChRs (McKay et al., 2007; González-Cestari et al., 2009). In the present studies, cell lines expressing either Hα4β2 or Hα3β4 nAChRs were used. The concentration-dependent responses to agonists (epibatidine and nicotine) and antagonists (mecamylamine and tubocurarine) are shown in Fig. 1 and Table 1. The EC50 values for epibatidine and nicotine on Hα4β2 nAChRs were 18.1 nM and 8.9 μM, respectively. The IC50 values for mecamylamine and tubocurarine on Hα4β2 nAChRs were 0.5 and 2.3 μM, respectively. The EC50 values for epibatidine and nicotine on Hα3β4 nAChRs were 37.4 nM and 9.1 μM, respectively. The IC50 values for mecamylamine and tubocurarine on Hα3β4 nAChRs were 0.2 and 1.6 μM, respectively. The pharmacology of these agonists and antagonists agree well with what has been reported previously (Wang et al., 1996; Olale et al., 1997; Stauderman et al., 1998; Nelson et al., 2001, 2003). Unlike these agonists and antagonists, our lead NAM, KAB-18, showed relative selectivity for Hα4β2 nAChRs. KAB-18 inhibited activation of Hα4β2 nAChRs (IC50 value 13.5 μM) and had no inhibitory activity on Hα3β4 nAChRs at concentrations up to 100 μM (Table 1; Fig. 1, C and D). The noncompetitive nature of this inhibitory activity is shown in Fig. 2; in the presence of KAB-18 at 30 μM (∼IC60), inhibition was not surmountable with increasing concentrations of epibatidine.
Fig. 1.
Concentration–response effects of agonists, antagonists, and KAB-18 on Hα4β2 and Hα3β4 nAChRs. A and B, the concentration–response effects of nicotine (□) and epibatidine (■) were investigated by using HEK ts201 cells expressing Hα4β2 and Hα3β4 nAChRs. Data are expressed as a percentage of peak fluorescence responses for either 3 μM epibatidine (epibatidine curves) or 100 μM nicotine (nicotine curves). C and D, tubocurarine (■), mecamylamine (♦), and KAB-18 (□) were investigated by using HEK ts201 cells expressing Hα4β2 and Hα3β4 nAChRs. Epibatidine (3 μM) was used as the agonist, and data are expressed as a percentage of peak fluorescence responses for 3 μM epibatidine. The dotted and dashed horizontal lines show the 100% control response and 100% inhibition, respectively. Values represent means ± S.E.M.s (n = 4–10).
Fig. 2.
Concentration–response effects of epibatidine in the absence or presence of KAB-18. The concentration-response effects of epibatidine were investigated in the absence (■) or presence (□) of 30 μM KAB-18 by using HEK ts201 cells expressing Hα4β2 nAChRs. Values represent means ± SEMs (n = 4).
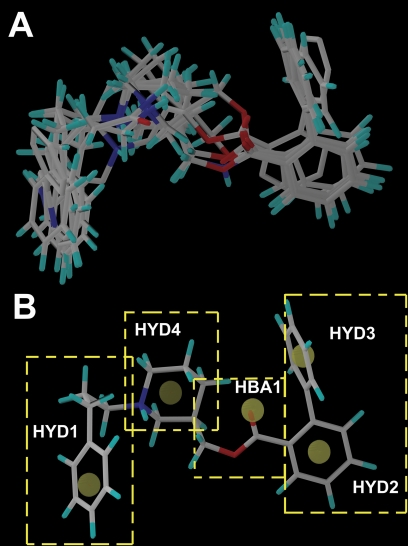
NAM Alignment and Pharmacophore.
Figure 3 illustrates the GASP alignment of the NAMs used for the model generation in these studies. The GASP-generated pharmacophore model (Fig. 3) featured four hydrophobic regions (HYD1, HYD2, HYD3, and HYD4) and one hydrogen bond acceptor region (HBA1). Using the information generated by our pharmacophore model, four regions were delineated to facilitate our SAR studies. Region 1 was defined as the substitution on the nitrogen moiety of the piperidine ring containing hydrophobic domain 1 (HYD1). Region 2 was defined as the ester acyl substitution containing HYD2 and HYD3. Region 3 was the piperidine ring that has been defined in the pharmacophore as the fourth hydrophobic region (HYD4). Region 4 was the linkage between regions 2 and 3, containing an ester bond with a hydrogen bond-accepting domain (HBA1).
Fig. 3.
Alignment of the molecules and the proposed pharmacophore targeting the negative allosteric site of the Hα4β2 nAChR. A, the alignment of the NAMs used in model generation is shown in capped stick representation. B, the features of the pharmacophore model generated using GASP are illustrated using KAB-18. Four hydrophobic features (HYD1, HYD2, HYD3, and HYD4) and a hydrogen bond acceptor (HBA1) feature of the pharmacophore are marked. The yellow dashed boxes outline the four regions used in the SAR studies.
Region 1 SAR.
In this first series of experiments, the functional effects of several compounds with chemical modifications in region 1 were investigated and compared with our lead molecule, KAB-18 (Table 2). Molecules with hydrogen, methyl, or dimethyl substitutions (COB-1, COB-2, and COB-3, respectively) retained inhibitory activity on Hα4β2 nAChRs (IC50 values 7.1, 6.9, and 2.4 μM, respectively) with small increases in potencies for COB-2 and COB-3. These modifications associated with COB-1, COB-2, and COB-3 resulted in a gain of activity on Hα3β4 nAChRs (IC50 values 8.1, 4.8, and 6.4 μM, respectively). A carbonyl substitution on the α carbon of KAB-18 (DDR-13) in region 1 resulted in an increase in potency on Hα4β2 nAChRs and no change in relative selectivity for Hα4β2 nAChRs (i.e., no inhibitory activity on Hα3β4). The azidopropyl derivative, DDR-15, was more potent than KAB-18 on Hα4β2 nAChRs but also inhibited activation of Hα3β4 nAChRs. Replacement of the phenyl group of KAB-18 with a phenyl-substituted triazole (DDR-18) resulted in a slight increase in potency (IC50 value, 6.4 μM), with no change in relative selectivity for Hα4β2 nAChRs. Elongation of the carbon chain of region 1 and addition of a methoxy group to the para position of the phenyl ring (APB-8) did not affect potency on Hα4β2 nAChRs; however, this modification resulted in a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 22.4 μM).
Region 2 SAR.
The functional effects of molecules with chemical modifications to the ester acyl component in region 2 were investigated and compared with our lead molecule, KAB-18 (Table 3). Addition of an oxygen molecule between the phenyl rings (DDR-1) resulted in a decrease in potency on Hα4β2 nAChRs and introduced inhibitory activity on Hα3β4 nAChRs (IC50 value 13.2 μM). Replacement of the ether oxygen of DDR-1 with an amine (DDR-2) resulted in a slight decrease in potency for Hα4β2 nAChRs and a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 19.5 μM). Moving the 2-phenyl substituent of KAB-18 to the 4-position (KAB-21) improved potency on Hα4β2 nAChRs and introduced inhibitory activity on Hα3β4 nAChRs (IC50 value 18.5 μM). Addition of a triazole ring between the two phenyls (DDR-19) resulted in a 5-fold increase in potency for the Hα4β2 nAChR (IC50 value 2.8 μM) but resulted in a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 4.3 μM). Replacement of the phenyl with a chloride (KAB-22), methoxy (KAB-23), acetamide (KAB-19), or trifluoromethyl group (KAB-24) did not affect potency for Hα4β2 nAChRs; however, these modifications resulted in a gain of inhibitory activity on Hα3β4 nAChRs (IC50 values of 23.1, 12.1, 4.3, and 5.5 μM, respectively).
Region 3 SAR.
The functional effects of compounds with chemical modifications in region 3 were investigated and compared with our lead molecule, KAB-18 (Table 4). Enantiomerically pure analogs of KAB-18 (S)-enantiomer and (R)-enantiomer showed no significant change in potency or relative selectivity for Hα4β2 nAChRs compared with KAB-18. Enantiomers of succinamide-containing compounds (IB-2 and IB-4) also showed no change in potency for either nAChR subtype (Table 4) compared with KAB-18. Moving the 3-hydroxymethyl group to the 4-position (COB-4) produced a slight increase in potency on Hα4β2 nAChRs and a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 10.5 μM).
Region 4 SAR.
The functional effects of molecules with chemical modifications in region 4 were investigated and compared with our lead molecule, KAB-18 (Table 5). The reverse ester (JHB-9) did not affect potency for Hα4β2 nAChR; however, this modification produced a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 10.2 μM). Replacement of the ester linkage with an amide linkage (DDR-5) did not affect potency on Hα4β2 nAChRs and maintained relative selectivity. The reverse ester of DDR-5 (DDR-3) did not affect potency for Hα4β2 nAChR; however, this modification produced a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 14.5 μM). Shortening the linker of region 4 by using hydroxypiperidine instead of hydroxymethylpiperidine (DDR-14) produced a 2-fold increase in potency for Hα4β2 nAChRs and a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 2.7 μM). Modification of the ester linkage to an ether linkage (APB-21) produced a slight decrease in potency for the Hα4β2 nAChR and resulted in a gain of inhibitory activity on Hα3β4 nAChRs (IC50 value 15.5 μM).
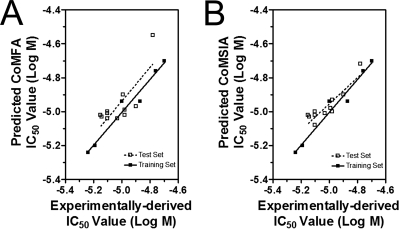
Computational Modeling.
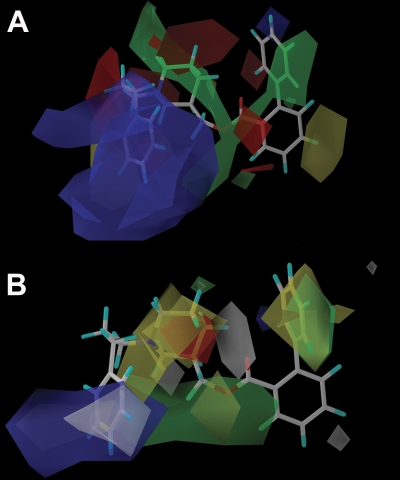
NAMs were aligned by using GASP-guided subgroup alignment (Fig. 3). Subsequent 3D-QSAR analyses (CoMFA and CoMSIA) were successfully carried out based on this alignment. The statistical significance of the CoMFA model was indicated by a cross-validated r2 (q2) value of 0.95 (Table 6) and a correlation coefficient (r2) of 0.97 (Table 6). The model's predictive power was further corroborated by the predictive value of 0.97 (r2) when applied to the training set (Tables 6 and 7; Fig. 4). The test set resulted in a correlation coefficient (r2) of 0.62 (Fig. 4; Table 8). The alignment of CoMFA fields with KAB-18 is shown in Fig. 5. Lead molecule KAB-18 is surrounded by yellow contours, which represents areas of steric hindrance. The green contour around the phenyl of region 2 emphasizes the importance of sterically bulky groups in this region of the binding domain, providing stabilizing hydrophobic interactions with the ligand. In a similar manner, the green contour surrounding regions 3 and 4 shows the importance of steric interactions with the piperidine ring and the phenylpropyl of region 1. The red contour surrounding the molecule emphasizes the role of a negative charge that is favorable in interaction with the positive nitrogen of the piperidine ring.
TABLE 6.
Statistics for CoMFA and CoMSIA models
| q2 | S | % Steric | % Electrostatic | r2a | r2b | |
|---|---|---|---|---|---|---|
| CoMFA | 0.95 | 0.10 | 80 | 20 | 0.97 | 0.62 |
| CoMSIA | 0.87 | 0.02 | 80c | 20d | 0.97 | 0.81 |
Training set prediction.
Test set prediction.
Percentage of hydrophobic interaction.
Percentage of H-bond acceptor interaction.
Fig. 4.
Predicted and experimentally derived functional IC50 values of training set and test set compounds using CoMFA model (A) or CoMSIA model (B) were compared and linear regression analysis was performed. Log IC50 values of training set compounds are listed in Table 6, and log IC50 values of test set compounds are listed in Table 7.
Fig. 5.
CoMFA and CoMSIA models of NAM binding. A, CoMFA coefficient contours map aligned with KAB-18. The contours of the steric map are shown in yellow and green, and the contours of the electrostatic map are shown in red and blue. Greater potency (lower IC50 values) is correlated with less bulk near yellow, more bulk near green, more negative charge near red, and more positive charge near blue. B, CoMSIA contours aligned with KAB-18. The contours of the hydrophobic map are shown in white, yellow, and green, and the contours of the HBA map are shown in red and blue. Greater potency (lower IC50 values) is correlated with less bulk near yellow, more bulk near green and white, more HBA near blue, and less HBA near red.
A CoMSIA model was also generated, and the predictive nature of this model was supported by the cross-validated r2 (q2) value of 0.87 (Table 6) and a correlation coefficient (r2) of 0.97 (Table 6). The molecules chosen for the training set and test set in CoMFA studies were repeated for CoMSIA (Tables 7 and 8). The correlation coefficient (r2) using the training set molecules was 0.97 (Fig. 4; Tables 6 and 7). The correlation coefficient (r2) using the test set molecules was 0.81 (Fig. 4). When CoMSIA coefficients are plotted with lead molecule KAB-18 (Fig. 5), the yellow contours surrounding the molecule correlate with unfavorable hydrophobic interactions. The green and white contours indicate the important hydrophobic interactions with the phenyl ring of region 2 and the phenylpropyl of region 1. The blue contours suggest favorable interactions between the aromatic rings of regions 1 and 2 with positive charges in the binding pocket. The red contour near region 3 suggests the relative selectivity of less HBA in this region for biological activity.
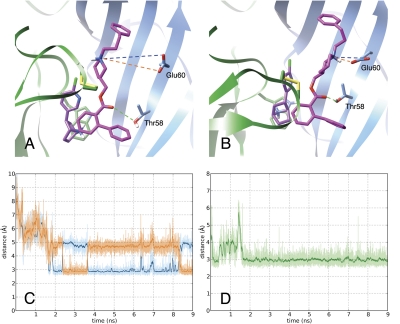
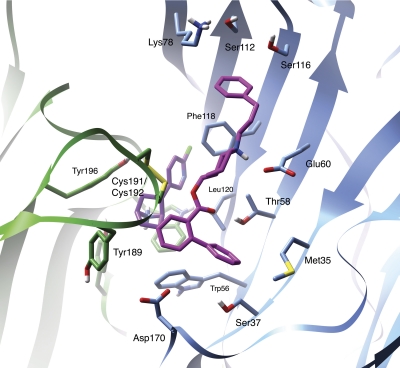
Previously, we described potential binding sites of our NAMs using blind docking to an ensemble of rat α3β4 nAChR conformations collected from an MD simulation (González-Cestari et al., 2009). A docking mode for our lead molecule, KAB-18, at the α/β interface of the Hα4β2 nAChR was evaluated more closely through an MD simulation. This site is adjacent to the agonist binding site (Figs. 6 and 7). KAB-18 remained stable at its docked position through the 9 ns of the simulation. Two important polar binding interactions were observed: 1) a hydrogen bond between the keto group of the ester linkage of KAB-18 and Thr58 of the β2 subunit and 2) an interaction between the positively charged nitrogen atom of the piperidine moiety of KAB-18 and Glu60 of the β2 subunit. After a slight induced fit in the earlier stages of the MD simulation, these interactions became stable (Fig. 6). The final binding mode of KAB-18 in the Hα4β2 nAChR with the hypothesized amino acid contacts is shown in Fig. 7.
Fig. 6.
Stability of KAB-18 at its proposed binding site. A and B, MD simulation of the Hα4β2 model was used to demonstrate initial docking mode (A) and induced binding mode at 7 ns (B) of KAB-18 (magenta) to the epibatidine-bound (purple) interface of the Hα4 (green ribbon) and Hβ2 (blue ribbon) subunits. Dotted lines identify key polar interactions between the ligand and the receptor. C and D, interatomic distances over time between the nitrogen of the positively charged piperidine and the two oxygen atoms of β2Glu60 (orange and blue labeling) and the keto group of the ester linkage and the hydroxyl-oxygen of β2Thr58 (green labeling), respectively.
Fig. 7.
Identification of ligand–receptor contacts of KAB-18 in the Hα4β2 nAChR model. A snapshot at 7.0 ns from MD simulation shows the induced binding mode of KAB-18 (magenta) to the epibatidine-bound (purple) interface of the Hα4 (green ribbon) and Hβ2 (blue ribbon) subunits. Key amino acids contributing to the binding of KAB-18 are shown as stick figures.
Blind docking identified a binding site for KAB-18 on Hα4β2 nAChRs. To support interaction of KAB-18 at the predicted site, Hα4β2 nAChRs with a mutation in the α4 subunit (R187I) and the β2 subunit (T58K) were transiently expressed in HEK ts201 cells (Hα4Mβ2M). These amino acids were chosen because they are proposed to be in contact with KAB-18 (Fig. 7). These mutations changed the amino acids in these positions to the cognate amino acids present in the human α3 and β4 subunits, respectively. Both epibatidine (agonist) and d-tubocurarine (competitive antagonist) showed no difference in apparent affinity between Hα4β2 WT and Hα4Mβ2M nAChRs (Table 9). The noncompetitive antagonist, mecamylamine, also showed no difference in apparent affinity between Hα4β2 WT and Hα4Mβ2M nAChRs (Table 9). KAB-18 showed a 10-fold reduction in apparent affinity (p < 0.005) in Hα4Mβ2M nAChRs compared with Hα4β2 wild-type nAChRs (Table 9). KAB-24, a negative control molecule, did not show a reduction in apparent affinity (Table 9).
Discussion
In this work, several of the effects of NAMs on human α4β2 nAChRs have been characterized. Our lead molecule, KAB-18, and many of our compounds, have functional IC50 values in the low micromolar range. The potencies for our NAMs are similar to potencies of other established nAChR antagonists including dihydro-β-erythroidine, d-tubocurarine, tetracaine, buproprion, and mecamylamine (McKay and Sanchez, 1990; McKay and Burkman, 1993; Jensen et al., 2005). In addition, the potency of these NAMs is similar to atypical antipsychotics that act as noncompetitive inhibitors of α4β2 and α7 nAChRs (Grinevich et al., 2009). A significant finding in this study is that several of these NAMs do not inhibit Hα3β4 nAChRs.
SAR studies defined chemical features that contribute to the potency of our lead molecule and its relative selectivity for Hα4β2 nAChRs. We found that the aromatic ring and carbon chain of region 1 are important for relative selectivity on Hα4β2 nAChRs. Molecules with small substitutions in region 1 (COB-1, COB-2, and COB-3) were equipotent on Hα4β2 and Hα3β4 nAChRs. The addition of a carbonyl at the α carbon in region 1 of KAB-18 (DDR-13) maintains relative selectivity for Hα4β2 nAChRs. This substitution also increased potency (2-fold) for Hα4β2 nAChRs, possibly caused by additional noncovalent contacts within the allosteric binding site. DDR-13 is also unique considering that the piperidine nitrogen is uncharged because of electron resonance with the carbonyl. The relative selectivity of KAB-18 for Hα4β2 nAChRs is also affected by substitutions in region 2; replacement of the phenyl group of KAB-18 for smaller substituents (methoxy, chloride, acetamide, and trifluoromethyl) results in molecules (KAB-23, KAB-22, KAB-19, and KAB-24) that are equipotent on Hα4β2 and Hα3β4 nAChRs. Substitution of the aromatic ring with a succinimide ring (KAB-30, KAB-20, IB-2, and IB-4) also results in molecules that are equipotent on Hα4β2 and Hα3β4 nAChRs. These data suggest that the aromatic ring in region 2 is necessary for relative selectivity.
The ester linkage in region 4 is of interest because of susceptibility to esterases, resulting in poor drugability. We found that replacement of this ester in region 3 with an amide (DDR-5) does not affect either potency or relative selectivity. Another finding is the importance of the orientation of the carbonyl group in region 4. Molecules with the carbonyl adjacent to the benzene ring of region 2 (KAB-18 and DDR-5) maintain relative selectivity for Hα4β2 nAChRs; however, molecules with the reverse ester (JHB-9 and DDR-3) are equipotent on Hα4β2 and Hα3β4 nAChRs. In addition, replacement of the ester linkage with an ether linkage (APB-21) does not affect potency but relative selectivity for Hα4β2 nAChRs is lost. This may be caused by additional geometries that are available in the ether that are not present in either the amide or ester or important hydrogen bonding interaction in the LBD. Regardless, these data support the importance of the carbonyl in region 4.
A clear disadvantage of targeting noncompetitive, allosteric sites is the limited knowledge of the binding site. To address this problem, ligand-based computational modeling was used to delineate information of this novel site on human nAChRs. CoMFA and CoMSIA models were constructed by using a set of molecules that showed relative selectivity for Hα4β2 nAChRs. A critical factor for proper implementation of CoMFA models lies in the precise structural alignment (Cramer, 2003). For several of the molecules in this study, precise alignment was made difficult because of structural differences. The results of the test set correlation show that CoMFA is not as suitable compared with our CoMSIA model. The CoMSIA model emphasizes the importance of both steric and hydrophobic interactions surrounding regions 1 and 2. These data are in agreement with data obtained from SAR studies, which showed that both regions were important for relative selectivity on Hα4β2 nAChRs.
In addition to the ligand-based approaches, structure-based approaches were used to help define the NAM binding site. Using docking and MD simulation on the Hα4β2 nAChR homology model, we identified a novel binding site for KAB-18 within 10 Å from the agonist binding site at the interface of the α and β subunits; however, the interacting amino acids are primarily on the β subunit. Other allosteric sites have been reported (i.e., galantamine) for nAChRs on non-α subunits (Iorga et al., 2006; Hansen and Taylor, 2007). The docking position of KAB-18 in the Hα4β2 nAChR model corresponds well to the predicted pharmacophore. The pharmacophore, CoMSIA, and CoMFA models predict four favorable hydrophobic interactions between the ligand and receptor that can be mapped as follows: 1) β4Phe118 and β4Lys78 for HYD1; 2) α4Cys191 and α4Cys192, β4Trp61 and β4Leu120 for HYD2; 3) β4Met35, β4Trp61, and β4Leu120 for HYD3; and 4) α4Cys191 and α4Cys192, β4Phe118 and β4Thr58 for HYD4. The β4Thr58 is the hydrogen bond donor. The pharmacophore, CoMFA, and CoMSIA models predict a favorable interaction between a positively charged region and the region 1 phenyl group of KAB-18. This interaction can be mapped to β4Lys78 in the predicted binding mode.
To determine the importance of the amino acids identified from the homology modeling studies, two amino acids of the Hα4β2 nAChR in close contact with KAB-18 were chosen for mutation to their cognate amino acids found in the Hα3β4 nAChR (α4 R187I + β2 T58K). We hypothesized that if the selected amino acids interacted with KAB-18, a loss of apparent affinity should be observed. Similar approaches have been reported to validate amino acids identified through models (Selent et al., 1992; Pepin et al., 1997). The functional studies showed a 10-fold reduction in apparent affinity of KAB-18 for the mutant nAChR. These data support the location of these amino acid residues within the allosteric LBD and that the homology model studies identified the NAM's binding site. To further validate the binding of this class of NAMs to this site, a molecule that was proposed to have no interaction with the mutated amino acids, KAB-24, was chosen. KAB-24 showed no difference in apparent affinity between the wild-type and mutant α4β2 nAChRs (Table 9).
The computed binding mode of KAB-18 to the α4β2 nAChR homolgy model may explain the selectivity for Hα4β2 nAChRs over Hα3β4 nAChRs. All pharmacophore features important for selectivity for Hα4β2 nAChRs are at positions that come into contact with amino acids that are not conserved between the two subtypes. All of these positions occur on the β subunit. In region 1, Ser112 and Lys78 of the β2 subunit exist as an arginine and isoleucine, respectively, at the corresponding site of the β4 subunit. In region 2, a Met35 exists as a glutamine at the corresponding site of the β4 subunit, whereas in regions 3 and 4 Thr58 and Phe118 exist as a lysine and leucine, respectively, at their corresponding sites of the β4 subunit. These residues may play a role in the preferential binding of particular compounds to the Hα4β2 nAChRs. Our computed binding mode may also explain the species differences observed with KAB-18 (González-Cestari et al., 2009). KAB-18 has been shown to inhibit rat α3β4 nAChRs while having no activity on Hα3β4 nAChRs. Our homology models show amino acid differences between human and rat in the NAM binding site on α3β4 nAChRs that potentially influence binding affinities: a Gln (human) versus Glu (rat) at β4 position 35 and a Leu (human) versus Gln (rat) at β4 position 118. Site-directed mutagenesis studies would need to be performed to validate this hypothesis, as done by Young et al. (2007) where similar observations were made with the compound TMAQ [5-(trifluoromethyl)-6-(1-methyl-azepan-4-yl)-methyl-1H-quinolin-2-one].
The discovery of selective antagonists of Hα4β2 nAChRs has important implications for the treatment of nicotine addiction. Worldwide, tobacco smoking is the leading cause of preventable death. Current pharmacotherapies [e.g., nicotine replacement therapy, antidepressant therapy and varenicline (Chantix)] have limited success and are associated with marked side effects, probably associated with off-target activities. Additional treatment strategies for nicotine addiction are needed, and the discovery of selective nAChR antagonists that target Hα4β2 nAChRs may provide a novel approach for smoking cessation. Mecamylamine (nonselective, noncompetitive antagonist) has shown clinical promise for nicotine cessation when coapplied with nicotine patches (Rose et al., 1994, 1998); however, mecamylamine's usage is limited because of side effects, probably linked to its nonselectivity for nAChR subtypes (Papke et al., 2001). Recently, NAMs have been shown to block nicotine reinforcement (Yoshimura et al., 2007; Hall et al., 2010). The studies presented here characterize functional activity of a novel NAM with selectivity for Hα4β2 nAChRs over Hα3β4 nAChRs and provide insight into important structural features and binding modes, contributing to the relative selectivity of our NAMs for Hα4β2 nAChRs. This work also documents the importance of SAR and ligand-based and structure-based computational modeling approaches for the discovery of NAMs targeting specific nAChR subtypes.
Acknowledgments
We thank Dr. Jon M. Lindstrom (Department of Neuroscience School of Medicine, University of Pennsylvania, Philadelphia) for stably transfected human cell lines and human nAChR α4 and β2 cDNAs and Dr. Rene Anand (Department of Neuroscience, College of Medicine, Ohio State University, Columbus, OH) for HEK ts201 cells.
This work was supported by the National Institutes of Health National Institute on Drug Abuse [Grant DA029433]. B.J.H. was supported by the National Institutes of Health National Institute on Drug Abuse Diversity Supplement. R.E.P. was supported by the American Foundation for Pharmaceutical Education. A grant of computational resources was received from the Ohio Supercomputer Center.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.168211.
- nAChR
- neuronal nicotinic acetylcholine receptor
- Hα4β2
- human α4β2
- Hα3β4
- human α3β4
- NAM
- negative allosteric modulator
- LBD
- ligand binding domain
- MD
- molecular dynamics
- CoMFA
- comparative molecular field analysis
- CoMSIA
- comparative molecular similarity indices analysis
- SAR
- structure–activity relationship
- QSAR
- quantitative SAR
- GASP
- genetic algorithm similarity program
- HBK
- HEPES-buffered Krebs
- HEK
- human embryonic kidney
- fluo-4-AM
- fluo-4-acetoxymethyl ester
- DMSO
- dimethyl sulfoxide
- HBA
- hydrogen bond acceptor.
References
- Anand R, Conroy WG, Schoepfer R, Whiting P, Lindstrom J. (1991) Neuronal nicotinic acetylcholine receptors expressed in Xenopus oocytes have a pentameric quaternary structure. J Biol Chem 266:11192–11198 [PubMed] [Google Scholar]
- Arias HR. (1998) Binding sites for exogenous and endogenous non-competitive inhibitors of the nicotinic acetylcholine receptor. Biochim Biophys Acta 1376:173–220 [DOI] [PubMed] [Google Scholar]
- Bergmeier SC, Ismail KA, Arason KM, McKay S, Bryant DL, McKay DB. (2004) Structure activity studies of ring E analogues of methyllycaconitine. Part 2: synthesis of antagonists to the α3β4 nicotinic acetylcholine receptors through modifications to the ester. Bioorg Med Chem Lett 14:3739–3742 [DOI] [PubMed] [Google Scholar]
- Bergmeier SC, Lapinsky DJ, Free RB, McKay DB. (1999) Ring E analogs of methyllycaconitine (MLA) as novel nicotinic antagonists. Bioorg Med Chem Lett 9:2263–2266 [DOI] [PubMed] [Google Scholar]
- Bryant DL, Free RB, Thomasy SM, Lapinsky DJ, Ismail KA, McKay SB, Bergmeier SC, McKay DB. (2002) Structure-activity studies with ring E analogues of methyllycaconitine on bovine adrenal α3β4* nicotinic receptors. Neurosci Res 42:57–63 [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changeux JP, Pinset C, Ribera AB. (1986) Effects of chlorpromazine and phencyclidine on mouse c2 acetylcholine receptor kinetics. J Physiol 378:497–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper E, Couturier S, Ballivet M. (1991) Pentameric structure and subunit stoichiometry of a neuronal nicotinic acetylcholine receptor. Nature 350:235–238 [DOI] [PubMed] [Google Scholar]
- Cramer RD. (2003) Topomer CoMFA: a design methodology for rapid lead optimization. J Med Chem 46:374–388 [DOI] [PubMed] [Google Scholar]
- Gallagher MJ, Chiara DC, Cohen JB. (2001) Interactions between 3-(trifluoromethyl)-3-(m-[125I] iodophenyl) diazirine and tetracaine, phencyclidine, or histrionicotoxin in the torpedo species nicotinic acetylcholine receptor ion channel. Mol Pharmacol 59:1514–1522 [DOI] [PubMed] [Google Scholar]
- González-Cestari TF, Henderson BJ, Pavlovicz RE, McKay SB, El-Hajj RA, Pulipaka AB, Orac CM, Reed DD, Boyd RT, Zhu MX, et al. (2009) Effect of novel negative allosteric modulators of neuronal nicotinic receptors on cells expressing native and recombinant nicotinic receptors: implications for drug discovery. J Pharmacol Exp Ther 328:504–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Riganti L, Vailati S, Clementi F. (2006) Brain neuronal nicotinic receptors as new targets for drug discovery. Curr Pharm Des 12:407–428 [DOI] [PubMed] [Google Scholar]
- Grinevich VP, Papke RL, Lippiello PM, Bencherif M. (2009) Atypical antipsychotics as noncompetitive inhibitors of α4β2 and α7 neuronal nicotinic receptors. Neuropharmacology 57:183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BJ, Pearson LS, Buccafusco JJ. (2010) Effect of administration of the nicotinic acetylcholine receptor antagonist BTMPS, during nicotine self-administration, on lever responding induced by context long after withdrawal. Neuropharmacology 58:429–435 [DOI] [PubMed] [Google Scholar]
- Hansen SB, Taylor P. (2007) Galanthamine and non-competitive inhibitor binding to ACh-binding protein: evidence for a binding site on non-α-subunit interfaces of heteromeric neuronal nicotinic receptors. J Mol Biol 369:895–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Orac CM, McKay S, McKay DB, Bergmeier SC. (2008) The synthesis of 5-substituted ring E analogs of methyllycaconitine via the Suzuki-Miyaura cross-coupling reaction. Bioorg Med Chem 16:3816–3824 [DOI] [PubMed] [Google Scholar]
- Iorga B, Herlem D, Barré E, Guillou C. (2006) Acetylcholine nicotinic receptors: finding the putative binding site of allosteric modulators using the “blind docking” approach. J Mol Model 12:366–372 [DOI] [PubMed] [Google Scholar]
- Jensen AA, Frølund B, Liljefors T, Krogsgaard-Larsen P. (2005) Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. J Med Chem 48:4705–4745 [DOI] [PubMed] [Google Scholar]
- Lloyd GK, Williams M. (2000) Neuronal nicotinic acetylcholine receptors as novel drug targets. J Pharmacol Exp Ther 292:461–467 [PubMed] [Google Scholar]
- Lukas RJ, Changeux JP, Le Novère N, Albuquerque EX, Balfour DJ, Berg DK, Bertrand D, Chiappinelli VA, Clarke PB, Collins AC, et al. (1999) International Union of Pharmacology. XX. Current status of the nomenclature for nicotinic acetylcholine receptors and their subunits. Pharmacol Rev 51:397–401 [PubMed] [Google Scholar]
- McKay DB, Burkman AM. (1993) Nicotinic and non-nicotinic receptor-mediated actions of vinblastine. Proc Soc Exp Biol Med 203:372–376 [DOI] [PubMed] [Google Scholar]
- McKay DB, Trent-Sanchez P. (1990) Effect of noncompetitive nicotinic receptor blockers on catecholamine release from cultured adrenal chromaffin cells. Pharmacology 40:224–230 [DOI] [PubMed] [Google Scholar]
- McKay DB, Chang C, González-Cestari TF, McKay SB, El-Hajj RA, Bryant DL, Zhu MX, Swaan PW, Arason KM, Pulipaka AB, et al. (2007) Analogs of methyllycaconitine as novel noncompetitive inhibitors of nicotinic receptors: pharmacological characterization, computational modeling, and pharmacophore development. Mol Pharmacol 71:1288–1297 [DOI] [PubMed] [Google Scholar]
- Moaddel R, Jozwiak K, Wainer IW. (2007) Allosteric modifiers of neuronal nicotinic acetylcholine receptors: new methods, new opportunities. Med Res Rev 27:723–753 [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. (1998) Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J Comput Chem 19:1639–1662 [Google Scholar]
- Nelson ME, Kuryatov A, Choi CH, Zhou Y, Lindstrom J. (2003) Alternate stoichiometries of α4β2 nicotinic acetylcholine receptors. Mol Pharmacol 63:332–341 [DOI] [PubMed] [Google Scholar]
- Nelson ME, Wang F, Kuryatov A, Choi CH, Gerzanich V, Lindstrom J. (2001) Functional properties of human nicotinic AChRs expressed by IMR-32 neuroblastoma cells resemble those of α3β4 AChRs expressed in permanently transfected HEK cells. J Gen Physiol 118:563–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olale F, Gerzanich V, Kuryatov A, Wang F, Lindstrom J. (1997) Chronic nicotine exposure differentially affects the function of human α3, α4, and α7 neuronal nicotinic receptor subtypes. J Pharmacol Exp Ther 283:675–683 [PubMed] [Google Scholar]
- Papke RL, Sanberg PR, Shytle RD. (2001) Analysis of mecamylamine stereoisomers on human nicotinic receptor subtypes. J Pharmacol Exp Ther 297:646–656 [PubMed] [Google Scholar]
- Pepin MC, Yue SY, Roberts E, Wahlestedt C, Walker P. (1997) Novel “restoration of function” mutagenesis strategy to identify amino acids of the δ-opioid receptor involved in ligand binding. J Biol Chem 272:9260–9267 [DOI] [PubMed] [Google Scholar]
- Pratt MB, Pedersen SE, Cohen JB. (2000) Identification of the sites of incorporation of [3H] ethidium diazide within the torpedo nicotinic acetylcholine receptor ion channel. Biochemistry 39:11452–11462 [DOI] [PubMed] [Google Scholar]
- Rose JE, Behm FM, Westman EC, Levin ED, Stein RM, Ripka GV. (1994) Mecamylamine combined with nicotine skin patch facilitates smoking cessation beyond nicotine patch treatment alone. Clin Pharmacol Ther 56:86–99 [DOI] [PubMed] [Google Scholar]
- Rose JE, Behm FM, Westman EC. (1998) Nicotine-mecamylamine treatment for smoking cessation: the role of pre-cessation therapy. Exp Clin Psychopharmacol 6:331–343 [DOI] [PubMed] [Google Scholar]
- Selent U, Rüter T, Köhler E, Liedtke M, Thielking V, Alves J, Oelgeschläger T, Wolfes H, Peters F, Pingoud A. (1992) A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry 31:4808–4815 [DOI] [PubMed] [Google Scholar]
- Stauderman KA, Mahaffy LS, Akong M, Veliçelebi G, Chavez-Noriega LE, Crona JH, Johnson EC, Elliott KJ, Gillespie A, Reid RT, et al. (1998) Characterization of human recombinant neuronal nicotinic acetylcholine receptors subunit combinations of α2β4, α3β4 and α4β4 stably expressed in HEK293 cells. J Pharmacol Exp Ther 284:777–789 [PubMed] [Google Scholar]
- Wang F, Gerzanich V, Wells GB, Anand R, Peng X, Keyser K, Lindstrom J. (1996) Assembly of human neuronal nicotinic receptor α 5 subunits with α3, β2, and β4 subunits. J Biol Chem 271:17656–17665 [DOI] [PubMed] [Google Scholar]
- Yoshimura RF, Hogenkamp DJ, Li WY, Tran MB, Belluzzi JD, Whittemore ER, Leslie FM, Gee KW. (2007) Negative allosteric modulation of nicotinic acetylcholine receptors blocks nicotine self-administration in rats. J Pharmacol Exp Ther 323:907–915 [DOI] [PubMed] [Google Scholar]
- Young GT, Broad LM, Zwart R, Astles PC, Bodkin M, Sher E, Millar NS. (2007) Species selectivity of a nicotinic acetylcholine receptor agonist is conferred by two adjacent extracellular β4 amino acids that are implicated in the coupling of binding to channel gating. Mol Pharmacol 71:389–397 [DOI] [PubMed] [Google Scholar]