Abstract
Differential regulation of drug-metabolizing enzymes (DMEs) is a common cause of adverse drug effects in cancer therapy. Due to the extremely important role of cytochrome P450 3A4 (CYP3A4) in drug metabolism and the dominant regulation of human pregnane X receptor (hPXR) on CYP3A4, finding inhibitors for hPXR could provide a unique tool to control drug efficacies in cancer therapy. Camptothecin (CPT) was demonstrated as a novel and potent inhibitor (IC50 = 0.58 μM) of an hPXR-mediated transcriptional regulation on CYP3A4 in this study. In contrast, one of its analogs, irinotecan (CPT-11), was found to be an hPXR agonist in the same tests. CPT disrupted the interaction of hPXR with steroid receptor coactivator-1 but had effects on neither the competition of ligand binding nor the formation of the hPXR and retinoid X receptor α heterodimer, nor the interaction between the regulatory complex and DNA-responsive elements. CPT treatment resulted in delayed metabolism of nifedipine in human hepatocytes treated with rifampicin, suggesting a potential prevention of drug-drug interactions between CYP3A4 inducers and CYP3A4-metabolized drugs. Because CPT is the leading compound of topoisomerase I inhibitors, which comprise a quickly developing class of anticancer agents, the findings indicate the potential of a new class of compounds to modify hPXR activity as agonists/inhibitors and are important in the development of CPT analogs.
Topoisomerase I inhibitors are a quickly developing class of anticancer agents. Clinical trials with the leading compound of this class of drugs, camptothecin (CPT), showed remarkable anticancer activity by interrupting DNA replication in cancer cells (Chen and Liu, 1994). Several semisynthetic derivatives of CPT, such as topotecan and irinotecan (CPT-11), have been used in cancer chemotherapy (Creemers et al., 1994). Although the main mechanism by which topoisomerase I inhibitors produce their antitumor effects has been widely investigated, both the mechanism of interindividual variability in the plasma disposition and the potential to regulate the efficacy of other compounds when used in combination are not fully understood (van Warmerdam et al., 1996).
Up-regulation/down-regulation of drug-metabolizing enzymes (DMEs) is a common cause of unpredictable drug effects during cancer therapy (Rochat, 2005), especially the alteration in the expression of cytochrome P450 3A4 (CYP3A4), an enzyme involved in the metabolism of 60% of drugs (Guengerich, 1999), including many anticancer drugs. Human pregnane X receptor (hPXR, also called steroid and xenobiotic receptor), a ligand-activated nuclear receptor, has been characterized as a dominant transcriptional factor of CYP3A4 (Chen and Nie, 2009). Evidence has shown that hPXR translocates from cytoplasm to the nucleus to increase the transcription of CYP3A4 as well as other DMEs and transporters after treatment with its agonists (Kawana et al., 2003; Chen et al., 2007). In contrast, the hPXR inhibitors block the biological actions of hPXR. Because many anticancer drugs have narrow therapeutic indexes, the modulation of the hPXR-CYP3A4 pathway may cause significant toxicity or unpredictable, ineffective therapy. Presently, although many hPXR agonists have been reported, few hPXR compounds antagonizing the hPXR-CYP3A4 pathway have been identified.
Because the CYP3A4-hPXR pathway is extremely important in drug efficacy (Bertilsson et al., 1998; Chen et al., 2009), establishing an interaction between the CYP3A4-hPXR pathway and CPT analogs is important for reducing drug-drug interactions (DDIs) and toxicity during drug development and clinical applications. In this study, the effects of CPT and its analog irinotecan on the hPXR-CYP3A4 pathway, and the underlying mechanisms, were characterized. CPT potently attenuated CYP3A4 induction by blocking the activation of nuclear receptors, especially hPXR. The clinical relevance of CYP3A4 inhibition was proven by attenuated nifedipine metabolism in CPT-pretreated primary cultures of human hepatocytes. Further mechanistic studies revealed that CPT inhibited hPXR by interrupting the binding of steroid receptor cofactor-1 (SRC-1) to hPXR. Conversely, irinotecan was a moderate agonist of hPXR, with no additive or inhibitive effects on rifampicin-induced hPXR activation.
Here, we established CPT as a novel and potent inhibitor of hPXR. On one hand, CPT and its analogs can be useful tools for modulating DME expression and drug efficacies. On the other hand, because CPT is the leading compound of a class of quickly developing anticancer drugs, the potential effects of CPT analogs on DMEs levels, especially the CYP3A4 level, should be noted in drug development.
Materials and Methods
Reagents and Cells.
CPT, irinotecan (Supplemental Fig. 1), 1α,25-dihydroxyvitamin D3 [1,25-(OH)2VD3], 6-(4-chlorophenyl) imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl) oxime (CITCO), pregnenolone-16α-carbonitrile (PCN), proteinase inhibitors, nifidipine, oxidized nifedipine, and nitrendipine were purchased from Sigma-Aldrich (St. Louis, MO). Rifampicin, SR12813, and ketoconazole were from BIOMOL Research Laboratories (Plymouth Meeting, PA). Dual-luciferase assay kit, β-galactosidase assay kit, CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay kit, and TNT T7 quick coupled transcription/translation system were purchased from Promega (Madison, WI). Escherichia coli Rosetta (DE3) and His·Bind Quick 300 cartridges were acquired from EMD Chemicals Inc. (Gibbstown, NJ). Zeba desalt spin column, EZ-Link sulfo-NHS-LC-biotin, and proFound pull-down biotinylated protein:protein interaction kit were obtained from Pierce Chemical (Rockford, IL). RNeasy mini kit was purchased from QIAGEN (Valencia, CA). TaqMan real-time PCR reagents and primers were obtained from Applied Biosystems (Foster City, CA). An Odyssey electrophoretic mobility shift assay (EMSA) buffer kit was from LI-COR Biosciences (Lincoln, NE). HepG2, 293T, and HCT116 cells were obtained from American Type Culture Collection (Manassas, VA). Fresh primary cultures of human hepatocytes from three donors and a hepatocyte culture media kit were obtained from BD Gentest (Woburn, MA). Donor demographics are shown in Supplemental Table 1. CBS cells were obtained from Dr. Subhas Chakrabarty (SimmonsCooper Cancer Institute, Southern Illinois University, Springfield, IL). All high-performance liquid chromatography (HPLC) grade reagents were from Acros Organics (Fairlawn, NJ).
Plasmids.
pGL3 vectors were purchased from Promega. PetDuet-1 and pet28a were from EMD Chemicals Inc. pGEM3-human vitamin D receptor (hVDR) was produced by American Type Culture Collection. The full-length hVDR cDNA was isolated from the pGEM3-hVDR plasmid and subcloned into a pcDNA3.1 vector. A pGL3-CYP3A4-XREM-luc plasmid was constructed by inserting two CYP3A4 regulatory elements, a distal enhancer (−7849 to −7219) and a proximate promoter (−369 to +37), into a pGL3 basic vector (Chen et al., 2007). Gal4DB-SRC-1-RID, TK-MH100×4-LUC, pSG5-mouse pregnane X receptor (mPXR), pVP-hPXR, pTracer-CMV2-human constitutive androstane receptor 3 (hCAR3), pTracer-CMV2, and pCMX-retinoid X receptor α (RXRα) were kind gifts from various laboratories (see Acknowledgments).
Cell Culture and Drug Treatments.
HepG2 cells were maintained in Eagle's minimal essential medium supplemented with 10% charcoal-stripped fetal bovine serum (Invitrogen, Carlsbad, CA). Only cells of passage 4 were used for transient transfections and drug treatments. Fresh human hepatocytes were available in 24-well collagen I plates. Cells were cultured in Hepato-STIM media supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 10 ng/ml epidermal growth factor and incubated in a humidified 37°C incubator with a 95% air, 5% CO2 atmosphere. Drugs were prepared in vehicle freshly on the day of use. Fresh dosing solutions were added every 24 h so that cells have a total of three exposures to the treatments over a 72-h time period.
Cell Toxicity Assessment.
Cells were plated into a 96-well plate and treated with drugs at different concentrations or vehicle controls for a certain time period. Cell viabilities after treatments were evaluated by an CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega) according to the manufacturer's instructions. Cytotoxicity for human hepatocytes was determined by a Vi-Cell XR cell viability analyzer (Beckman Coulter, Fullerton, CA).
Quantitative RT-PCR.
Total RNA was prepared using an RNeasy mini kit (QIAGEN, Valencia, CA) with on-column DNase treatment. RNA quantitation and quality were determined by a biophotometer (Eppendorf North America, Hauppauge, NY). TaqMan one-step RT-PCR assays were performed with 100 ng of RNA sample using an ABI Prism 7500 fast real-time PCR system (Applied Biosystems). An initial RT step occurred for 15 min at 48°C and was subsequently followed by heating to 95°C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 1 min. Primers for CYP3A4 and hPXR are as follows: CYP3A4 forward, 5′-TCAGCCTG GTGCTCCTCTATCTAT-3′; CYP3A4 reverse, 5′-AAGCCCTTATGGTAGGACAAAATATTT-3′; and CYP3A4 TaqMan probe, 5′-5-carboxyfluorescein-TCCAGGGCC CACACCTCTGCCT-5-carboxytetramethylrhodamine-3′; and hPXR forward, 5′-GCAGGAGCAATTCGCCATT-3′; hPXR reverse, 5′-TCGGTGAGCATAGCCATGATC-3′; and hPXR TaqMan probe, 5′-5-carboxyfluorescein-CCAGCCTGCTC ATAGGTTCTTGTTCCTGAA-5-carboxytetramethylrhodamine-3′. The sets of primers or probes for CYP3A4 and hPXR were designed to span exon junctions to prevent detection of any possible contamination of genomic DNA. Each assay contained 0.9 μM each of forward and reverse primer and 0.25 μM TaqMan probe. SYBR Green amplification of human 28S RNA was used as an internal control (28S forward, 5′-GGTATGGGCCCGACGCT-3′; 28S reverse, 5′-CCGATGCCGACGCTCAT-3′). Standard curves were constructed by serial 10-fold dilutions, ranging from 30 copies to 300,000 copies, of interest genes. Quantitations of gene expression levels were determined by interpolation of threshold cycle values to a standard curve. Gene-specific mRNA was subsequently normalized to 28S.
Western Blot.
Cells were lysed and sonicated in SDS running buffer. The lysate was centrifuged, and the supernatant was separated by 10% SDS-polyacrylamide gel electrophoresis. Proteins were then electroblotted onto a polyvinylidene fluoride membrane. After blocking, the membrane was incubated with a primary antibody for 1 h at room temperature followed by incubation with a secondary antibody for another hour. The antibodies used are listed in Supplemental Table 2. β-Actin was used as a loading control. The proteins were visualized by an Odyssey infrared imaging system, and the intensities of protein bands were determined using Odyssey 2.0 software (both from LI-COR Biosciences).
Luciferase Assay.
Cells were seeded into a 12-well plate. After attachment, cells at 80% confluence were cotransfected with 0.15 μg of pcDNA3.1-hPXR, pTracer-CMV2-CAR3, pSG5-mPXR, pcDNA3.1-hVDR, or vector control; 0.15 μg of pGL3-CYP3A4-XREM; and 10 ng of pGL3-CMV-Renilla (each well). Twenty-four hours after transfection, the cells were treated with drugs for 2 h followed by a fresh medium treatment for another 22 h. Then, cells were harvested and lysed. The luciferase activity was measured and normalized for transfection efficiency with Renilla activity.
Mammalian Two-Hybrid Assays.
HepG2 cells were seeded into a 12-well plate. After attachment, cells at 80% confluence were cotransfected with 500 ng of Gal4DB-SRC-1-RID, 100 ng of pVP-hPXR, 500 ng of TK-MH100×4-LUC, and 200 ng of pCMV-LacZ (each well). Twenty-four hours after transfection, the cells were treated with drugs. Then, the cells were harvested and lysed. The luciferase activity was measured and normalized for transfection efficiency with β-galactosidase activity.
Cofactor Binding Assay.
Residues 130 to 434 of PXR ligand binding domain (PXRLBD) were engineered as a C-terminal His-tagged fusion protein and subcloned into multiple cloning site 2 of petDuet-1. Residues 623 to 710 of the human SRC-1 protein were subcloned into multiple cloning site 1 of petDuet-1 to improve the stability and solubility of PXRLBD. The soluble PXRLBD fragment was obtained in Rosetta (DE3) with the induction of 0.05 mM isopropyl β-d-thiogalactoside in a shaker at 22°C. The protein was purified by His·Bind Quick 300 cartridges and changed into Tris-buffered saline buffer by a Zeba desalt spin column for pull-down assays. The 186-amino acid fragment of human SRC-1 protein with a C-terminal His-tag was amplified from an SRC-1-containing plasmid, subcloned into pet28a plasmid, and transformed into Rosetta (DE3). The expression was induced by isopropyl β-d-thiogalactoside overnight at 18°C. The SRC186 fragment was purified by His·Bind Quick 300 cartridges and changed buffered into phosphate-buffered saline. The purified SRC186 fragment was labeled with EZ-Link sulfo-NHS-LC-biotin reagent using a 3-fold molar excess of biotin reagent. Then, the peptide was purified and changed into Tris-buffered saline buffer. The pull-down study was performed using a proFound pull-down biotinylated protein:protein interaction kit (Pierce Chemical) according to the manufacturer's instructions. The biotinylated SRC186 fragment was immobilized on streptavidin-agarose beads. Then, the purified PXRLBD peptide and tested compounds were added to the beads. After prey capture, the beads were washed and eluted. The eluates were used for SDS-polyacrylamide gel electrophoresis analysis.
hPXR Competitive Binding Assay.
This assay was performed by biochemical nuclear receptor profiling service (Invitrogen). In brief, the assay was performed in 96-well noncoated polystyrene assay plate. Each well contained 20 μl of reaction mixture (n = 4): tested compounds/control compound (SR12813), 40 nM Fluormone hPXR Green (a green fluorescent hPXR ligand), 50 μM dithiothreitol, 10 nM purified glutathione transferase-tagged hPXR LBD, 10 nM terbium-labeled anti-glutathione transferase-tag antibody, and 1% DMSO. Plates were incubated at room temperature without light and evaporation for 1 to 2 h. Binding of Fluormone tracer was then measured by monitoring fluorescence resonance energy transfer (FRET) from the terbium-labeled antibody to the Fluormone tracer, resulting in a high TR-FRET ratio (520 nm fluorescent emission of Fluormone: 490 nm fluorescent emission of terbium). Competitors displace the Fluormone tracer from the receptor and disrupt FRET, resulting in a lower TR-FRET ratio. To read a LanthaScreen TR-FRET assay, the instrument is configured to excite the terbium donor at approximately 340 nm (30-nm bandwidth), and to separately read the terbium emission peak that is centered at approximately 490 nm (10-nm bandwidth), and the fluorescein emission that is centered at approximately 520 nm (25-nm bandwidth). The 520/490 TR-FRET ratio was measured using an EnVision fluorescent plate reader (PerkinElmer Life and Analytical Sciences, Boston, MA). A 100-μs delay followed by a 200-μs integration time was used.
Metabolism of Nifedipine by Human Hepatocytes.
Nifedipine was dissolved in 400 μl of medium to give a final concentration of 50 μM. Preliminary experiments were performed to determine the nifedipine concentration and incubation time in the linear ranges. The hepatocytes were incubated in a 37°C incubator (with CO2) for 30 min. At the end of the incubation period, the assay was stopped by adding 400 μl of acetonitrile and 1 μl of 40 μM nitrendipine to each well. The mixtures were centrifuged at 13,000 rpm for 5 min. A 20-μl aliquot of supernatants was used for HPLC.
HPLC Analyses.
The HPLC analyses of nifedipine, oxidized nifedipine (CYP3A4 metabolite), and nitrendipine (internal control) were performed on an LC-20AT HPLC system equipped with an SPD 20AV UV detector (both from Shimadzu, Kyoto, Japan). A Hypersil gold column (4.6 × 150 mm, 3 μm; Thermo Fisher Scientific, Waltham, MA), preceded by a C18 precolumn cartridge, was used in all assays. The solvent system at a flow rate of 0.5 ml/min consisted of 58% solvent A (0.1 M sodium phosphate, pH 3.0) and 42% solvent B (100% acetonitrile). The compounds were detected by monitoring of UV absorbance at 230 nm and compared with authentic standard compounds. The concentration was quantified by comparing the ratio of peak area of oxidized nifedipine to that of nitrendipine. A standard curve was constructed with the range of oxidized nifedipine used being from 500 pg to 200 ng. The acceptance standard for the calibration curves was a regression coefficient >0.99. The limit of quantification was defined as a signal-to-noise ratio of 10:1, with an acceptable level of variation (<10%).
In Vitro Translation and EMSAs.
EMSAs were performed using full-length hPXR and human RXRα synthesized in vitro. A pCMX-hRXRα plasmid and a pcDNA3.1-hPXR plasmid were used in a TNT T7 quick coupled transcription/translation system (Promega). Single-strand DNA oligonucleotides were labeled with IRDye 800 CM phosphoramidite at the 5′ end by Integrated DNA Technologies, Inc. (Coralville, IA) and then annealed to form double-stranded DNA fragments. The following IRDye-labeled oligonucleotides were used as probes (only the sense strand is shown, with consensus sequence in bold): CYP3A4-pER6, 5′-IRDye 800-TAG AATATGAACTCAAAGGAGGTCAGTGAGT-3′ and CYP3A4-dER6, 5′-IRDye 800-CCCTTGAAATCATGTCGGTTCAAGCA-3′. Proteins were incubated for 20 min at room temperature in darkness with 2.5 nM IRDye-labeled oligonucleotides in 10 mM Tris, pH 7.5, 50 mM KCl, 0.25% Tween 20, 5% glycerol, 3.5 mM dithiothreitol, 50 ng/μl poly(dI-dC), and 1× proteinase inhibitor cocktail. The mixture was then subjected to electrophoresis on a native Tris-glycine-EDTA polyacrylamide gel in a buffer containing 25 mM Tris base, 190 mM glycine, and 1 mM EDTA. Images were generated by scanning the plates directly in an Odyssey infrared scanner (LI-COR Biosciences) at the channel of 800 CM. The quantification was performed by Odyssey 2.0 software (LI-COR Biosciences). For competition bindings, unlabeled oligonucleotides at a 100-fold molar excess were coincubated. All shift assays were performed in the dark.
Statistical Analysis.
Student's t test (two-tailed) was used to analyze the difference between two groups. For comparisons among three or more groups, analysis of variance was used. A P value between groups, if smaller than 0.05, is considered statistically significant.
Results
Attenuation of CYP3A4 Induction in Primary Cultures of Human Hepatocytes.
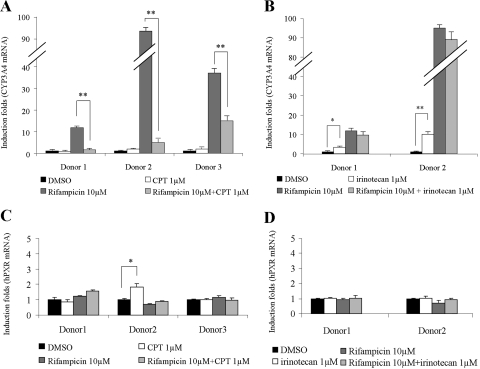
To assess the regulation of CPT on CYP3A4, primary cultures of human hepatocytes from three donors (Supplemental Table 1) were treated with CPT alone or in the combination with compounds that had previously been identified as positive (rifampicin; Goodwin et al., 1999) or negative (ketoconazole; Huang et al., 2007) for CYP3A4 induction for 72 h. Hepatocytes had no visible morphological changes or cell death after CPT treatment alone or in combination with rifampicin at the concentrations used. During treatment with single drugs, rifampicin significantly induced CYP3A4 in all three donors, with high interindividual variability (Fig. 1, A and B). CPT had no significant effects on basal CYP3A4 mRNA level (Fig. 1A). However, the induction of CYP3A4 by rifampicin was decreased by the coadministration with CPT by 60 to 90% (Fig. 1A). The CPT analog irinotecan was tested in parallel in experiments. It showed a moderate induction of CYP3A4 and had no synergetic or inhibitory effects on rifampicin-induced CYP3A4 levels in donor 1 and donor 2 (Fig. 1B). As a dominant regulator of CYP3A4, the hPXR mRNA levels were also evaluated in the same samples. Real-time PCR results showed hPXR levels were unchanged in all treatments compared with significantly changed CYP3A4 effects (Fig. 1, C and D).
Fig. 1.
CYP3A4/hPXR mRNA levels in primary cultures of human hepatocytes after drug treatments. Columns, mean induction folds (±S.D.) compared with mRNA levels in DMSO-treated group from four repeats. Bars are S.D. *, P < 0.05 and **, P < 0.01, comparing groups treated with CPT or irinotecan alone with DMSO-treated groups, or comparing groups treated with CPT or irinotecan in combination with rifampicin with rifampicin-treated groups. A, CYP3A4 mRNA levels in hepatocytes treated with CPT alone or in combination with rifampicin. B, CYP3A4 mRNA levels in hepatocytes treated with irinotecan alone or in combination with rifampicin. C, hPXR mRNA levels in hepatocytes treated with CPT alone or in combination with rifampicin. D, hPXR mRNA levels in hepatocytes treated with irinotecan alone or in combination with rifampicin.
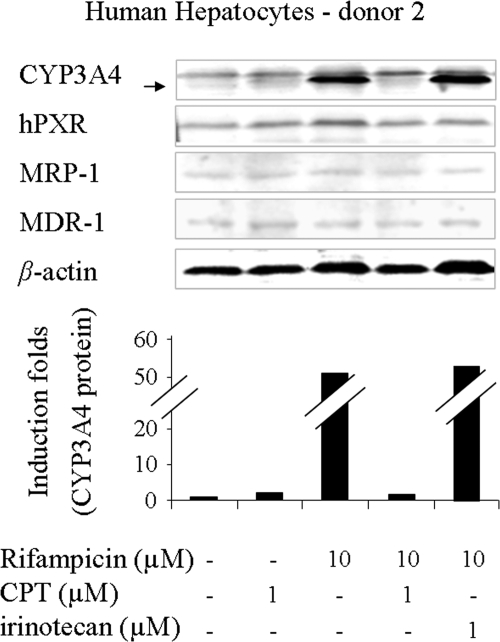
Western blots were also performed to evaluate protein levels. The results agreed with mRNA data. CPT reduced the ability of rifampicin to increase CYP3A4 protein levels in hepatocytes markedly in donors 1 and 2 (Fig. 2; Table 1) and moderately in donor 3 (Table 1). However, its analog irinotecan alone induced CYP3A4 protein levels in donor 1 (Table 1). No further CYP3A4 inductions were observed in rifampicin-treated hepatocytes from donors 1 and 2 when rifampicin was coadministered with irinotecan (Table 1; Fig. 2). The unusual down-regulation of CYP3A4 induction by CPT led us to investigate whether CPT could attenuate the inductions of other hPXR target genes caused by known inducers. However, a Western blot performed on hepatocytes treated with CPT/irinotecan alone or in combination with rifampicin did not detect significant induction/inhibition of hPXR, multidrug resistance gene-1 (MDR-1), or multiple drug resistance protein-1 (MRP-1) levels (Fig. 2). The induction/inhibition folds of CYP3A4 protein in all three donors after drug treatments are shown in Table 1. CPT also caused a remarkable decrease in the induction of CYP3A4 mRNA and protein levels in HepG2 cells, a hepatocarcinoma cell line, and CBS cells, a colon caner cell line. Other CYP3A4 inducers, such as SR12813 and paclitaxel, also were tested. The results showed it was not a rifampicin-specific effect (data not shown).
Fig. 2.
Western blot of proteins in primary cultures of human hepatocytes (donor 2) after drug treatments. β-Actin was used as a loading control. CYP3A4 protein levels were normalized to the vehicle control and expressed as fold induction.
TABLE 1.
Induction of CYP3A4 protein levels in primary cultures of human hepatocytes
The data are indicated as folds of CYP3A4 levels compared with the DMSO-treated group in each donor.
| Donor | Drug Treatment |
|||||
|---|---|---|---|---|---|---|
| DMSO | CPT (1 μM) | Iri (1 μM) | Rif (10 μM) | Rif (10 μM) + CPT (1 μM) | Rif (10 μM) + Iri (1 μM) | |
| 1 | 1 | 1.5 | 4.1 | 8.8 | 1.9 | 10 |
| 2 | 1 | 1.8 | — a | 53 | 1.7 | 56 |
| 3 | 1 | 1.2 | — | 31 | 12 | — |
Iri, irinotecan; Rif, rifampicin.
—, not determined.
Inhibition of Nuclear Receptor-Mediated CYP3A4 Induction.
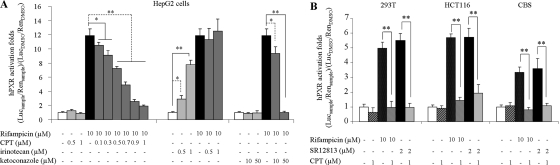
Previous reports have revealed that hPXR is a dominant regulator of CYP3A4 expression. Although the hPXR levels remained constant during CPT treatments (Figs. 1 and 2), the inhibition of hPXR transactivation activity on CYP3A4 promoter also could suppress CYP3A4 induction. An hPXR-based reporter assay was used to test this hypothesis. Cells were cotransfected with a Renilla expression vector, a reporter gene construct containing a responsive element for hPXR in CYP3A4 promoter, and an hPXR expression plasmid/vector control. Cells were treated with drugs at different concentrations (Fig. 3A), and the induction potential was then measured by the ability of a test compound to activate transcription of the reporter gene. The results are shown in Fig. 3. CPT strongly attenuated the transactivation of CYP3A4 promoter induced by rifampicin in a concentration-dependent manner (Fig. 3A). The IC50 value was 0.58 μM. In contrast, irinotecan alone induced hPXR transactivation in a dose-dependent manner and showed no synergetic effects on rifampicin-induced hPXR activity (Fig. 3A). These effects were observed only in cells cotransfected with the hPXR expression vector, suggesting an hPXR-dependent mechanism of regulation. Similar experiments were then carried out in different cell lines to exclude cell line-specific effects (Fig. 3B). Another hPXR agonist, SR12813, was used in evaluations for the purpose of excluding rifampicin-specific effects. The presence of CPT resulted in 60 to 80% inhibition of transcriptional activities induced by SR12813 or rifampicin in other cell lines (Fig. 3B).
Fig. 3.
Transactivation of the CYP3A4 promoter by the hPXR. The luciferase activities were normalized to the Renilla signals. The induction potential was then indicated as induction folds compared with vehicle control group (0.1% DMSO). Columns, mean hPXR induction potential determined in triplicate independent experiments; *, P < 0.05 and **, P < 0.01, comparing groups treated with CPT or irinotecan or ketoconazole alone with DMSO-treated groups, or comparing groups treated with CPT or irinotecan or ketoconazole in combination with rifampicin with rifampicin-treated groups. A, assays in HepG2 cells. Rifampicin, a classic hPXR agonist, was used as a positive control. Ketoconazole, a known hPXR inhibitor, was used as an inhibition control. B, assays in other cell lines. Both rifampicin and SR12813 (another hPXR agonist) were used to induce hPXR transactivation.
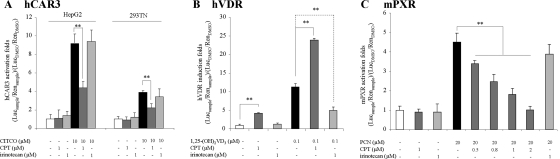
The CYP3A4 expression is also regulated by the nuclear receptors hCAR (Tirona et al., 2003) and hVDR (Thompson et al., 2002). Similar reporter assays were performed in HepG2 cells to evaluate the CPT effects on hCAR- and hVDR-mediated CYP3A4 transactivations. In contrast to hPXR, hCAR has a constitutive activation nature in immortalized cells. To avoid problems in in vitro assessment, a splicing variant hCAR3 was used in assay, which displays a low basal activity and still exhibits chemical-mediated activation in published studies (Faucette et al., 2007). CPT blocked 50% of CITCO-induced CYP3A4 transactivation through hCAR, whereas irinotecan only slightly affected the induction in both HepG2 cells and 293T cells (Fig. 4A). It is of interest to note that CPT and irinotecan showed opposite results in hVDR assays. CPT itself worked as an agonist in hVDR-mediated CYP3A4 transactivation and produced a synergetic induction with a strong hVDR agonist, 1,25-(OH)2VD3. However, irinotecan showed inhibitory effects on 1,25-(OH)2VD3-induced activation in this case (Fig. 4B). In control experiments without overexpressions of nuclear receptors, neither hVDR nor hCAR activity was increased/inhibited by drug treatments. In sum, these results show that CPT inhibits both hPXR- and hCAR-mediated transactivations of the CYP3A4 promoter but that it induced hVDR activity. The total effect is the inhibition on CYP3A4 levels because of the dominant modulation of hPXR on CYP3A4 expression in the context of cooperative modulations of all three nuclear receptors.
Fig. 4.
Transactivations of the CYP3A4 promoter by the hCAR, hVDR, and mPXR. The luciferase activities were normalized to the Renilla signals. The induction potential was then indicated as induction folds compared with vehicle control group (0.1% DMSO). Columns, mean induction potentials determined in triplicate independent experiments. Bars are S.D. *, P < 0.05 and **, P < 0.01, comparing groups treated with CPT or irinotecan alone with DMSO-treated groups, or comparing groups treated with CPT or irinotecan in combination with agonist with agonist-treated groups. A, transactivation of the CYP3A4 promoter by hCAR3 after drug treatments. CITCO, a selective agonist for human CAR, was used as positive control. B, transactivation of the CYP3A4 promoter by hVDR after drug treatments. 1,25-(OH)2VD3, a strong agonist of hVDR, was used as a positive control. C, transactivation of the CYP3A4 promoter by the mPXR after drug treatments. PCN, a strong activator of mPXR, was used as positive control.
Finally, previous studies have reported the notable species variance in the LBD of hPXR in different species. The role of CPT in mouse PXR activation also was examined. The results in Fig. 4C indicate that CPT blocks the activation of mPXR by PCN, an mPXR agonist, with an IC50 value of 0.87 μM, suggesting CPT is also an inhibitor of mPXR.
Cytotoxicity assays showed cell viabilities greater than 80% for all cell lines under the testing conditions (data not shown).
Effects on Formation of hPXR:hRXRα:hPXR-Responsive Elements Complex.
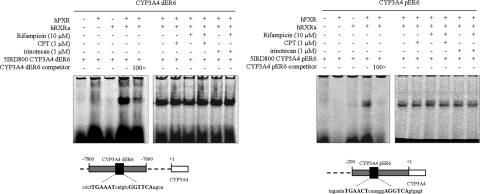
To elucidate the underlying mechanisms of the observed inhibitory effects on hPXR activation, we examined effects of CPT on key steps of hPXR activation. Upon activation, hPXR forms a heterodimer with hRXRα and then binds to the hPXRE present in the regulatory region of target genes (Chen and Nie, 2009). The hPXRE that have been identified in the regulatory regions of CYP3A genes consist of direct repeats of the consensus sequence (A/G)G(T/G)TCA separated by three nucleotides (DR3) (Kliewer et al., 1998), everted repeats separated by six bases (ER6) (Liu et al., 2008), and inverted repeats with a six-base spacer (IR6). An EMSA was performed to determine whether CPT interferes with the binding of hPXR:hRXRα complex to CYP3A4 hPXRE. Two important ER6 oligonucleotides were used in assays. One ER6 located at the proximal CYP3A4 promoter is hereafter referred to as CYP3A4 pER6, and the other ER6 located at distal CYP3A4 promoter is hereafter referred to as CYP3A4 dER6 (Fig. 5). A single retarded band was observed when both full-length hPXR and hRXRα were incubated with IRDye 800 CM phosphoramidite-labeled CYP3A4 pER6 or CYP3A4 dER6, but not when hPXR or hRXRα was incubated alone. An 100 M excess of unlabeled CYP3A4 pER6 or CYP3A4 dER6 suppressed the specific hPXR:hRXRα:hPXRE complex. The treatments with CPT/irinotecan alone or in the combination with rifampicin had no significant effects on either hPXR:hRXRα heterodimer formation or the binding of the heterodimer to both ER6 hPXRE (Fig. 5).
Fig. 5.
Analyses of the formation of hPXR:hRXRα:hPXRE complex by EMSA. Images were generated by scanning the plates directly in an Odyssey infrared scanner at the channel of 800 CM. The quantification was performed by Odyssey 2.0 software.
Inhibition on the Binding of Cofactor to hPXR.
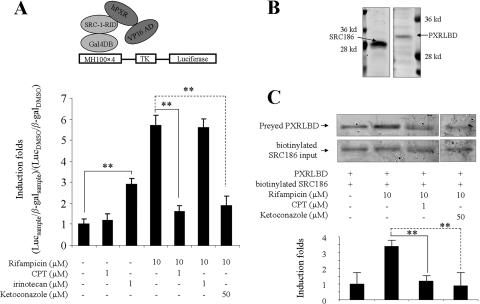
The interaction of hPXR with coactivator is a critical step of hPXR Signaling (Rosenfeld et al., 2006). Recently, the hPXR inhibitor ketoconazole has been shown to disrupt the interaction of hPXR with SRC-1 (Huang et al., 2007). In this study, we first used the mammalian two-hybrid assay to evaluate whether CPT individually or in combination with rifampicin could affect the interaction of hPXR with SRC-1 in cells. Two fusion plasmids, Gal4DB-SRC-1-RID and pVP-hPXR, were used in assay. Gal4DB-SRC-1-RID was constructed by fusing GAL4-DBD (yeast DNA binding domain) to SRC-1 receptor interacting domain. pVP-hPXR was a plasmid containing full-length hPXR fused to Herpes simplex virus VP16 activation domain. The reporter vector TK-MH100×4-LUC was constructed by inserting four copies of the yeast upstream activating sequences UASG enhancer (MH100) into the luciferase reporter plasmid TK-LUC. As shown in Fig. 6A, rifampicin significantly promoted the specific interaction of SRC-1 with hPXR in HepG2 cells. Both CPT and ketoconazole individually inhibited rifampicin-induced SRC-1 recruitment to hPXR. As expected, irinotecan alone increased the interaction between SRC-1 and hPXR but had no interference on rifampicin-induced SRC-1:hPXR binding.
Fig. 6.
Recruitment of coactivator SRC-1 to hPXR. A, analyses of the recruitment of SRC-1 to hPXR by a mammalian two-hybrid system. Rifampicin treatment and ketoconazole treatment were used as controls. The luciferase activity was measured and normalized for transfection efficiency with β-galactosidase activity. The interaction potentials are indicated as luciferase induction folds compared with vehicle control group (0.1% DMSO). Columns, mean induction folds determined in triplicate independent experiments. Bars are S.D. *, P < 0.05 and **, P < 0.01, comparing groups treated with CPT or irinotecan in combination with rifampicin with rifampicin-treated groups. B, SDS-polyacrylamide gel electrophoresis analyses of purified proteins obtained from E. coli expression. SRC186 was overexpressed in E. coli Rosetta (DE3). The expected protein size is 28 kDa. Residues 130 to 434 of PXRLBD were obtained from E. coli Rosetta (DE3), with a coexpression with residues 623 to 710 of the human SRC-1. The expected size of PXRLBD is 32 kDa. C, analysis of hPXR and SRC-1 binding in vitro by a pull-down assay. Preyed PXRLBD was normalized by inputted SRC186. Rifampicin treatment and ketoconazole treatment were used as controls. The interaction potential was indicated as induction fold compared with the vehicle control group (0.1% DMSO).
To confirm the results of mammalian two-hybrid system, we further performed an in vitro coactivator binding assay. Soluble SRC186 (186-amino acid residue of the human SRC-1 protein, which contains all three motifs interacting with hPXR) was labeled by biotin, immobilized on streptavidin-agarose beads to pull down the soluble fragment of PXRLBD in the absence or presence of drugs (Fig. 6B). Consistent with mammalian two-hybrid system, there was a basal interaction between SRC186 and PXRLBD in the absence of drugs. The addition of rifampicin induced a moderate but significant augment on the pulling down of PXRLBD. Cotreatment of CPT or ketoconazole with rifampicin suppressed the recruitment of SRC-1 to hPXR (Fig. 6C).
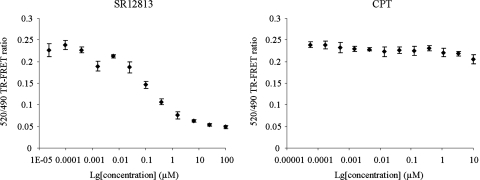
Competitive Ligand Binding Assay.
We then determined whether CPT exerted its antagonism effects through the competition of LBD binding with agonists. Test compounds were screened using the LanthaScreen TR-FRET hPXR competitive binding assay. In this assay, ligands are identified by their ability to compete with and displace the green fluorescent hPXR ligand Fluormone PXR Green from hPXR. When the positive control SR12813 was tested, an IC50 value of 0.14 μM was observed. In contrast, at the testing concentrations ranging from 57 pM to 10 μM, CPT did not compete with Fluormone PXR Green (Fig. 7). These results suggest that CPT at the concentrations used for the transcription assays might act outside the pocket to prevent coactivator recruitments.
Fig. 7.
Competition of binding to PXRLBD in a LanthaScreen TR-FRET competitive binding assay. The assay was performed in 96-well noncoated polystyrene assay plate. The 520/490 TR-FRET ratio was measured using an EnVision fluorescent plate reader with laser excitation and emission filters. Negative control without competitor (1% DMSO) and positive control with SR12813 were performed in parallel.
Suppression of CYP3A4-Mediated Nifedipine Metabolism in Primary Cultures of Human Hepatocytes.
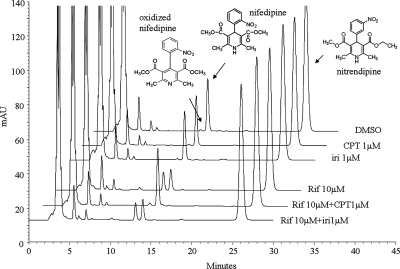
The above-mentioned data demonstrate that CPT can interfere with the induction of CYP3A4 expression in humans by limiting the activation of nuclear receptors. To determine whether such inhibitions can interfere with the metabolism of coadministered drugs, especially of those metabolized by CYP3A4, and lead to potential DDIs, we performed metabolism assays of nifedipine in CPT-treated primary cultures of human hepatocytes from three donors. Nifedipine is a commonly used CYP3A4 probe substrate and has been successfully used both in vivo and in vitro to measure CYP3A4 metabolic activity (Zhou et al., 2005). The authentic compounds were used in an HPLC-UV system for substrate and metabolite determination. The peak area ratio of oxidized nifedipine (metabolite from CYP3A4) to nitrendipine (internal control) was used for quantitation.
The preliminary chromatographic data showed that the retention times for oxidized nifedipine, nifedipine, and nitrendipine were 13.2, 14.1, and 26.4 min, respectively. The limit of quantification of oxidized nifedipine was 45 pg. Rifampicin effectively increased nifedipine metabolism in all three donors (Table 2). In donor 2, codosing CPT with rifampicin almost completely attenuated the metabolism of nifedipine (Fig. 8). A moderate suppression was also observed in donors 1 and 3 (Table 2). Irinotecan coadministration had no effect on rifampicin-induced production of oxidized nifedipine in donor 2. Assays treated with CPT or irinotecan alone did not show notable changes in nifedipine metabolism (Fig. 8; Table 2). As demonstrated in primary cultures of human hepatocytes, CPT also inhibited the induction of nifedipine metabolism by rifampicin in HepG2 cells (data not shown).
TABLE 2.
Metabolism of nifedipine in primary cultures of human hepatocytes
Metabolism of nifedipine was quantified by comparing the ratio of peak area of oxidized nifedipine to that of nitrendipine in HPLC analyses. The data are indicated as induction folds of the production of oxidized nifedipine (CYP3A4 metabolite) compared with the DMSO-treated group in each donor.
| Donor | Drug Treatment |
|||||
|---|---|---|---|---|---|---|
| DMSO | CPT (1 μM) | Iri (1 μM) | Rif (10 μM) | Rif (10 μM) + CPT (1 μM) | Rif (10 μM) + Iri (1 μM) | |
| 1 | 1 | — a | — | 7.6 | 3.9 | — |
| 2 | 1 | 2 | 1.6 | 26 | 2 | 21 |
| 3 | 1 | 1.5 | — | 17 | 12 | — |
Iri, irinotecan; Rif, rifampicin.
—, not determined.
Fig. 8.
HPLC analyses of the metabolism of nifedipine in primary cultures of human hepatocytes from donor 2. The oxidized nifedipine, nifedipine, and nitrendipine were eluted at 13.2, 14.1, and 26.4 min, respectively. The metabolisms of nifedipine were quantified by comparing the ratio of peak area of oxidized nifedipine to that of nitrendipine. Rif, rifampicin; Iri, irinotecan.
Discussion
In this study, we demonstrated by quantitative gene expression analyses that CPT, the leading compound of a class of topoisomerase I inhibitors, could antagonize the induction of CYP3A4 in primary cultures of human hepatocytes. CPT was later identified as an inhibitor of hPXR and mPXR in nuclear receptor transactivation assays. Additional receptor transactivation assays showed that CPT also could suppress hCAR activations but had inductive effects on hVDR activation. Furthermore, the underlying mechanisms of hPXR inhibition were explored. CPT could inhibit the recruitment of its coactivator SRC-1 to hPXR but had no effects on the ligand binding and hPXR:hRXRα:hPXRE complex formation. The inductive effects of rifampicin on nifedipine metabolism could be attenuated by coadministration with CPT in primary cultures of human hepatocytes, suggesting a potential prevention on DDIs between CYP3A4 inducers and drugs metabolized by the CYP3A4 in human body. The CPT analog irinotecan was tested in the study in parallel. No suppression of rifampicin-induced CYP3A4 expression or function was observed. Although numerous hPXR activators have been identified, to date there are few reports of potent inhibitors of hPXR. Here, we establish CPT as a novel functional inhibitor of hPXR, which can be a useful chemical tool for modulating PXR-regulated gene expression in vitro or in vivo.
CPT is a cytotoxic quinoline alkaloid that inhibits the DNA enzyme topoisomerase I. CPT showed remarkable anticancer activity but also low solubility and severe adverse drug effects. Numerous derivatives of CPT have been developed to increase the benefits of the chemical (Wall et al., 1993). In this study, one of the CPT analogs, irinotecan, showed itself as a moderate hPXR inducer and had no inhibitory effects on the induction of CYP3A4 by rifampicin. Topotecan (Supplemental Fig. 1), another semisynthetic analog of CPT used for the treatment of cisplatin refractory ovarian carcinoma, was reported as a CYP3A4 inducer (Schuetz et al., 2002). It suggests the potential of CPT analogs, which have similar structures, to modify hPXR activity, although they might be in opposite directions—agonism or antagonism. At present, although the induction of CYP3A4 level via hPXR is fully understood, the suppression of CYP3A4 level via hPXR is less well characterized. The fact that the receptor has conformational flexibility makes studies somewhat inefficient in recognizing key structures in inhibitors (Xue et al., 2007). Computational approaches combined with experimental data starting from a class of compounds have achieved some advances in revealing the key pharmacophores and binding regions for inhibitor activities of ketoconazole and its derivatives, the azoles (Ekins et al., 2007). Differently from other reports on single hPXR inhibitors, our study provides a new class of compounds with similar structures but opposite activities. Strategies starting from a class of compounds with similar structures but opposite activities have significant advantages over other studies, because it is easier to detect the key structure after comparison. Further structure-activity study is needed to understand the molecular determinants of receptor-compound interactions with the intent of creating inhibitors.
In this study, more than one hPXR agonist was tested with CPT. The inductive effects of both rifampicin and SR12813 were attenuated by coadministration with CPT (Fig. 3B), indicating it was not a drug-specific effect. In the study, hPXR and several other hPXR target genes also were tested (Figs. 1 and 2). It has been suggested that hPXR ligands enhanced hPXR-mediated transcription in a ligand- and promoter-dependent manner, which in turn differentially regulated the expression of individual hPXR targets, especially CYP3A4 and MDR-1 (Masuyama et al., 2005). Although MDR-1 and MRP-1 have been reported to be up-regulated by rifampicin (Geick et al., 2001), the treatment of rifampicin failed to induce MDR-1 and MRP-1 levels in hepatocytes after 72-h treatment in the current study. So far, we cannot determine whether the antagonism of agonist-induced hPXR activation is CYP3A4-specific.
Extensive cross-talk exists among the nuclear receptors. CYP3A4 expression also is regulated by hCAR and hVDR, aside from hPXR (Thompson et al., 2002; Tirona et al., 2003). Results from cell-based nuclear receptor transactivation assays indicate that CPT selectively antagonizes hPXR and hCAR but agonizes hVDR. Our study showed that CPT exerted its inhibitory effects on hPXR by inhibiting cofactor, SRC-1, recruitment. hCAR works in a mechanism similar to hPXR by recruiting SRC-1 in CYP3A4 regulation (Auerbach et al., 2005). Both receptors exhibit promiscuous, low-affinity ligand binding characteristics (Kliewer and Willson, 2002). It is reasonable to hypothesize that the binding of CPT on hCAR is unable to allosterically conform the liganded hCAR complex into a complex that attracts transcriptional coactivators as in hPXR. Of all receptors, hVDR is the receptor with the highest sequence identity to hPXR (Kliewer et al., 1998). hPXR and hVDR both possess unusually large ligand binding pockets (Watkins et al., 2001) and recognize similar response element motifs in the CYP3A4 promoter (Thompson et al., 2002). However, hVDR has been thought of as a high-affinity nuclear receptor with a different agonist/inhibitor selection mechanism from that of hPXR (Schmiedlin-Ren et al., 2001). Moreover, ligand-induced hVDR transactivation of CYP3A4 in its classical target tissue, the intestine, differs from hPXR-mediated CYP3A4 regulation in hepatocytes (Schmiedlin-Ren et al., 2001; Pavek et al., 2010). Studies are ongoing in our laboratory to clarify the differential regulation mechanisms of different nuclear receptors.
The regulation of DMEs/drug transporters is a common cause of adverse drug effects/drug resistance. Because cancer patients are generally coadministered multiple drugs and supplements, it is understandable that unpredictable therapeutic outcomes emerge due to the hPXR-mediated transcriptional effects on drug pharmacokinetics and pharmacodynamics. CPT was discovered in 1966 by its remarkable anticancer activity but also low solubility. In preliminary trails, the mean concentration of CPT in plasma was 0.1 μM (Natelson et al., 1996). Multiple new drug delivery techniques have been used to improve the concentration of CPT in vivo (Schluep et al., 2006), which is comparable with the effective CPT concentration in the current study. In addition, some findings in human studies are consistent with our conclusion that CPT is an hPXR inhibitor. Lockhart et al. (2004) showed that CPT coadministration attenuated drug-induced hPXR activation. Chen et al. (1991) found the use of CPT could overcome MDR-1-mediated drug resistance in colon cancer cells. In 2004, Yoshikawa et al. (2004) revealed the ability of camptothecin analogs to combat drug resistance mediated by the drug resistance gene ABCG2. The references mentioned above generally establish that CPT may function as an hPXR inhibitor. Considering the extremely important role of DMEs/transporters in drug metabolism and the dominant regulation of hPXR on DMEs/transporters, finding inhibitors for hPXR could provide a unique tool for controlling drug metabolism in cancer therapy.
Aside from its important role in xenobiotic defense, hPXR is involved in many physiological and pathophysiological processes, such as the homeostasis of bone (Tabb et al., 2003), lipid (Zhou et al., 2006), and mineralocorticoid (Zhai et al., 2007), glucocorticoid (Zhai et al., 2007), and thyroid hormones (Wong et al., 2005); inflammatory responses (Gu et al., 2006); cell growth and apoptosis (Zucchini et al., 2005); and uterine contractility (Mitchell et al., 2005). Hence, hPXR inhibitors may have utility as chemical modulators to interrogate the physiological roles of hPXR. Studies have shown that the antineoplastic agent ET-743 (Synold et al., 2001), the antifungal agent ketoconazole(s) (Huang et al., 2007), the dietary isothiocyanate sulforaphane (Zhou et al., 2007), the human immunodeficiency virus protease inhibitor A-792611 (Healan-Greenberg et al., 2008), and the phytoestrogen coumestrol (Wang et al., 2008) are all functional inhibitors of hPXR. Here, we describe a novel, effective hPXR inhibitor, among the topoisomerase I class of drugs, that is a potent hPXR inhibitors described, with an IC50 value of 0.58 μM toward rifampin-mediated receptor activation. Although the CPT has innate toxic action as an antineoplastic drug, its use as an hPXR modulator will not be limited because it is less toxic to normal cells. Furthermore, because CPT is a leading compound of a class of topoisomerase I inhibitors, chemists are working on developing more potent compounds. CPT analogs present the promising prospect of finding an hPXR inhibitor with higher efficiency but lower toxicity.
In conclusion, we have identified a novel and potent inhibitor of hPXR in this study. We have found that CPT attenuates the induction of CYP3A4 by inhibiting agonist-activated hPXR and hCAR. CPT disrupts the interaction of liganded hPXR with a coactivator, but it exerts effects on neither the ligand binding nor the formation of hPXR:hRXRα:hPXRE complex. In addition, our study and previous reports have shown that two CPT analogs, irinotecan and topotecan (Schuetz et al., 2002), are hPXR agonists, which suggest the potential of CPT analogs to modify hPXR activities in opposite directions: agonism or antagonism. Such information may have utility in the development of new CPT derivatives with minimal propensity for causing induction/inhibition-type drug interactions. Finally, because CPT is the leading compound in a group of quickly developing small molecules, the findings offer promise to obtain better hPXR modulators that may be used to tune the efficacy of therapeutics that serve as hPXR agonists in clinic.
Supplementary Material
Acknowledgments
We thank the following people and affiliations for providing plasmids: Gal4DB-SRC-1-RID and TK-MH100×4-LUC (Dr. Sridhar Mani, Albert Einstein College of Medicine, Bronx, NY), pSG5-mPXR and pVP-hPXR (Dr. Jeff L. Staudinger, Department of Pharmacology and Toxicology, University of Kansas, Lawrence, KS), pTracer-CMV2-CAR3 (Curtis J. Omiecinski, Center for Molecular Toxicology and Carcinogenesis, The Pennsylvania State University, University Park, PA), and pTracer-CMV2 pCMX-RXRα (Dr. David John Mangelsdorf, Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, TX).
This work was supported by the Fiscal Year 2009 Penny Severn postdoctoral fellowship from Offices of Women's Health, Illinois Department of Public Health; the U.S. Department of Defense postdoctoral prostate cancer training award [Grant PC081012]; and the National Institutes of Health National Cancer Institute [Grants R15-CA133776, R01-CA13345].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.168294.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- CPT
- camptothecin
- DME
- drug-metabolizing enzyme
- hPXR
- human pregnane X receptor
- DDI
- drug-drug interaction
- SRC-1
- steroid receptor cofactor-1
- 1,25-(OH)2VD3
- 1α 25-dihydroxyvitamin D3
- CITCO
- 6-(4-chlorophenyl)imidazo[2,1-b][1,3]thiazole-5-carbaldehyde O-(3,4-dichlorobenzyl)oxime
- PCN
- pregnenolone-16α-carbonitrile
- SRC186
- 186 amino acid residues of the human SRC-1 protein
- SR12813
- [[3,5-bis(1,1-dimethylethyl)-4-hydroxyphenyl]ethenylidene]bis-phosphonic acid tetraethyl ester
- PCR
- polymerase chain reaction
- EMSA
- electrophoretic mobility shift assay
- HPLC
- high-performance liquid chromatography
- hVDR
- human vitamin D receptor
- hCAR3
- human constitutive androstane receptor 3
- mPXR
- mouse pregnane X receptor
- RXRα
- retinoid X receptor α
- RT
- reverse transcription
- CMV
- cytomegalovirus
- LBD
- ligand binding domain
- DSMO
- dimethyl sulfoxide
- FRET
- fluorescence resonance energy transfer
- TR
- time resolved
- MDR-1
- multidrug resistance gene-1
- MRP-1
- multiple drug resistance protein-1
- hPXRE
- human pregnane X receptor-responsive element
- ER
- everted repeat
- A-792611
- methyl 1-(5-(2-(3-benzyl-2-oxoimidazolidin-1-yl)-3,3-dimethylbutanamido)-4-hydroxy-6-phenyl-1-(4-(pyridin-2-yl)phenyl)hexan-2-ylamino)-3,3-dimethyl-1-oxobutan-2-ylcarbamate
- ET-743
- (1′R,6R,6aR,7R,13S,14S,16R)-6′,8,14-trihydroxy-7′,9-dimethoxy-4,10,23-trimethyl-19-oxo-3′,4′,6,7,12,13,14,16-octahydrospiro[6,16-(epithiopropano oxymethano)-7,13-imino-6aH-1,3-dioxolo[7,8]isoquino [3,2-b][3]benzazocine-20,1′(2′H)-isoquinolin]-5-yl acetate
- IR
- inverted repeat.
References
- Auerbach SS, Stoner MA, Su S, Omiecinski CJ. (2005) Retinoid X receptor-alpha-dependent transactivation by a naturally occurring structural variant of human constitutive androstane receptor (NR1I3). Mol Pharmacol 68:1239–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Bäckman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. (1998) Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA 95:12208–12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AY, Liu LF. (1994) DNA topoisomerases: essential enzymes and lethal targets. Annu Rev Pharmacol Toxicol 34:191–218 [DOI] [PubMed] [Google Scholar]
- Chen AY, Yu C, Potmesil M, Wall ME, Wani MC, Liu LF. (1991) Camptothecin overcomes MDR1-mediated resistance in human KB carcinoma cells. Cancer Res 51:6039–6044 [PubMed] [Google Scholar]
- Chen Y, Nie D. (2009) Pregnane X receptor and its potential role in drug resistance in cancer treatment. Recent Pat Anticancer Drug Discov 4:19–27 [DOI] [PubMed] [Google Scholar]
- Chen Y, Tang Y, Chen S, Nie D. (2009) Regulation of drug resistance by human pregnane X receptor in breast cancer. Cancer Biol Ther 8:1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Tang Y, Wang MT, Zeng S, Nie D. (2007) Human pregnane X receptor and resistance to chemotherapy in prostate cancer. Cancer Res 67:10361–10367 [DOI] [PubMed] [Google Scholar]
- Creemers GJ, Lund B, Verweij J. (1994) Topoisomerase I inhibitors: topotecan and irenotecan. Cancer Treat Rev 20:73–96 [DOI] [PubMed] [Google Scholar]
- Ekins S, Chang C, Mani S, Krasowski MD, Reschly EJ, Iyer M, Kholodovych V, Ai N, Welsh WJ, Sinz M, et al. (2007) Human pregnane X receptor antagonists and agonists define molecular requirements for different binding sites. Mol Pharmacol 72:592–603 [DOI] [PubMed] [Google Scholar]
- Faucette SR, Zhang TC, Moore R, Sueyoshi T, Omiecinski CJ, LeCluyse EL, Negishi M, Wang H. (2007) Relative activation of human pregnane X receptor versus constitutive androstane receptor defines distinct classes of CYP2B6 and CYP3A4 inducers. J Pharmacol Exp Ther 320:72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geick A, Eichelbaum M, Burk O. (2001) Nuclear receptor response elements mediate induction of intestinal MDR1 by rifampin. J Biol Chem 276:14581–14587 [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C. (1999) The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol Pharmacol 56:1329–1339 [DOI] [PubMed] [Google Scholar]
- Gu X, Ke S, Liu D, Sheng T, Thomas PE, Rabson AB, Gallo MA, Xie W, Tian Y. (2006) Role of NF-kappaB in regulation of PXR-mediated gene expression: a mechanism for the suppression of cytochrome P-450 3A4 by proinflammatory agents. J Biol Chem 281:17882–17889 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (1999) Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol 39:1–17 [DOI] [PubMed] [Google Scholar]
- Healan-Greenberg C, Waring JF, Kempf DJ, Blomme EA, Tirona RG, Kim RB. (2008) A human immunodeficiency virus protease inhibitor is a novel functional inhibitor of human pregnane X receptor. Drug Metab Dispos 36:500–507 [DOI] [PubMed] [Google Scholar]
- Huang H, Wang H, Sinz M, Zoeckler M, Staudinger J, Redinbo MR, Teotico DG, Locker J, Kalpana GV, Mani S. (2007) Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene 26:258–268 [DOI] [PubMed] [Google Scholar]
- Kawana K, Ikuta T, Kobayashi Y, Gotoh O, Takeda K, Kawajiri K. (2003) Molecular mechanism of nuclear translocation of an orphan nuclear receptor, SXR. Mol Pharmacol 63:524–531 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterström RH, et al. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Willson TM. (2002) Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J Lipid Res 43:359–364 [PubMed] [Google Scholar]
- Liu FJ, Song X, Yang D, Deng R, Yan B. (2008) The far and distal enhancers in the CYP3A4 gene co-ordinate the proximal promoter in responding similarly to the pregnane X receptor but differentially to hepatocyte nuclear factor-4alpha. Biochem J 409:243–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart AC, Lee W, Ingram AD, Lee KA, Kim RB. (2004) Modulation of adopted nuclear receptors, pregnane X receptor (PXR) and constitutive androstane receptor (CAR), by commonly prescribed chemotherapy agents. Clin Pharmacol Ther 75:51 [Google Scholar]
- Masuyama H, Suwaki N, Tateishi Y, Nakatsukasa H, Segawa T, Hiramatsu Y. (2005) The pregnane X receptor regulates gene expression in a ligand- and promoter-selective fashion. Mol Endocrinol 19:1170–1180 [DOI] [PubMed] [Google Scholar]
- Mitchell BF, Mitchell JM, Chowdhury J, Tougas M, Engelen SM, Senff N, Heijnen I, Moore JT, Goodwin B, Wong S, et al. (2005) Metabolites of progesterone and the pregnane X receptor: a novel pathway regulating uterine contractility in pregnancy? Am J Obstet Gynecol 192:1304–1313, discussion 1313–1305 [DOI] [PubMed] [Google Scholar]
- Natelson EA, Giovanella BC, Verschraegen CF, Fehir KM, De Ipolyi PD, Harris N, Stehlin JS. (1996) Phase I clinical and pharmacological studies of 20-(S)-camptothecin and 20-(S)-9-nitrocamptothecin as anticancer agents. Ann NY Acad Sci 803:224–230 [DOI] [PubMed] [Google Scholar]
- Pavek P, Pospechova K, Svecova L, Syrova Z, Stejskalova L, Blazkova J, Dvorak Z, Blahos J. (2010) Intestinal cell-specific vitamin D receptor (VDR)-mediated transcriptional regulation of CYP3A4 gene. Biochem Pharmacol 79:277–287 [DOI] [PubMed] [Google Scholar]
- Rochat B. (2005) Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism. Clin Pharmacokinet 44:349–366 [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20:1405–1428 [DOI] [PubMed] [Google Scholar]
- Schluep T, Cheng J, Khin KT, Davis ME. (2006) Pharmacokinetics and biodistribution of the camptothecin-polymer conjugate IT-101 in rats and tumor-bearing mice. Cancer Chemother Pharmacol 57:654–662 [DOI] [PubMed] [Google Scholar]
- Schmiedlin-Ren P, Thummel KE, Fisher JM, Paine MF, Watkins PB. (2001) Induction of CYP3A4 by 1 alpha,25-dihydroxyvitamin D3 is human cell line-specific and is unlikely to involve pregnane X receptor. Drug Metab Dispos 29:1446–1453 [PubMed] [Google Scholar]
- Schuetz E, Lan L, Yasuda K, Kim R, Kocarek TA, Schuetz J, Strom S. (2002) Development of a real-time in vivo transcription assay: application reveals pregnane X receptor-mediated induction of CYP3A4 by cancer chemotherapeutic agents. Mol Pharmacol 62:439–445 [DOI] [PubMed] [Google Scholar]
- Synold TW, Dussault I, Forman BM. (2001) The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med 7:584–590 [DOI] [PubMed] [Google Scholar]
- Tabb MM, Sun A, Zhou C, Grün F, Errandi J, Romero K, Pham H, Inoue S, Mallick S, Lin M, et al. (2003) Vitamin K2 regulation of bone homeostasis is mediated by the steroid and xenobiotic receptor SXR. J Biol Chem 278:43919–43927 [DOI] [PubMed] [Google Scholar]
- Thompson PD, Jurutka PW, Whitfield GK, Myskowski SM, Eichhorst KR, Dominguez CE, Haussler CA, Haussler MR. (2002) Liganded VDR induces CYP3A4 in small intestinal and colon cancer cells via DR3 and ER6 vitamin D responsive elements. Biochem Biophys Res Commun 299:730–738 [DOI] [PubMed] [Google Scholar]
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. (2003) The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 9:220–224 [DOI] [PubMed] [Google Scholar]
- van Warmerdam LJ, Creemers GJ, Rodenhuis S, Rosing H, de Boer-Dennert M, Schellens JH, ten Bokkel Huinink WW, Davies BE, Maes RA, Verweij J, et al. (1996) Pharmacokinetics and pharmacodynamics of topotecan given on a daily-times-five schedule in phase II clinical trials using a limited-sampling procedure. Cancer Chemother Pharmacol 38:254–260 [DOI] [PubMed] [Google Scholar]
- Wall ME, Wani MC, Nicholas AW, Manikumar G, Tele C, Moore L, Truesdale A, Leitner P, Besterman JM. (1993) Plant antitumor agents. 30. Synthesis and structure activity of novel camptothecin analogs. J Med Chem 36:2689–2700 [DOI] [PubMed] [Google Scholar]
- Wang H, Li H, Moore LB, Johnson MD, Maglich JM, Goodwin B, Ittoop OR, Wisely B, Creech K, Parks DJ, et al. (2008) The phytoestrogen coumestrol is a naturally occurring antagonist of the human pregnane X receptor. Mol Endocrinol 22:838–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, Redinbo MR. (2001) The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292:2329–2333 [DOI] [PubMed] [Google Scholar]
- Wong H, Lehman-McKeeman LD, Grubb MF, Grossman SJ, Bhaskaran VM, Solon EG, Shen HS, Gerson RJ, Car BD, Zhao B, et al. (2005) Increased hepatobiliary clearance of unconjugated thyroxine determines DMP 904-induced alterations in thyroid hormone homeostasis in rats. Toxicol Sci 84:232–242 [DOI] [PubMed] [Google Scholar]
- Xue Y, Chao E, Zuercher WJ, Willson TM, Collins JL, Redinbo MR. (2007) Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg Med Chem 15:2156–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Ikegami Y, Hayasaka S, Ishii K, Ito A, Sano K, Suzuki T, Togawa T, Yoshida H, Soda H, et al. (2004) Novel camptothecin analogues that circumvent ABCG2-associated drug resistance in human tumor cells. Int J Cancer 110:921–927 [DOI] [PubMed] [Google Scholar]
- Zhai Y, Pai HV, Zhou J, Amico JA, Vollmer RR, Xie W. (2007) Activation of pregnane X receptor disrupts glucocorticoid and mineralocorticoid homeostasis. Mol Endocrinol 21:138–147 [DOI] [PubMed] [Google Scholar]
- Zhou C, Poulton EJ, Grün F, Bammler TK, Blumberg B, Thummel KE, Eaton DL. (2007) The dietary isothiocyanate sulforaphane is an antagonist of the human steroid and xenobiotic nuclear receptor. Mol Pharmacol 71:220–229 [DOI] [PubMed] [Google Scholar]
- Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. (2006) A novel pregnane X receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem 281:15013–15020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Yung Chan S, Cher Goh B, Chan E, Duan W, Huang M, McLeod HL. (2005) Mechanism-based inhibition of cytochrome P450 3A4 by therapeutic drugs. Clin Pharmacokinet 44:279–304 [DOI] [PubMed] [Google Scholar]
- Zucchini N, de Sousa G, Bailly-Maitre B, Gugenheim J, Bars R, Lemaire G, Rahmani R. (2005) Regulation of Bcl-2 and Bcl-xL anti-apoptotic protein expression by nuclear receptor PXR in primary cultures of human and rat hepatocytes. Biochim Biophys Acta 1745:48–58 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.