Abstract
Structural genomics discovery projects require ready access to both X-ray and NMR instrumentation which support the collection of experimental data needed to solve large numbers of novel protein structures. The most productive X-ray crystal structure determination laboratories make extensive frequent use of tunable synchrotron X-ray light to solve novel structures by anomalous diffraction methods. This requires that frozen cryo-protected crystals be shipped to large government-run synchrotron facilities for data collection. In an effort to eliminate the need to ship crystals for data collection, we have developed the first laboratory-scale synchrotron light source capable of performing many of the state-of-the-art synchrotron applications in X-ray science. This Compact Light Source is a first-in-class device that uses inverse Compton scattering to generate X-rays of sufficient flux, tunable wavelength and beam size to allow high-resolution X-ray diffraction data collection from protein crystals. We report on benchmarking tests of X-ray diffraction data collection with hen egg white lysozyme, and the successful high-resolution X-ray structure determination of the Glycine cleavage system protein H from Mycobacterium tuberculosis using diffraction data collected with the Compact Light Source X-ray beam.
1. Introduction
1.1 Protein Crystallography with the Compact Light Source
Over the past 30 to 40 years, synchrotron X-ray sources have had a rapidly growing impact on many fields of science. Their role within on the biological community has been very broad and includes many novel techniques that take advantage of the coherence and intensity of these X-ray sources, such as phase contrast imaging of soft tissue and macromolecular crystallography (Hendrickson, 1985).
The Protein Structure Initiative (PSI), led by the National Institute of General Medical Sciences (NIGMS), has placed an extraordinary emphasis on the role of synchrotron light sources in collecting the vast amounts of data necessary for determining the three dimensional structure of a wide variety of proteins and macromolecules. To further extend the influence of the special techniques developed at the large synchrotrons, we have developed a laboratory-scale synchrotron light source, called the Compact Light Source (CLS) capable of performing many of the state-of-the-art synchrotron applications in X-ray science.
The initial early development of the prototype CLS was funded by the NIGMS through Small Business Innovative Research (SBIR) grants to Lyncean Technologies, Inc. (LTI). Over the last four years the development of the CLS for protein crystallography applications has been funded by the Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), a PSI-2 Specialized Center that is supported by both the NIGMS and the National Center for Research Resources (NCRR). The ATCG3D consortium is focused on the accelerated development, integration, and deployment of emerging technologies that have high potential to improve the economics of protein structure determination by X-ray crystallographic methods.
Here we report on the first X-ray crystal structures from X-ray diffraction data collected from the NIH-funded, commercially developed, miniature synchrotron the Compact Light Source (CLS). The CLS was first used to collect X-ray diffraction data for crystals of hen egg white lysozyme, a benchmarking protein commonly used in the development of new technologies related to protein X-ray crystallography. We then went on to use the CLS to collect X-ray diffraction data for crystals of the Glycine cleavage system protein H from Mycobacterium tuberculosis (MytuGCSPH) whose structure was solved in collaboration with the Seattle Structural Genomics Center for Infectious Disease. The resulting 2.0 Å resolution protein structure was published to the RCSB Protein Data Bank (PDB) (PDBID: 3IFT) together with a sister PDB entry, a second crystal of the same protein solved at 1.75 Å using X-ray diffraction data collected using current state-of-the-art in-house (non synchrotron) X-ray source (PDB ID:3HGB). The Seattle Structural Genomics Center for Infectious Disease (SSGCID) project is funded by the National Institute of Allergy and Infectious Disease (NIAID) with the aim of solving protein structures that are relevant for potential structure based drug design of new antimicrobial agents.
1.2 Background
While the total flux from a typical home laboratory (homelab, rotating anode) X-ray source may be quite large, the necessary filtering and focusing results in limited monochromatic flux at prescribed, non-tunable energies. As a consequence, long exposure times are required to record a crystallographic data set in comparison to the speed with which a high resolution data set can be collected using intense X-rays generated at large national synchrotron facilities. In addition, the lack of tenability limits homelab sources from fully exploiting techniques such as SAD and MAD. Homelab X-ray sources have therefore been used heavily for screening crystals for diffraction quality prior to study at a synchrotron where cryopreserved crystals are sent by “FedEx” shipment in dry-shipper containers held at liquid nitrogen temperatures for data collection.
However, the difficulties of conducting research at a remote location continue to pose challenges for the structural biologist, many of whom would still prefer to conduct research in their own laboratories. In fact a common sentiment voiced among synchrotron users is the wish for a local, tunable source, with suitable properties to perform their favorite synchrotron-developed applications. This sentiment has driven synchrotron beamlines to invest heavily in remote, web-enabled data collection services using software such as web-ICE.
The idea for the Compact Light Source (CLS) is a by-product of high-energy physics research at the Stanford Linear Accelerator Center (SLAC) aimed at producing low-emittance electron beams (Huang, 1998). Later, further research at SLAC explored the viability of using a miniature electron storage ring and a high-finesse optical cavity optimized for creating high average flux in the hard X-ray regime (Loewen, 2003). These early studies led to the formation of Lyncean Technologies, Inc., which developed the Compact Light Source (CLS) based on the prior concept but completely re-designed and engineered for commercialization. By combining a “low-energy” (MeV scale) electron storage ring and a high-power laser cavity, X-rays are generated when the electrons and photons collide through an interaction known as inverse Compton scattering. While this phenomenon had been originally studied by colliding high-power laser pulses with electrons from a linear accelerator, the average flux was always too low to be relevant for most applications. However, using an electron storage-ring, together with the optical cavity, can provide several orders of magnitude higher X-ray flux; enough to yield a practical X-ray source.
Existing synchrotron light sources employ multi-GeV electron beams stored in large rings of magnets. Special insertion devices, such as undulator magnets, cause the electron beam to wiggle and produce the characteristic radiation of synchrotron light. For example, to produce 1 Å wavelength radiation with a 2 cm-period undulator magnet requires an electron beam with an energy of several GeV stored in a ring that is about 300 m in diameter. However, a laser beam colliding with a electron beam has the same effect as an undulator magnet. The electric and magnetic fields of the laser pulse cause the electrons to wiggle inducing a radiation spectrum similar to a long undulator magnet. To produce 1 Å radiation with a laser wavelength of 1 micron one requires an electron beam energy of only 25 MeV, reducing the scale of the storage ring by a factor of about 200; a 25 MeV electron storage ring is small enough to fit within the footprint of a large desk.
The CLS was designed by LTI to operate over a wide range of electron energies and can be tuned to produce X-rays in the range between 10 keV (1.23 Å) and 35 keV (0.35 Å). The X-rays are directed in a narrow cone in the direction of the electron beam. The spot size of the X-ray beam is largely determined by the focused spot of the laser beam, which has a radius of 50μm rms, while the divergence of the X-ray beam, a few mrad, is largely determined by the divergence of the electron beam at the interaction point. The electron beam divergence and energy spread, together with the angular acceptance of the X-ray optics, determine the natural energy bandwidth of the X-rays of about 3–4%.
For crystallography experiments, a set of 1:1 multilayer X-ray mirrors are placed near the X-ray output of the CLS to focus the emitted cone beam back to its original source size at a focal point located approximately 2 m from the optics.
1.3 The Compact Light Source -Technology
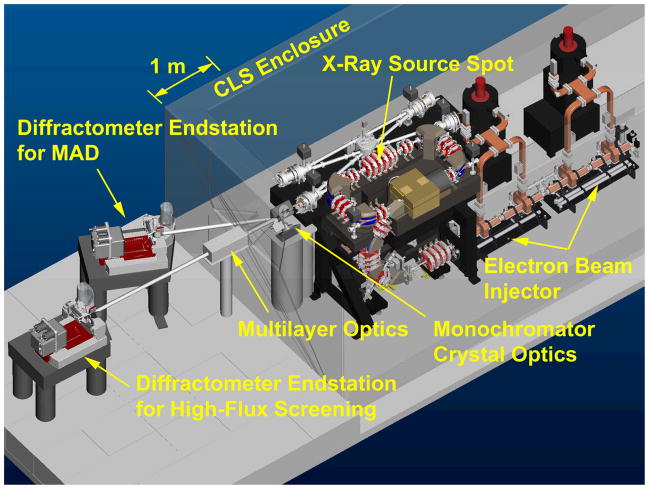
A CAD engineering model of the CLS system, including an experimental area for crystallography, is shown to scale in Figure 1. The CLS itself consists of an electron beam injector, a storage ring, and an integrated optical cavity system. The injector is shown with two commercial high-power microwave sources that power the accelerator sections. The electron storage ring and bowtie optical cavity circulate beams which synchronously interact at one particular location to produce X-rays. The entire CLS is contained within a shielded enclosure, shown partially transparent in Figure 1. The narrow cone of X-rays exits through a window towards sets of X-ray optics which can monochromatize and focus the beam. The X-ray beam is transported through evacuated chambers to end stations (shown with MarDTB and Rayonix MX-225 CCD) where it can be used for data collection.
Figure 1.
CAD drawing of the Compact Light Source together with endstation layout. The experimental area is separated by a shielding wall (shown mostly transparent).
A more detailed view of the CLS electron storage ring and optical cavity is shown in Figure 2. The electron source produces a single electron bunch using an RF gun with a laser photocathode. A short linear accelerator then accelerates the electron bunch to the full energy desired in the ring—20 to 45 MeV. This electron bunch is injected and stored in the miniature storage ring for about one million turns, and the injector periodically refreshes the electron bunch in the storage ring to maintain high beam quality. On one side of the ring is a straight section in which the electron beam is transversely focused to a small spot. This straight section also serves as one leg of the optical gain-enhancement cavity for the laser pulse. The electron bunch and the laser pulse collide each turn at the interaction point producing a burst of X-rays. The high flux of the CLS results from the high circulation rate of the storage ring and cavity.
Figure 2.
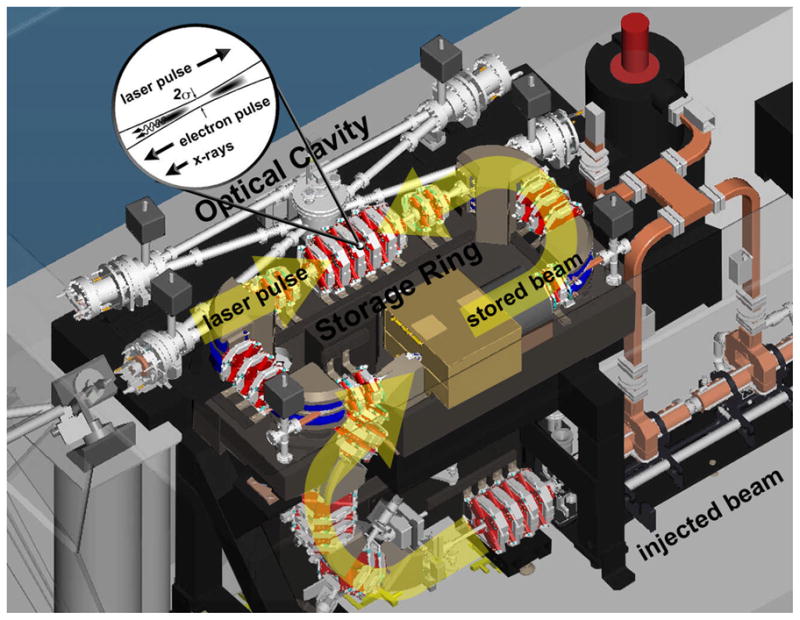
CAD drawing of the Compact Light Source illustrating laser-electron pulse interaction. Major components are the injector (electron gun not shown), the electron storage ring, and the integrated optical cavity. Electron-photon scattering at the interaction point produces naturally collimated, narrow bandwidth X-rays that exit a window at left of figure. The ring is a rectangle of approximately 1 m by 2 m.
The native CLS X-ray output is summarized in Table 1. Present flux has not yet reached design values, although the intensity has been already sufficient to demonstrate published results for Differential Phase Contrast Imaging (Bech, 2009b) (Bech, 2009a) in addition to crystallography. Ongoing development and funded hardware upgrades of the CLS are targeting the future performance levels listed in Table 1.
Table 1.
CLS X-ray output parameters
| Parameter | Present | Planned | Notes |
|---|---|---|---|
| Total Flux | ~1011 ph/s | ~1013 ph/s | Full bandwidth |
| Total Flux (output BW) | ~109 ph/s | ~1011 ph/s | 3–4% bandwidth |
| Source Spot Size | 50 μm rms | 30 μm rms | Also image size for 1:1 optics |
| Source Divergence | ~2.5 mrad | ~2.0 mrad | |
| X-ray Energy Range | 10–20 keV | 7–35 keV | Tunable |
External X-ray optics provide further manipulation of the CLS X-ray output. The cone beam can be intercepted just past the output window of the device and collimated or focused, with varying energy bandwidths depending on the type of optics employed. The reduction in flux should be proportional to the reduction in bandwidth of the output beam, limited only by the efficiency of the X-ray optic design. Although the prototype optics that were used for the experiments described here were very inefficient, new X-ray optics are currently under development which will provide close to optimal performance for bandwidths ranging from 0.5% to 1.5%.
Images of selected components of the CLS and the native X-ray spectrum are shown in Figure 3.
Figure 3.
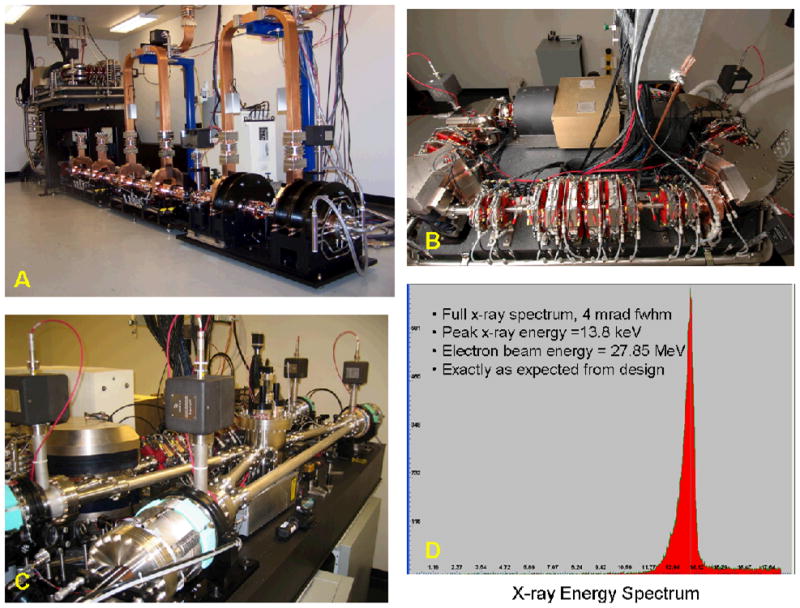
A. CLS Injector from prototype installed at LTI, B) electron storage ring, C) CLS optical cavity system, D) energy spectrum of X-rays from CLS output.
1.4 The Glycine cleavage system
The glycine cleavage system (Kikuchi, 2008)is the major route for glycine catabolism. The four-enzyme complex catalyzes in a reversible manner the conversion of glycine, tetrahydrofolate and NAD+ to 5, 10-methylene-tetrahydro-folate, carbon dioxide, ammonia, and NADH. The four proteins of the complex include three enzymes and one carrier protein: The P-protein (200kDa, multimer) is a glycine dehydrogenase which contains pyridoxal phosphate; the T-protein (40kDa) is an aminomethyl-transferase and carries out the tetrahydrofolate-dependent reaction; the L-protein is a dihydrolipamide dehydrogenase; the H-protein (14kDa) shuttles reaction intermediates and reduction equivalents between the different components of the system.
The first crystal structure of a Glycine cleavage system protein H was determined at 2.6 Å resolution by Pares et al. (Pares, 1994), PDBID code 1HPC. The protein was extracted from pea leaves and has a lipoate moiety bound to Lys63, called the lipoyl-lysine arm. Since, the structure of the protein from pea leaves has been determined in various forms: oxidized (Cohen-Addad, 1997)(PDBID:1HPC), reduced (Faure, 2000)(PDBID:1DXM), apo (Macherel, 1996) and bound to the reaction intermediate methylamine (Cohen-Addad, 1997) (PDBID:1HTP). Crystal structures from Themus thermophilus (Nakai, 2003) (PDBID:1ONL) and Thermotoga maritima (PDBID:1ZKO) have been determined in the recent years.
The Seattle Structural Genomics Center for Infectious Disease (SSGCID) is a collaborative center funded by the National Institute for Allergy and Infectious Disease (NIAID) to solve numerous protein structures of infectious disease targets from NIAID category A-C threat agents. The SSGCID targets are all selected with the long term goal that they can represent a blueprint for future structure guided drug design. The glycine cleavage system in bacteria has been selected by SSGCID as a pathway that could potentially be disrupted by a small molecule anti-bacterial agent that targets one or another of the components of the glycine cleavage system, with the goal that such a compound would compromise cell viability. In order to provide a variety of comparative structural information on selected targets, the SSGCID has drafted numerous homologous targets from a variety of our selected pathogen organisms, which includes Mycobacterium tuberculosis, the causative agent of tuberculosis (TB). The SSGCID strives to avoid duplication of effort with other structural genomics centers by coordinating target selection through the PSI funded TargetDB, a database of targets that allow centers to communicate progress towards structure determination of selected targets (http://tagetdb.PDB.org). At the time of initiating our SSGCID project, the glycine cleavage protein H of Mycobacterium tuberculosis was not being worked on by other structural genomics centers, and therefore it was selected for structure determination by SSGCID.
2. Materials and Methods
2.1 Protein expression and purification
The target gene was PCR amplified from purified Mycobacterium tuberculosis strain H37Rv genomic DNA (gift from David Sherman, SBRI) using the following primers: MytuD.01046.a.A1_FWD (5′GGGTCCTGGTTCGATGGTGAGCGATATCCCGTCCG) and MytuD.01046.a.A1_REV (5′CTTGTTCGTGCTGTTTATTACTCGGTCAGTGTGCCGCGGT). The purified amplicon was subsequently cloned into the bacterial expression vector AVA0421 using a ligation-independent cloning (LIC) methodology (Aslanidis, 1990) and transformed into the amplification host NovaBlue (Novagen). The expression vector AVA0421 is derived from pET14b, regulated by the T7 promoter, and contains the bla gene conferring ampicillin resistance. AVA0421 yields protein constructs with an N-terminal His6-Tag and a 3C protease cleavage site: MAHHHHHHMGTLEAQTQGPGSM-ORF. Cleavage of the His6-Tag by 3C protease yields proteins with an N-terminal sequence: GPGSM-ORF. The resultant plasmid was transformed into the Rosetta Oxford bacterial host (gift from Ray Hue, Structural Genomics Consortium), grown in auto-induction medium (Studier, 2005), the cells lysed, the supernatant passed over Ni2+ beads, and soluble protein quantified by SDS-PAGE to evaluate expression. Glycerol stocks were made at this stage and DNA prepared for sequencing to confirm that the correct target has been cloned and does not contain frame-shifts or premature stop codons.
The frozen cells were defrosted and resuspended in 200 ml of Lysis Buffer (20 mM HEPES, pH 7.2–7.4, 300 mM NaCl, 5% glycerol, 30 mM Imidazole, 0.5% CHAPS, 10 mM MgCl2, 3 mM β-mercaptoethanol, 25 units/ml of Benzonase® nuclease, and 0.05 mg/ml lysozyme). The resuspended cell pellet was then disrupted on ice for 15 minutes with a Branson Digital Sonifier 450D (settings at 70% amplitude, with alternating cycles of five seconds of pulse-on and ten seconds of pulse-off). The cell debris was clarified by centrifugation on a Sorvall RC5 at 6,000 RPM for 60 min at 4°C. The hexa-histidine tagged protein of interest was purified from the clarified cell lysate by immobilized metal affinity chromatography binding on Ni Sepharose High Performance resin (GE Biosciences, Piscataway, NJ) equilibrated with Binding Buffer (20 mM HEPES, pH 7.2–7.4, 300 mM NaCl, 5% glycerol, 30 mM Imidazole). The recombinant protein was eluted in 500 mM imidazole plus 5 mM β-mercaptoethanol and was further resolved by size-exclusion gel chromatography (SEC, Superdex 75 26/60; GE Biosciences, Piscataway, NJ). Pure fractions collected in SEC Buffer (20 mM HEPES pH 7.0, 300 μM NaCl, 2 mM DTT, and 5% glycerol) as a single peak were analyzed by sodium dodecyl sulfate-polyacrylamide (SDS) gel electrophoresis and Invitrogen Corp. SimplyBlue Safestain. The protein was then pooled, concentrated, flash frozen and stored in −80°C in SEC Buffer.
2.2 Protein crystallization
Lysozyme
Hen egg white lysozyme (Sigma) was crystallized using the hanging drop vapor diffusion method at room temperature. Protein solution contained 100 mg/mL protein in 100 mM sodium acetate trihydrate (Fluka) at pH 4.8. Precipitant solution contained 30% (w/v) polyethylene glycol monomethyl ether 5,000 (Fluka), 1.0 M sodium chloride (Fisher Scientific), and 50 mM sodium acetate trihydrate at pH 4.8. Hanging drops comprised 2 uL protein solution added to 2 uL precipitant solution, suspended over 1.0 mL precipitant solution. Crystals formed within minutes and were harvested after overnight growth. The 0.6 × 0.6 × 0.6 mm crystalline sample used for data collection was transferred to a cryogenic solution containing the precipitant solution components with added 20% (v/v) glycerol (Fisher Scientific), flash frozen in liquid nitrogen, and immediately mounted on the goniometer head in cryo stream at 100 K prior to diffraction data collection.
MytuGCSPH
The purified MytuGCSPH protein was concentrated to 27 mg/ml, and various commercial crystallization screens were set up using 0.4ul+ 0.4ul drops and Emerald BioSystems Compact Junior 96 well crystallization plates. Initial crystal hits were obtained in the JCSG+ screen (Emerald BioSystems) (Newman, 2005), condition B11: 1.6M Na-citrate. The crystals for the data set collected in-house were obtained directly from the screen and vitrified without further cryo-protection. Optimized crystals were obtained with the following reservoir solution: 1.5M Na-citrate, 50mM Bis-Tris pH 6.3. The crystals grew to a size of up to 0.3mm in each direction, see Figure 4. The optimized crystals were cryopreserved by freezing the crystals in liquid nitrogen following their transfer into a cryopreservation solution comprised of three parts of the reservoir crystallization cocktail mixed with one part of ethylene glycol as a cryoprotectant such that it had a final concentration of 25% v/v ethylene glycol. The cryopreserved crystals, mounted in nylon loops were used for data collection at the CLS.
Figure 4.
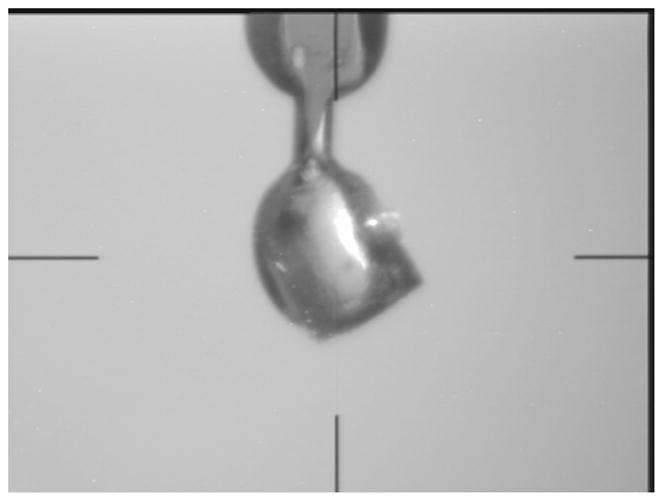
Crystal of MytuGCSPH cryo-protected and mounted to the goniometer head of the Compact Light Source. The size of the crystal is about 250μm in each direction.
2.3 Data collection
Experimental Set Up
For crystallography experiments, a set of 1:1 crystal or multilayer X-ray mirrors can be placed near the X-ray output of the CLS which focuses the emitted cone beam back to its original source size at a focal point located approximately 2 m from the optics. Because the X-ray radiation from the CLS is produced in only a narrow bandwidth, there are no thermal or other shielding issues associated with the optics. The focused beam can then be directly aligned onto a commercial goniostat providing extremely low background radiation on the detector.
Lysozyme data collection-CLS
For the lysozyme crystal experiment, the CLS X-ray beam was focused by two bent silicon crystals (Si 111). These were arranged in a K-B configuration following a design by Prof. Jens Als-Nielsen, and manufactured by JJ X-ray (Lyngby, Denmark). The end station was a MarDTB with a Rayonix SX-165 CCD detector (Rayonix LLC, Evanston IL, USA). The beam spot size on the detector was dominated by the point spread function. Due to figure errors in the optics, the flux was decreased from 109 down to 3×105, a reduction of about 3000. This loss is far more than expected from bandwidth reduction. These X-ray optics will not be used in future experiments.
X-ray diffraction data collection occurred over a two-day period while the lysozyme crystal was held in the cryo-stream of nitrogen gas at 100 K. Ninety images were collected using a 1.0 degree oscillation range per frame and an exposure time of 8 minutes. Additional data collection information and statistics are reported in Table 2.
Table 2.
Diffraction data and refinement statistics
| data collection | Lysozyme | MytuGCSPH | MytuGCSPH |
|---|---|---|---|
| beamline | CLS | CLS | rotating anode |
| wavelength [Å] | 0.92939 | 0.81836 | 1.5418 |
| space group | P43212 | C2 | C2 |
| a [Å] | 78.61 | 86.45 | 86.86 |
| b [Å] | 78.61 | 51.01 | 51.47 |
| c [Å] | 37.30 | 32.57 | 32.53 |
| beta [°] | 90 | 95.10 | 90 |
| resolution [Å] | 50 – 2.80 (2.87– 2.80) | 20 – 2.00 (2.05 – 2.00) | 20 – 1.75 (1.80 – 1.75) |
| reflections (unique) | 3,131 (156) | 9,647 (705) | 14,091 (1,067) |
| redundancy | 4.0 (3.9) | 3.1 (3.1) | 2.3 (1.4) |
| completeness [%] | 98.9 (99.6) | 98.8 (99.3) | 96.9 (97.7) |
| I/sigma | 12.7 (4.2) | 10.2 (3.3) | 23.8 (6.2) |
| Rsym | 0.105 (0.268) | 0.095 (0.385) | 0.027 (0.088) |
| Refinement | |||
| Rwork | 0.209 | 0.181 | 0.181 |
| Rfree | 0.278 | 0.253 | 0.220 |
| RMSD bonds [Å] | 0.012 | 0.016 | 0.008 |
| RMSD angles [°] | 1.37 | 1.59 | 1.19 |
| RMSD chirals [Å3] | 0.104 | 0.076 | 0.073 |
| Ramachandran plot | |||
| preferred | 119 (94.4%) | 129 (97.0%) | 121 (96.8%) |
| allowed | 7 (5.6%) | 3 (2.3%) | 4 (3.2%) |
| outliers | 0 (0%) | 1 (0.7%) | 0 (0%) |
| PDB ID code | n/a | 3IFT | 3HGB |
The benchmark data set for lysozyme was not fully refined and the structure not deposited.
(1) Numbers in parenthesis represent highest resolution shell of data.
(2) Rmerge = (|ΣIhkl −<I>|/(ΣIhkl), where the average intensity <I> is taken over all symmetry equivalent measurements and Ihkl is the measured intensity for any given reflection.
(3) I/σI is the mean reflection intensity divided by the estimated error.
(4) Rwork = ||Fo| − |Fc||/|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively.
(5) Rfree is equivalent to Rcryst but calculated for 5% of the reflections chosen at random and omitted from the refinement process.
MytuGCSPH (MytuD.01046.a) data collection - CLS
For the MytuGCSPH crystal experiment the CLS X-ray beam was focused by two bent multilayers on loan from the Niels Bohr Institute, University of Copenhagen, Denmark. These were developed by Anette Jensen and Jens Als-Nielsen (Jensen, 2007). For this experiment, the end station location was shifted to the angle appropriate for the optics, but otherwise the set up was identical to that for the lysozyme experiment. The multilayer X-ray optics resulted in a factor of 200 reduction in flux (from 109 down to 5×106 ph/s), once again far more than expected from simple bandwidth reduction. This is understandable because they were developed for a rotating anode source. In order to reduce ice rings, cryopreserved mounted crystal was annealed twice by allowing the crystal to thaw at room temperature followed by re-application of the cryo-stream nitrogen gas to rapidly freeze and maintain the crystal at 100 K.
MytuGCSPH (MytuD.01046.a) data collection -rotating anode
A data set for a MytuGCSPH crystal was initially collected using a conventional rotating anode (Rigaku FR-E+ Superbright) equipped with a Rigaku Saturn 944+ CCD detector. In order to include high angle spots, the detector was swung in 2θ by 10°. 180 images were collected in 1° wedges with 10 seconds exposure per image.
2.4 Structure solution and refinement
Lysozyme
Diffraction images for Lysozyme were analyzed and processed in HKL2000 (Otwinowski, 1997). The best 60 images from the 90-image data collection were selected to yield a data set up to 2.8 Å resolution.
The Molecular Replacement was done with PHASER (McCoy, 2007), using published a 1.65 Å resolution structure of tetragonal lysozyme (PDB code 2HU1) with all non-protein atoms removed as the search model. Rotation and translation function values for the molecular replacement were 12.2 and 29.3, respectively. The resulting initial structure was fit using COOT (Emsley, 2010) and refined using REFMAC5 (Murshudov, 1997)n CCP4, to yield the refinement statistics reported in Table 2.
MytuGCSPH
The diffraction data sets for the MytuGCSPH crystals were reduced with the XDS (Kabsch, 1988) package and scaled with XSCALE(Kabsch, 1988)to 1.75Å (rotating anode) and 2.0Å (CLS), respectively. The diffraction statistics are summarized in Table 2.
The Molecular Replacement for both data sets was done with PHASER (McCoy, 2007)using the PDB-entry 1ONL (in-house data) or 3HGB (in-house structure for the CLS data set) as the search model. The models were iteratively built with COOT (Emsley, 2010)and refined with REFMAC5.5.0088 (Murshudov, 1997). The structures were refined to Rwork=0.181 and Rfree=0.220 (in-house data) and R Rwork=0.181 and Rfree=0.253 (CLS data) with good stereochemistry (see Table 2). Coordinates and structure factors were deposited in the PDB with the PDB-code 3HGB (rotating anode data) and 3IFT (CLS data), respectively.
3. Results and Discussion
As of mid-2009, the Compact Light Source prototype had reached a number of milestones in both protein crystallography and biological imaging. The CLS prototype development began with initial funding in 2002 and after one year of design work and two years of construction, the prototype electron storage ring first achieved a stored electron beam in June of 2005. The optical cavity system required further development, but it was installed 6 months later, and on February 23, 2006 the CLS prototype achieved its first X-ray beam. The first test of prototype crystal optics followed in June of 2006 when the CLS achieved its first focused X-ray beam. The next year focused on studies of the X-ray flux and spectrum. In June of 2007, the CLS prototype was used in its first science experiment to study Differential Phase Contrast Imaging (DPCI) (Pfeiffer, 2008). One year later in June of 2008, the first crystal diffraction was achieved with a small molecule. Several experiments followed in 2008 including the second DPCI experiment and the first protein diffraction and partial data set in November and December of 2008. The next year, 2009, began with the first scientific publication from the CLS which reported on the imaging results from the DPCI experiment(Bech, 2009b). In March 2009 a trial lysozyme data set was collected and later in July, data for the first novel protein was collected and reported herein.
Before the detailed discussion of the last two experiments, it is important to note that in parallel to the CLS prototype experimental development, the ATCG3D ‘Beta’ CLS was under construction starting in 2005. This Beta CLS took advantage of the lessons learned from the prototype, and the prototype took advantage of the Beta development. Several ATCG3D subsystems were installed on the CLS prototype and used for the experiments described below.
After the successful completion of a trial lysozyme data set in March 2009, the CLS prototype at Lyncean Technologies was prepared to collect data on a novel, unsolved protein. The ATCG3D/SSGCID collaboration proposed a suitable target (MytuGCSPH, target ID MytuD.01046.a), samples of which were sent to Lyncean Technologies in July of 2009. After screening a dozen crystals, a candidate crystal was selected and mounted to a MAR desktop beamline, outfitted with a Rayonix SX-165 detector. The crystal size (~250×250×100 microns) was well matched to the focused X-ray beam (~200 micron FWHM).
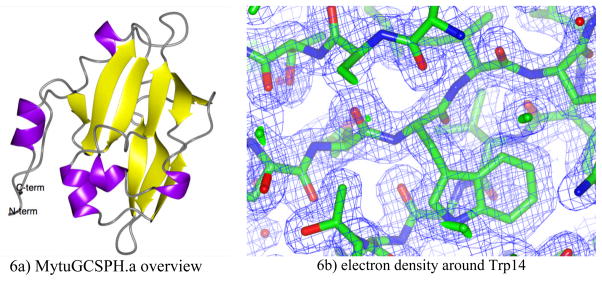
The CLS was tuned-up for X-rays at 15.1 keV (0.818 A) with a bandwidth of 1.4% for this data collection, comprising 152 images. Each image was taken with 6 min exposure times, with times-two dezingering. Diffraction spots extended to 2.0 Å resolution. The entire data set was taken during normal daytime working hours over a 3-day span. The structure was solved using Molecular Replacement with 3HGB; the sister structure was solved using data collected from a homelab source. The electron density for both data sets is very clear: an example for the CLS data set is shown in Figure 6b. Residues from the N-term including two residues from the His6 purification tag could be modeled without any gaps throughout the C-terminus of the construct. MytuGCSPH has a very compact fold in which two central anti-parallel b-sheets are flanked by a few α-helices (Figure 6a). The fold is member of the biotinyl-lipoyl-domain superfamily. Only few protein structures with significant sequence homology are known, all of them are members of the Glycine cleavage system. A search for structural homologues using SSM (Krissinel, 2004) yields the same set of structures. The two structures determined with data from the CLS and a conventional rotating anode are virtually identical: all Ca atoms superimpose with a RMSD of 0.23 Å. The lysine arm (Lys65) which is lipoate-bound in the protein extracted from pea leaves, is ordered however unmodified in both structures of MutuGCSPH. In fact, a modification of this residue might disturb the crystallographic packing of this crystal form.
Figure 6. Structure and electron density of MytuGCSPH.
6a: Overview of the structure for MytuGCSPH in ribbon representation.
6b: Electron density from the MytuGCSPH CLS data set at 2.0 Å resolution centered around Trp14
Although the focused X-ray flux for this experiment was significantly lower than the design specifications of the CLS, its stable operation and inherently low radiation background provided a high signal-to-noise ratio for the diffraction data. Likewise, the 1.4% energy bandwidth of the X-rays did not substantially affect the accuracy of the data reduction.
4. Summary and Future Plans
Herein, we have described the first protein crystal structure determination using X-ray diffraction data collected using inverse Compton X-ray light from the CLS. In the spring of 2010, we anticipate to commission the new beta version of the CLS for ATCG3D, which will include several improvements to performance. Ongoing technical development of the CLS is targeted to achieve another factor of 100 in native intensity. The X-ray optics used for the crystallography experiments described herein were extremely inefficient and will be replaced with new optics currently under development to improve the flux on sample by another factor of ten.
Together, these flux enhancements will make the CLS a useful tool for protein crystallography and other synchrotron applications. In particular, the crystallography community has shown interest in the CLS for rapid turnaround of results from crystallization trials, to X-ray screening and data collection. The synchrotron imaging community has been hoping for a CLS-like source for more than 20 years. Ultimately, we believe that the CLS can fill the role of a local synchrotron X-ray source, expanding the breadth and impact of X-ray science on biology and human health by opening avenues for exploration that might not be considered or possible at a large facility.
Figure 5.
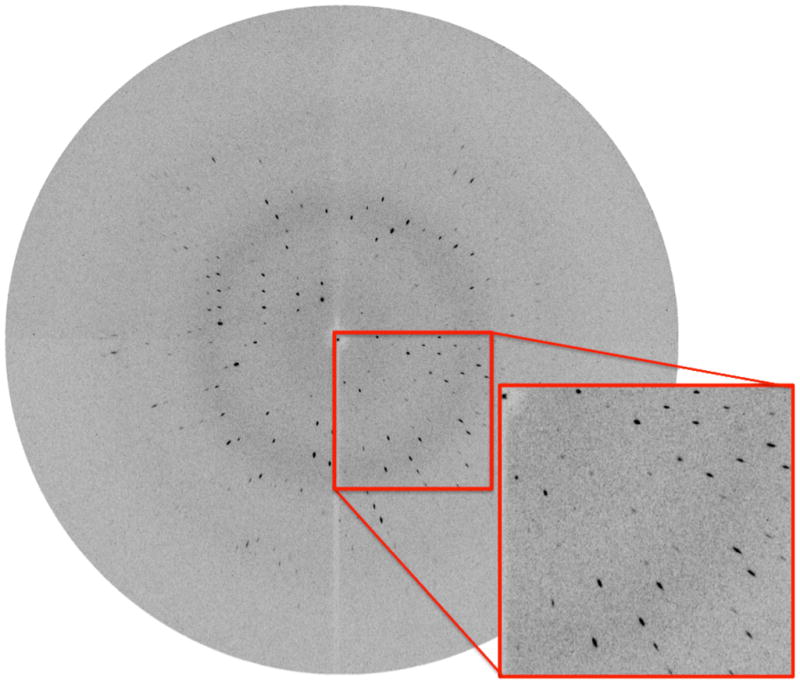
Diffraction image of MytuGCSPH from the Compact Light Source. The lower right part is magnified. The enhanced radial portion of the mosaic spread is caused by the 1.4% spread of wavelengths used for data collection.
Acknowledgments
We gratefully acknowledge NIH funding for the developments included in this paper. The prototype CLS was funded by the NIGMS under grant R44-GM66511. The X-ray station and optics development was funded by the NIGMS and NCRR under grant R44-GM074437. The development of the ‘Beta” CLS was funded by the NIGMS and NCRR with have co-sponsored the Accelerated Technologies Center for Gene to 3D Structure (www.atcg3d.org), a PSI-2 Specialized Center, under Grant U54 GM074961. The production of MytuGCSPH protein and crystals was funded by NIAID under Federal Contract No. HHSN272200700057C which supports the Seattle Structural Genomics Center for Infectious Disease (www.SSGCID.org). Special thanks to Shellie Dieterich, Becky Poplawski, and Jeff Christensen at Emerald BioStructures for their support in molecular biology and protein crystallization. The authors also wish to thank the organizing committee of the PSI organized Enabling Technologies Meetings held each spring on the NIH campus. LTI collaborator wish to thank the staff at LTI, Michael Blum, Jens Als-Nielsen, Anette Jensen and Bill Weis.
References
- ASLANIDIS C, DE JONG PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acid Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BECH M. PhD Dissertation. University of Copenhagen; Denmark: 2009a. X-ray imaging with a grating interferometer. [Google Scholar]
- BECH M, BUNK O, DAVID C, RUTH R, RIFKIN J, LOEWEN R, FEIDENHANS’L R, PFEIFFER F. Hard X-ray phase-contrast imaging with the Compact Light Source based on inverse Compton X-rays. J Synchrotron Rad. 2009b;16:43–47. doi: 10.1107/S090904950803464X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COHEN-ADDAD C, FAURE M, NEUBERGER M, OBER R, SIEKER L, BOURGUIGNON J, MACHEREL D, DOUCE R. Structural studies of the glycine decarboxylation complex from pea leaf mitrochondria. Biochimie. 1997;79:637–644. doi: 10.1016/s0300-9084(97)83496-7. [DOI] [PubMed] [Google Scholar]
- EMSLEY P, LOHKAMP B, SCOTT W, COWTAN K. Features and Development of Coot. Acta Crystallographica Section D-Biological Crystallography. 2010 doi: 10.1107/S0907444910007493. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAURE M, BOURGUIGNON J, NEUBURGER M, MACHEREL D, SIEKER L, OBER R, KAHN R, COHEN-ADDAD C, DOUCE R. Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the glycine decarboxylase multienzyme system. Eur J Biochem. 2000;267:2890–2898. doi: 10.1046/j.1432-1033.2000.01330.x. [DOI] [PubMed] [Google Scholar]
- HENDRICKSON WA, SMITH JL, SHERIFF S. Direct phase determination based on anomalous scattering. Methods Enzymol. 1985;115:41–55. doi: 10.1016/0076-6879(85)15006-8. [DOI] [PubMed] [Google Scholar]
- HUANG Z, RUTH RD. Laser-Electron Storage Ring. Phys Rev Lett. 1998;80:976–979. [Google Scholar]
- JENSEN A. PhD Dissertation. University of Copenhagen; Denmark: 2007. Optical elements for hard X-ray radiation. [Google Scholar]
- KABSCH W. Automatic indexing of rotation diffraction patterns. J Appl Cryst. 1988;21:67–72. [Google Scholar]
- KIKUCHI G, MOTOKAWA Y, TOSHIDA T, HIRAGA K. Glycine cleavage system: reaction mechanism, pysiological significance, and hyperglycinemia. Proc Jpn Acad, Ser B. 2008;84:246–262. doi: 10.2183/pjab/84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KRISSINEL E, HENRICK K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2256–2658. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- LOEWEN R. PhD Dissertation. SLAC/Stanford University, SLAC-R-632; 2003. A compact light source: design and technical feasibility study of a laser electron storage ring x-ray source. [Google Scholar]
- MACHEREL DBJ, FOREST E, FUARE M, COHEN-ADDAD C, DOUCE R. Expression, lypoylation and structure determination of recombinant pea H-protein in Eschericia coli. Eur J Biochem. 1996;236:27–33. doi: 10.1111/j.1432-1033.1996.00027.x. [DOI] [PubMed] [Google Scholar]
- MCCOY AJ, GROSSE-KUNSTLEVE RW, ADAMS PD, WINN MD, STORONI LC, READ RJ. Phaser Crystallographic Software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURSHUDOV GN, VAGIN AA, DODSON EJ. Refinement of macromolecuar structures by the Maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- NAKAI T, ISHIJIMA J, MASUI R, KURAMITSU S, KAMIYA N. Structure of Thermus thermophilus HB8 H-protein of the glycine-cleavage system, resolved by a six-dimensional molecular-replacement method. Acta Crystallographica Section D-Biological Crystallography. 2003;59:1610–1618. doi: 10.1107/s0907444903014975. [DOI] [PubMed] [Google Scholar]
- NEWMAN J, EGAN D, WALTER TS, MEGED R, BERRY I, BEN JELLOUL M, STUART DI, PERRAKIS A. Towards rationalization of crystallization screening for small-to medium-sized academic larboartories: the PACT/JCSG+ strategy. Acta Crystallographica Section D-Biological Crystallography. 2005;61:1426–1431. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- OTWINOWSKI Z, MINOR W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- PARES S, COHEN-ADDAD C, SIEKER L, NEUBURGER M, DOUCE R. X-ray structure determination at 2.6AA resolution of a lipoate-containing protein: The H-protein of the glycine decarboxylase complex from pea leaves. Proc Natl Acad Sci USA. 1994;91:4850–4853. doi: 10.1073/pnas.91.11.4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PFEIFFER F, BECH M, BUNK O, KRAFT P, EIKENBERRY EF, BRÖNNIMANN C, GRÜNZWEIG C, DAVID C. Hard-X-ray dark-field imaging using a grating interferometer. Nature Materials. 2008;7:134–137. doi: 10.1038/nmat2096. [DOI] [PubMed] [Google Scholar]
- STUDIER FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]