Abstract
Transcription of protein-coding genes in Leishmania major and other trypanosomatids differs from that in most eukaryotes and bioinformatic analyses have failed to identify several components of the RNA polymerase (RNAP) complexes. To increase our knowledge about this basic cellular process, we used tandem affinity purification (TAP) to identify subunits of RNAP II and III. Mass spectrometric analysis of the complexes co-purified with TAP-tagged LmRPB2 (encoded by LmjF31.0160) identified seven RNAP II subunits: RPB1, RPB2, RPB3, RPB5, RPB7, RPB10 and RPB11. With the exception of RPB10 and RPB11, and the addition of RPB8, these were also identified using TAP-tagged constructs of one (encoded by LmjF34.0890) of the two LmRPB6 orthologues. The latter experiments also identified the RNAP III subunits RPC1 (C160), RPC2 (C128), RPC3 (C82), RPC4 (C53), RPC5 (C37), RPC6 (C34), RPC9 (C17), RPAC1 (AC40) and RPAC2 (AC19). Significantly, the complexes precipitated by TAP-tagged LmRPB6 did not contain any RNAP I-specific subunits, suggesting that, unlike in other eukaryotes, LmRPB6 is not shared by all three polymerases but is restricted to RNAP II and III, while the LmRPB6z (encoded by LmjF25.0140) isoform is limited to RNAP I. Similarly, we identified peptides from only one (encoded by LmjF18.0780) of the two RPB5 orthologues and one (LmjF13.1120) of the two RPB10 orthologues, suggesting that LmRPB5z (LmjF18.0790) and LmRPB10z (LmjF13.1120) are also restricted to RNAP I. In addition to these RNAP subunits, we also identified a number of other proteins that co-purified with the RNAP II and III complexes, including a potential transcription factor, several histones, an ATPase involved in chromosome segregation, an endonuclease, four helicases, RNA splicing factor PTSR-1, at least two RNA binding proteins and several proteins of unknown function.
Keywords: Leishmania, Transcription, RNA polymerase, Protein purification
1. Introduction
Leishmania is a parasitic protozoan and a member of the Trypanosomatidae family, which includes other parasites such as Trypanosoma and Leptomonas. The numerous human-infective Leishmania species cause a spectrum of disease ranging from asymptomatic to lethal, resulting in widespread human suffering and death, as well as considerable economic loss (Murray et al., 2005). Leishmania and other trypanosomatids possess unique mechanisms of gene expression (Clayton, 2002; Campbell et al., 2003). Transcription in these organisms initiates at only a few regions on each chromosome (Martinez-Calvillo et al., 2003, 2004) and mature nuclear mRNAs are generated from the polycistronic transcripts by trans-splicing, a process that adjoins a 39-nucleotide capped spliced-leader (SL) to the 5′ end of all the mRNAs (Parsons et al., 1984). Post-transcriptional mechanisms appear to regulate the steady-state levels of most of the mRNAs (Clayton, 2002).
In eukaryotic cells there are three distinct classes of nuclear RNA polymerase (RNAP): RNAP I, II and III. Each class of polymerase is responsible for the synthesis of a different kind of RNA: RNAP I is involved in the production of 18S, 5.8S and 28S rRNAs; RNAP II participates in the generation of mRNAs and most of the small nuclear RNAs (snRNAs); while RNAP III synthesizes small essential RNAs, such as tRNAs, 5S rRNA and some snRNAs (Lee and Young, 2000; Paule and White, 2000). RNAP II is the least complex of the RNA polymerases as it contains only 12 subunits in yeast, compared with 14 in RNAP I and 17 in RNAP III. Five subunits (RPB5, RPB6, RPB8, RPB10 and RPB12) are shared between all three RNAPs; two (RPAC1 and RPAC2, also known as AC40 and AC19, respectively) are shared between RNAP I and III, with homologues (RPB3 and RPB11, respectively) in RNAP II; and another five (RPA1/RPB1/RPC1, RPA2/RPB2/RPC2, RPA43/RPB7/RPC8, RPA14/RPB4/RPC9 and RPA12/RPB9/RPC10) are homologous subunits. In addition, two subunits (RPA49 and RPA34) are RNAP I-specific, while five (RPC3, RPC4, RPC5, RPC6 and RPC7) are exclusive to RNAP III (Geiduschek and Kassavetis, 2001; Hu et al., 2002). The five core subunits are homologous to the bacterial β′ (RPB1), β (RPB2), α (RPB3 and RPB11) and ω (RPB6) subunits, and correspond to the archaeal A′+A′′, B′+B′′, D, L and K subunits, respectively. The remaining shared and homologous subunits (except for RPB8) all have archaeal, but not bacterial, homologs.
The presence of all three RNAPs in Trypanosoma brucei has been demonstrated by Mono-Q anion exchange chromatography and nuclear run-on experiments with polymerase inhibitors (Grondal et al., 1989), while in Leishmania, they have been separated by a combination of carboxymethyl-sephadex and diethylaminoethyl-sephadex chromatography (Sadhukhan et al., 1997). BLAST analysis of the recently sequenced Leishmania major, T. brucei and Trypanosoma cruzi (Tritryp) genomes (Ivens et al., 2005) revealed the presence of all the shared and homologous subunits, with the exception of RPB12, RPA43 and RPA14/RPB4/RPC9, but most of the RNAP-specific subunits were not identified. Subsequent, more detailed analysis (Kelly et al., 2005) identified likely RPB4 and RPB12 orthologues. Interestingly, the Tritryp genomes possess at least two copies of the genes encoding RPB5, RPB6 and RPB10 (Ivens et al., 2005; Kelly et al., 2005; Nguyen et al., 2006). The paralogous copies are widely divergent, suggesting that the subunits they encode may not be shared by the different RNAP complexes, as they are in other organisms. Immunoprecipitation using protein C-tagged RPA1 (Schimanski et al., 2003; Nguyen et al., 2006) and protein A-tagged RPA12 (Walgraffe et al., 2005) confirmed the presence of RPA1, RPA2, RPA12, RPAC1, RPAC2, RPB5z (also called 1RPB5), RPB6z (1RPB6), RPB10z (1RPB10) and RPB8 in the T. brucei RNAP I. Similar experiments using tandem affinity purification (TAP)-tagged RPB9 (Devaux et al., 2006) and RPB4 (Das et al., 2006) have recently identified the T. brucei RPB1, RPB2, RPB3, RPB4, RPB5, RPB6, RPB7, RPB8, RPB9 and RPB11 subunits and confirmed that the RNAP I and RNAP II complexes contain different subunits of RPB5.
To isolate RNAP complexes in L. major, we have performed TAP (Puig et al., 2001) using RPB2 and one of the RPB6 orthologues. Mass spectrometry analyses of the complexes co-purified with TAP-tagged RPB2 confirmed the presence of RPB1, RPB3, RPB5, RPB7, RPB10 and RPB11 subunits in the RNAP II complex. TAP-tagged RPB6 precipitated an RNAP II complex containing RPB1, RPB2, RPB3, RPB5, RPB7 and RPB8 subunits, and an RNAP III complex containing the RPC1, RPC2, RPC6, RPAC1 and RPAC2 subunits, as well as probable orthologues of the RPC3, RPC4, RPC5 and RPC9 subunits. Interestingly, we did not identify any RNAP I-specific subunits, which confirms that this RPB6 paralogue (encoded by LmjF34.0890) is restricted to RNAP II and RNAP III, while the other paralogue (LmRPB6z, encoded by LmjF25.0140) is limited to RNAP I. In addition, several other proteins, including splicing factor PTSR-1, RNA binding proteins, as well as four helicases, an ATPase involved in chromosome segregation, an endonuclease, a chromatin-specific transcription elongation factor, histones, and a number of proteins with unknown function, co-precipitated with one or both TAP-tagged proteins, suggesting that they may be physically associated with the RNAP II complex.
2. Materials and methods
2.1. Plasmid constructs
To generate vectors pB2-TAP and pB6-TAP, the REL1 gene present in plasmid pREL1Lt-TAPmod (Aphasizhev et al., 2003a) was replaced with LmjF31.0160 and LmjF34.0890, respectively. To prepare the pREL1Lt-TAPmod backbone, the vector was digested with BamHI and XbaI, and the 1.4 kb fragment (containing the REL1 gene) was eliminated by agarose gel electrophoresis. LmjF31.0160 was PCR-amplified from genomic DNA with primers LmRPB2-BamHI-5′ (5′ATATGGATCCGAGACGCACGCCTCCATGAGGTC) and LmRPB2-XbaI-3′ (5′ATATTCTAGACAGAGGACCGGTACCGAGGCGCG), and LmjF34.0890 was amplified with oligonucleotides LmRPB6-BamHI-5′ (5′ATATGGATCCTAGTCACGGCTGCAGACTTGTGG) and LmRPB6-XbaI-3′ (5′ATATTCTAGAGATGTTCGTGTAGCGCTCATCCG). The PCR products were cloned into pGEM-T Easy vector (Promega), digested with BamHI and XbaI and ligated into the pREL1Lt-TAPmod backbone. The constructs were verified by sequencing.
2.2. Culture and transfection of Leishmania
Promastigotes of L. major MHOM/IL/81/Friedlin (LmjF) were grown in supplemented RPMI 1640 medium at 26°C (Yan et al., 2002) and harvested in the mid-log phase. Electroporation with plasmid constructs and cell plating were performed as previously described (Martinez-Calvillo et al., 2005).
2.3. Southern and Western blotting
Southern blot analysis of transfectant clones was carried out with genomic DNA digested with XbaI (which cuts once in the plasmids) and gene-specific probes (radiolabeled with High Prime Labeling System from Amersham). Protein samples from transfectant clones were fractionated by SDS-PAGE, blotted, probed with PAP reagent (Sigma-Aldrich) (which is specific for the Protein A domain of the TAP-tag) and developed using the ECL system (Amersham).
2.4. Extract preparation
To obtain whole-cell extracts, mid-log phase (3 L at 3-4 × 107 cells per ml) were harvested, washed once with PBS and resuspended in 14 ml of IPP-150 (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% NP40) containing two tablets of Complete EDTA-free protease inhibitors (Roche Molecular). Two ml of 10% Triton-X100 were then added and the samples incubated on ice until cells were completely lysed (∼ 20 min) and the cleared lysate was obtained by centrifugation at 12,096 g for 15 min at 4°C. Nuclear and cytoplasmic extracts were obtained by rinsing 1010 promastigotes with ice-cold PBS and resuspending the cells in 10 ml 10 mM N-2-Hydroxyehtylpiperazine-N′-2-ethanesulfonic acid (HEPES, pH 7.9), 10 mM KCl, 100 mM EDTA, 100 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA) and 1 mM dithiothreitol (DTT) along with a tablet of protease inhibitor. The cells were allowed to swell on ice for 30 min, before addition of 625 μl of the same buffer containing 10% NonidetP-40 (NP-40) and vortexing at full speed for 10 s. The samples were sonicated in an ice bath for 5 min using a microtip probe of Vcx 600 Vibra cell sonicator with pulse cycles of 5 s on and 9.9 s off at 40% amplitude. The cells were checked for lysis under the microscope and the cytoplasmic fraction was collected by centrifugation at 23,700 g for 10 min at 4°C. The nuclear pellet was resuspended in 20 mM HEPES, pH 7.9, 25% glycerol, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT with a tablet of protease inhibitors. The extract was agitated for 15 min on ice and sonicated as above. The nuclear extract was recovered by centrifugation at 12,096 g for 10 min at 4°C
2.5. TAPs
The extract (whole cell, nuclear or cytoplasmic) was added to a 20-ml disposable column containing 200 μl IgG-Sepharose beads (Pharmacia) and incubated at 4°C for 2 h with rotation. The column was drained by gravity flow and washed three times with 20 ml of cold IPP-150. The sample was then digested with 100 units of acTEV protease (Invitrogen) for 2 h at room temperature in 1 ml of TEV cleavage buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1 % NP40, 0.5 mM EDTA, 1 mM DTT). The IgG-Sepharose was drained and the eluted material mixed with 3.5 ml of CBB (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1 % NP40, 10 mM β-ME, 1 mM Mg acetate, 1 mM imidazole, 2 mM CaCl2) and 10 μl 1 M CaCl2, before transfer to a 5-ml column containing 200 μl of Calmodulin resin (Strategene). Samples were incubated at 4°C for 2 h with rotation, washed six times with 5 ml of CBB and eluted with 1 ml of CEB (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% NP40, 10 mM β-ME, 1 mM Mg acetate, 1 mM imidazole, 2 mM EGTA).
2.6. Mass spectrometry analysis
Eluted proteins from TAP-tag purification were concentrated by centrifugation under vacuum and analyzed by SDS-PAGE and SYPRO Ruby (Molecular Probes) staining. Individual lanes from the gels were sliced into three to six pieces and proteins subjected to in-gel tryptic digestion (Shevchenko et al., 1996) prior to liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS). Alternatively, concentrated protein eluates were digested with trypsin without prior SDS-PAGE fractionation. Peptides were submitted for mass spectrometry analysis using either a Micromass Q-TOF Tandem Hybrid Mass Spectrometer (at the Department of Medical Chemistry of the University of Washington), a Thermo Electron LCQ DECA XP or an LTQ Linear Ion Trap (SBRI Proteomics Core). The collision induced dissociation (CID) spectra were compared with a L. major protein database (version 5.2, downloaded from GeneDB) using TurboSequest software, and protein matches determined using PeptideProphet and ProteinProphet (Keller et al., 2002; Nesvizhskii et al., 2003).
3. Results
3.1. Tritryp RPB2 and RPB6 orthologues
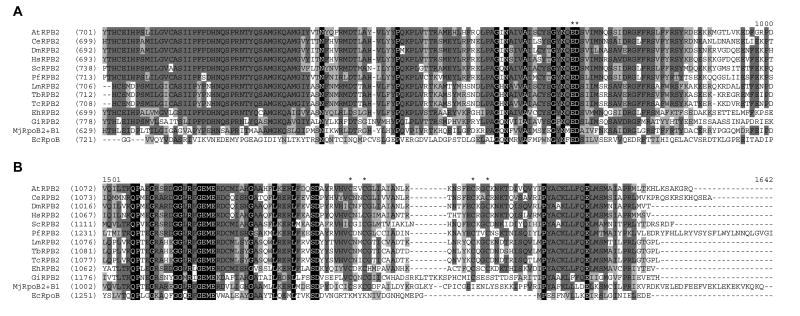
The second largest subunit of RNAP II, RPB2, is involved not only in initiation and elongation of transcription, but also in RNAP II processivity (Mason and Struhl, 2005). Blast searches of the Tritryp genomes revealed a clear orthologue in all three species (LmjF31.0160, Tb927.4.3810, and Tc00.1047053509951.49), with 33-46% sequence identity (50-66% similarity) spread throughout the entire length of the protein (Supplementary Fig. S1). The ED residues that correspond to part of the RNAP II active site are conserved in the Tritryp RPB2, as are the four cysteine residues forming the C-terminal zinc-binding domain (Fig. 1), and the nine regions (A to I) that are well-conserved in RPB2 from many different species (Sweetser et al., 1987) are also conserved in the Tritryp proteins (Supplementary Fig. S1A). The L. major and T. brucei/T. cruzi orthologues show ∼80% sequence identity, with an additional 10% similarity, and phylogenetic analyses indicate that the Tritryp orthologues branched from the eukaryotic crown lineage before the split between fungi and metazoa, as did those in other parasitic protozoa, such as Giardia and Entamoeba (Fig. 2A).
Fig. 1.
Sequence conservation of RPB2 subunits from several species. Protein sequences of RPB2 homologues from Arabidopsis thaliana (At), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs), Saccharomyces cerevisiae (Sc), Plasmodium falciparum (Pf), Leishmania major (Lm), Trypanosoma brucei (Tb), Trypanosoma cruzi (Tc), Entamoeba histolytica (Eh), Giardia intestinalis (Gi), Methanococcus jannaschii subunits B2 and B1 (MjRpoB2+B1) and Escherichia coli RNA polymerase subunit β (EcRpoB) were aligned using the default setting of AlignX module of VectorNTI (Invitrogen). Multiple sequence alignment of the regions surrounding the active site and C-terminal zinc-binding domain are showing in panels A and B, respectively. Asterisks above the sequence indicate conserved amino acids implicated in these functions. Amino acid residues conserved in all sequences are shaded in black; those conserved in more than 50% of the sequences are shaded in dark grey, while those that which show conservative replacement are shaded in light grey.
Fig. 2.
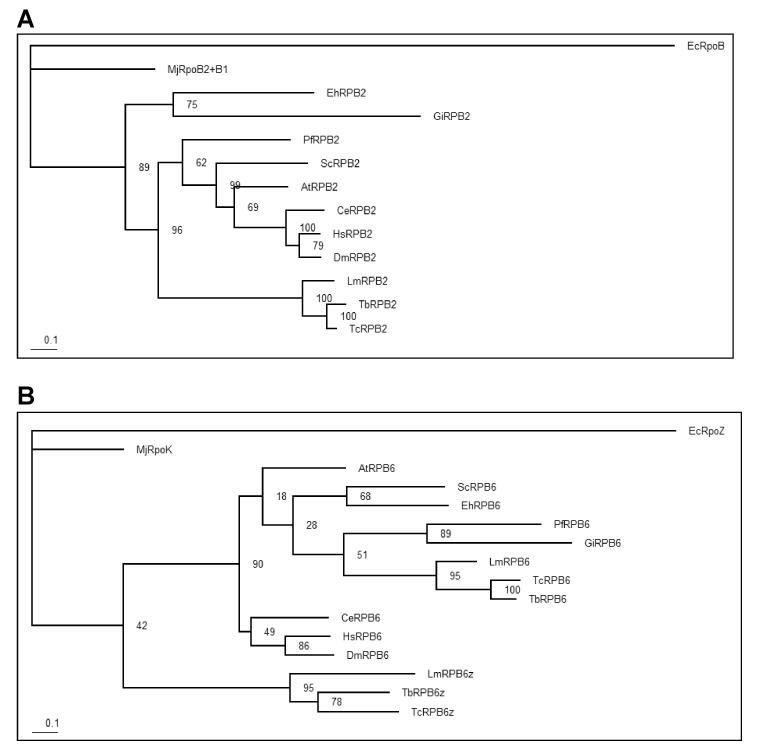
Phylogenetic analysis of RPB2 and RPB6 orthologues. Phylogenetic trees were constructed from the RPB2 (panel A) and RPB6 (panel B) sequence alignments shown in Supplementary Figs. 1 and 2, respectively, and rooted with the corresponding Escherichia coli sequence. Bootstrap values were obtained from 100 replicates on the Phyml web-site (http://atgc.lirmm.fr/phyml/)(Guindon et al., 2005) using the default settings.
The L. major genome contains two genes at different loci (LmjF25.0140 and LmjF34.0890) encoding distinct isoforms of RPB6 (Ivens et al., 2005). The T. brucei genome contains a single syntenic orthologue of the former but has two copies of the latter as part of tandem duplication with an adjacent gene. The situation in T. cruzi is similar to that in T. brucei, but one haplotype may contain three copies of the second RPB6 gene. In a previous report describing these isoforms in T. brucei (Kelly et al., 2005), the smaller isoform (corresponding to LmjF25.0140) was referred to as TbRBP6z and we shall continue to use that terminology, although in a more recent publication (Nguyen et al., 2006) the RPB6z and RPB6 paralogues have been called 1RPB6 and 2RPB6, respectively. Eukaryotic RPB6 is structurally and functionally equivalent to bacterial RNAP subunit ω and archaeal subunit K, with the greatest similarity falling within three regions referred to as CR1, CR2 and CR3 (Minakhin et al., 2001) (Supplementary Fig. S2A). Sequence identity of LmRPB6 ranges from 66% and 67% for the T. brucei and T. cruzi orthologues, respectively; 39-53% for other eukaryotes; 35% for the archaeal subunit K; and 18% for the Escherichia coli subunit ω (Fig. 2B). For LmRPB6z, these values range from 46% for T. brucei and T. cruzi, to 26-33% for other eukaryotes, 34% for the archaeal subunit K and 13% for the E. coli subunit ω. Phylogenetic analysis revealed that LmRPB6 (encoded by LmjF34.0890) has greater similarity to the RPB6 subunits from most other eukaryotes than LmRPB6z (LmjF25.0140), although the bootstrap evidence for this was somewhat marginal (Fig. 2B). Indeed, the LmRPB6 and LmRPB6z isoforms show only 26% identity, indicating that their divergence long precedes that of the trypanosomatid speciation and appears to have taken place near the base of the eukaryotic lineage. RPB6z has a shorter N-terminal region and an insertion of charged amino acids between CR2 and CR3 (Supplementary Fig. S2A). Interestingly, a similar insertion was previously observed in RPB6 from Plasmodium falciparum (Minakhin et al., 2001). The crystallographic structure of subunit ω from Thermus aquaticus revealed that conserved regions CR1 and CR2 correspond to specific secondary-structure elements; CR1 corresponds, almost precisely, to α-helix 2; the first half of CR3 correspond to α-helix 3 and the second half correspond to β-strand 1 (Minakhin et al., 2001). Similar structures are also present in the eukaryotic RPB6 subunits, including both Tritryp orthologues (Fig. 2A). A putative leucine-zipper domain, previously observed in RPB6 from yeast (Acker et al., 1994), was found in the Tritryp RPB6 but not in RPB6z. However, this sequence was not conserved in several other species, raising questions about its functional significance.
3.2. Identification of RNAP II subunits with TAP-tagged LmRPB2
In order to isolate transcriptional complexes from promastigotes of L. major, we carried out the TAP strategy (Puig et al., 2001), which allows the purification of proteins interacting with a given target protein under native conditions. Two cloned recombinant cell lines were obtained after transformation of L. major with the construct pB2-TAP, which contains the LmRPB2 gene with a C-terminal TAP tag (see Materials and methods). Southern blot analysis using the LmRPB2 probe showed the expected 10.5 kb XbaI fragment from multicopy episomal DNA in both clones, as well as the 77 kb fragment from chr31 that was also present in the parental (WT) cells (Fig. 3A). Western blotting using the PAP reagent, which is specific for the Protein A domain of the TAP-tag, showed a protein of ∼156 kDa in both cell lines, as expected from fusion of the 22 kDa TAP-tag to the 134 kDa LmRPB2 (Fig. 3B).
Fig. 3.
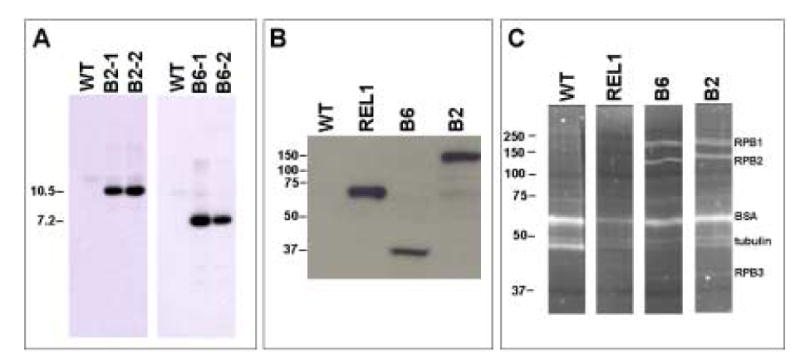
Molecular characterization of pB2-tandem assisted purification (TAP) and pB6-TAP transfectants. A) Southern blot analysis of the parental cell line (WT) and two clones each obtained after transfection of Leishmania major with pB2-TAP (B2-1 and B2-2) and pB6-TAP (B6-1 and B6-2). The genomic DNA was digested with XbaI, which cuts once in the vector, and the probe corresponded to part of the coding regions of LmRPB2 and LmRPB6, respectively. The sizes of the XbaI fragments (in kb) from the constructs are indicated to the left. B) Western blot analysis of cloned cell lines expressing TAP-tagged LmRPB2, LmRPB6, LtREL1, as well as the parental cell line (WT). The TAP-tag was detected with PAP reagent (which is specific for the Protein A domain of the TAP-tag). Molecular size markers (in kDa) are indicated to the left. C) SDS-PAGE analysis of the affinity purified complexes from nuclear extracts of clones expressing LmRPB2-TAP and LmRPB6-TAP, as well as the parental cell line (WT) and LtREL1-TAP. Proteins were stained with Sypro Ruby and the molecular size markers (in kDa) are indicated to the left. Proteins identified by liquid chromatography tandem mass spectrometry analysis are indicated on the right.
SDS-PAGE analysis of the TAP-tagged complexes isolated from total cell lysates of clone B2-1 showed several proteins that co-eluted with the LmRPB2-TAP fusion protein (Fig. 3C). LC-MS/MS analysis identified the major bands as LmRPB1 (LmjF31.2610), LmRPB2-TAP (LmjF31.0160) and LmRPB3 (LmjF29.0140), in addition to BSA and tubulin contaminants that were also present in the control (mock TAP using wild type nuclear extract). Interestingly, LmRPB1 appeared to migrate as a doublet (Fig. 3C). We also identified peptides from another four RNAP II subunits: LmRPB5 (LmjF18.0780), LmRPB7 (LmjF32.1280), LmRPB10 (LmjF13.1120) and LmRPB11 (LmjF27.1550) (see Table 1). Several other proteins were also identified in the TAP-tagged samples. While most of these represented likely contaminants such as heat shock protein, ribosomal proteins and translation factors, as well as several mitochondrial proteins; at least five other proteins appeared to co-purify with the RNAP II complex, since they were each represented by two or more different peptides (Table 2). The most interesting was a protein with sequence similarity to the large subunit of the metazoan FACT (facilitates chromatin transcription) complex, encoded by LmjF29.0020. LmjF32.0750 encodes a protein that contains two RNA recognition motifs and has weak similarity to poly (A)-binding proteins in other organisms and LmjF07.0870 encodes splicing factor PTSR-1. Another two probable RNA binding proteins (LmjF35.2200 and LmjF34.2700) were also identified but by only a single peptide each. The proteins encoded by LmjF17.0010 and LmjF34.0930 have no distinguishing features and the former appears to be trypanosomatid-specific.
Table 1. Protein components of RNA polymerase II and III complexes.
| Complex | Saccharomyces cerevisiae | Sequence conservation | Leishmania major geneb | Predicted sizea | Peptides identifiedc | |||
|---|---|---|---|---|---|---|---|---|
| subunit | sizea | name | systematic ID | LmRPB2 | LmRPB6 | |||
| RNAP II | B220 | 192 | +++ | RPB1 | LmjF31.2610 | 184.7 | 29(6) | 17(6) |
| B150 | 139 | +++ | RPB2 | LmjF31.0160 | 133.8 | 21(6) | 15(5) | |
| B44 | 35 | ++ | RPB3 | LmjF29.0140 | 37.5 | 7(6) | 5(3) | |
| B32 | 25 | + | RPB4 | (LmjF29.0400) | 18.0 | |||
| B16 | 19 | ++ | RPB7 | LmjF32.1280 | 21.9 | 2(2) | 1(1) | |
| B12.6 | 14 | ++ | RPB9 | LmjF28.0120 | 15.6 | |||
| B12.5 | 14 | ++ | RPB11 | LmjF27.1550 | 14.8 | 4(3) | ||
| Sharedd | ABC27 | 25 | ++ | RPB5 | LmjF18.0780 | 27.2 | 4(4) | 4(3) |
| RPB5z | LmjF18.0790 | 37.6 | ||||||
| ABC23 | 18 | ++ | RPB6 | LmjF34.0890 | 17.5 | 1(1) | ||
| RPB6z | LmjF25.0140 | 14.2 | ||||||
| ABC14.5 | 17 | + | RPB8 | LmjF28.0810 | 16.5 | 1(1)e | ||
| ABC10α | 8 | ++ | RPB12 | (LmjF20.0490) | 17.1 | |||
| ABC10β | 8 | ++ | RPB10 | LmjF13.1120 | 9.6 | 1(1) | ||
| RPB10z | LmjF25.1315f | 9.8 | ||||||
| RNAP III | C160 | 162 | +++ | RPC1 | LmjF34.0360 | 173.6 | 8(1) | |
| C128 | 129 | +++ | RPC2 | LmjF20.0010 | 126.8 | 4(1) | ||
| C82 | 74 | + | RPC3 | (LmjF27.2600) | 65.4 | 3(1) | ||
| (LmjF02.0680g) | 41.4 | |||||||
| C53 | 46 | + | RPC4 | (LmjF35.4170) | 37.9 | 1(1)h | ||
| C37 | 32 | + | RPC5 | (LmjF13.1370) | 76.5 | 4(1) | ||
| C34 | 36 | ++ | RPC6 | LmjF29.1320 | 48.1 | 1(1) | ||
| C31 | 28 | + | RPC7 | NFi | ||||
| C25 | 24 | ++ | RPC8 | LmjF09.1060 | 27.0 | |||
| C17 | 18 | + | RPC9 | (LmjF03.0790) | 45.4 | 2(1) | ||
| C11 | 12 | ++ | RPC10 | LmjF19.0420 | 16.3 | |||
| Sharedj | AC40 | 37 | +++ | RPAC1 | LmjF19.0660 | 47.3 | 2(1) | |
| AC19 | 16 | ++ | RPAC2 | LmjF28.2060 | 15.2 | 1(1) | ||
Molecular weight in kDa
Genes in parenthesis show weak similarity by Blast
The first number indicates the number of different peptides detected with each of the tandem affinity purification-tagged constructs, the number in parenthesis shows the number of experiments in which peptides were detected
Subunits shared between RNAPI, II and III in yeast and human
This peptide was identified by manual analysis of the Sequest output files, not by PeptideProphet.
Not in original L. major genome annotation; found by tBlastN analysis using Tb927.3.1250
LmjF02.0680 is probably a truncated pseudogene
Only a non-tryptic peptide was detected
Leishmania major orthologue not yet identified
Subunits shared between RNAPI and III in yeast and human
Table 2. Proteins co-purifying with RNA polymerase II and III complexesa.
| Leishmania major gene | Predicted sizeb | Peptides detectedc | Possible function | |
|---|---|---|---|---|
| LmRPB2 | LMRPB6 | |||
| LmjF32.0750 | 25.1 | 6(2) | 3(3) | RNA binding protein |
| LmjF17.0010 | 87.6 | 3(2) | 3(4) | Hypothetical protein; Tryp-specific |
| LmjF07.0870 | 41.8 | 3(2) | 2(2) | Splicing factor PTSR-1 |
| LmjF29.0020 | 114.8 | 3(1) | 3(2) | Transcription elongation factor (FACT 140) |
| LmjF10.0870d | 14.6 | 4(1) | 3(1) | Histone H3 |
| LmjF34.0930 | 12.5 | 2(2) | 3(1) | Hypothetical protein; conserved |
| LmjF06.0010 | 11.3 | 1(1) | 4(2) | Histone H4 |
| LmjF14.1440 | 103.9 | 1(1) | 3(2) | Hypothetical protein; Tryp-specific |
| LmjF25.0590 | 19.7 | 1(1) | 3(2) | Hypothetical protein; Tryp-specific |
| LmjF08.0430 | 100.1 | 1(1) | 2(1) | Hypothetical protein; Tryp-specific |
| LmjF30.2620 | 77.9 | 1(2) | 1(1) | Hypothetical protein; Tryp-specific |
| LmjF08.1222e | 11.4 | 1(1) | 1(1) | Hypothetical protein; Tryp-specific |
| LmjF32.0400 | 66.8 | 1(1) | 1(1) | DEAD/H box RNA helicase |
| LmjF29.2090 | 175.9 | 7(1) | Helicase | |
| LmjF35.1310f | 11.4 | 4(2) | Histone H4 | |
| LmjF09.1340g | 12.2 | 4(1) | Histone H2B | |
| LmjF10.0160 | 54.6 | 3(1) | Chromosome segregation ATPase? | |
| LmjF35.0370 | 46.4 | 2(1) | DEAD box RNA Helicase | |
| LmjF27.0840 | 95.3 | 2(2) | Hypothetical protein; conserved | |
| LmjF25.1550 | 11.6 | 2(2) | Hypothetical protein; Tryp-specific | |
| LmjF36.3050 | 15.8 | 2(2) | Hypothetical protein; Tryp-specific | |
| LmjF34.4100 | 35.2 | 2(2) | Hypothetical protein; Tryp-specific | |
| LmjF07.1110 | 40.5 | 2(2) | Hypothetical protein; Tryp-specific | |
| LmjF09.0050 | 29.4 | 2(1) | Endonuclease III | |
| LmjF07.0220 | 67.1 | 2(1) | Hypothetical protein; Tryp-specific | |
| LmjF26.1560 | 177.1 | 1(2) | Helicase | |
| LmjF32.0150 | 111.0 | 1(2) | Small nucleolar ribonucleoprotein? | |
Only proteins with more than one unique peptide detected or the same peptide detected in more than one experiment are shown. Proteins likely to be contaminants (including several ribosomal proteins, translation factors, heat shock proteins, α- and β-tubulin, paraflagellar rod protein and a number of mitochondrial proteins) were not included.
in kDa
The first number indicates the number of unique peptides detected with each of the tandem affinity purification-tagged constructs, the number in parenthesis shows the total number of experiments the peptide was observed in.
Also matches LmjF10.0990, LmjF16.0575, LmjF16.0570 and LmjF16.0610
Also matches LmjF08.1260
Also matches LmjF31.3180, LmjF36.0020, LmjF02.0020 and LmjF25.2450
Also matches LmjF19.0050, LmjF19.0030 and LmjF19.0040
3.3. Identification of RNAP II and III subunits with TAP-tagged LmRPB6
Southern blot analysis of two cloned recombinant cell lines obtained after transformation of L. major with the construct pB6-TAP (see Materials and methods) showed the 7.2 kb XbaI fragment expected from the episomal copy of LmRPB6, as well as the 35 kb chromosomal fragment (Fig. 3A). Western blot analysis confirmed the presence of the expected 39.8 kDa LmRPB6-TAP fusion protein in both cell lines (Fig. 3B). SDS-PAGE analysis of the protein complexes co-purified with this protein from the B6-1 cell line revealed a similar pattern to that seen for LmRPB2-TAP (Fig. 3C). Once again, LC-MS/MS analysis of these samples identified the major bands as LmRPB1 (LmjF31.2610), LmRPB2 (LmjF31.0160) and LmRPB3 (LmjF29.0140). In this case, the LmRPB2 band migrated with a lower molecular weight due to the absence of the TAP-tag. These analyses also revealed peptides for LmRPB5 (LmjF18.0780), LmRPB7 (LmjF32.1280) and LmRPB8 (LmjF28.0810), in addition to LmRPB6-TAP (LmjF34.0890). In addition, we also identified peptides from the RNAP III-specific subunits, LmRPC1 (LmjF34.0360), LmRPC2 (LmjF20.0010) and RPC6 (LmjF29.1320), as well as two subunits shared between RNAP I and RNAP III, LmRPAC1 (LmjF19.0660) and LmRPAC2 (LmjF28.2060) (Table 1).
Peptides were also detected from several additional proteins that appear to be excellent candidates for orthologues of RPC3 (encoded by LmjF27.2600), RPC5 (LmjF13.1370) and RPC9 (LmjF03.0790), none of which had been previously identified in the Tritryps by Blast analysis. LmjF27.2600 encodes a 605-amino acid protein, with orthologues in T. brucei (Tb927.2.2990) and T. cruzi (Tc00.1047053508737.140 and Tc00.1047053509127.40) that have 28-32% sequence identity and 50-53% similarity (Supplementary Fig. S3). Interestingly, L. major has a second copy of this gene on chr2 (LmjF02.0680) which encodes a smaller protein (377-amino acids) than LmjF27.2600 (and the trypanosome orthologues) due to a substantial N-terminal deletion, and also contains a C-terminal extension due to a frameshift (Supplementary Fig. S3A). Thus, LmjF02.0680 is probably a pseudogene, even though the T. brucei and T. cruzi orthologues are syntenic with this copy, rather than LmjF27.2600. These genes appear to be a site of recombination between chr2 and chr27, since the 19 kb (containing five protein-coding genes) from this region to the telomere are virtually identical on both chromosomes. The protein encoded by LmjF27.2600 shows 10-12% identity and 28-33% similarity to RPC3 subunits from a number of eukaryotes (Supplementary Fig. S3B). This is not surprising, since the sequence conservation of eukaryotic RPC3 is quite low, with only 16% identity and 34% similarity between the human and yeast (Saccharomyces cerevisiae) proteins (Supplementary Fig. S3B). Indeed, only four amino acids are identical in all six non-trypanosomatid species for which orthologues have been identified and only one (a glutamine at position 423 in LmRPC3) is conserved in the Tritryps.
The sequence conservation of RPC5 is similarly low, with the putative L. major orthologue (encoded by LmjF13.1370) showing only 11% identity and 27% similarity to human RPC5 and 11% identity and 36% similarity to S. cerevisiae RPC37 (Fig. S4). This sequence conservation is largely confined to the N-terminal portion of the proteins, with the C-terminal portion showing very little sequence conservation. Indeed, there is also considerable C-terminal size variation, with the Tritryp sequences being longer than those in other species. Only two amino acids are conserved in all eight non-trypanosomatid species and both are also conserved in the Tritryps (Supplementary Fig. S4A). As expected, the T. brucei (Tb11.02.0970) and T. cruzi (Tc00.1047053507629.10) orthologues showed higher sequence conservation to LmRPC5 (41-46% identity and 62-63% similarity).
Sequence conservation of RPC9 is also low, with the yeast and human orthologues showing only 39% identity and 58% similarity, and orthologues of other species having even lower similarity (Supplementary Fig. S5). The putative L. major orthologue (encoded by LmjF03.0790) has only 8-16% overall identity and 26-31% similarity to other eukaryotic orthologues (Supplementary Fig. S5B), but sequence conservation within the N-terminal region is higher (Supplementary Fig. S5A). The low overall level of sequence conservation is perhaps not surprising considering that the L. major and T. brucei (Tb927.2.2700) orthologues (which are syntenic) show only 29% identity and 50% similarity (Supplementary Fig. S5B). While the Caenorhabditis elegans RPC9 sequence appears to have the most divergent sequence in the N-terminal region, it contains a large C-terminal extension that is absent in the other eukaryote proteins, but is conserved in the Tritryp proteins.
A number of other proteins were identified in the LmRB6-TAP samples, including those encoded by LmjF32.0750 (RNA binding protein), LmjF17.0010 (Tryp-specific protein of unknown function), LmjF07.0870 (PTSR-1), LmjF29.0020 (FACT140) and LmjF34.0930 (conserved protein of unknown function) which were described above for LmRPB2-TAP (Table 2). Several other proteins that also co-purified with LmRPB2-TAP were identified; including histone H4 (encoded by LmjF06.0010), an RNA helicase (encoded by LmjF32.0400) and several Tryp-specific proteins of unknown function (encoded by LmjF14.1440, LmjF25.0590, LmjF08.0430, LmjF30.2620, and LmjF08.1222/LmjF08.1260). In addition, multiple peptides were identified for other histone H4 paralogues, as well as histones H2B and H3, three helicases (encoded by LmjF29.2090, LmjF25.0370, and LmjF26.1560), a small nucleolar ribonucleoprotein (LmjF32.0150), a putative ATPase involved in chromosome segregation (LmjF10.0160), an endonuclease (LmjF09.0050) and six proteins with unknown function (encoded by LmjF27.0840, LmjF25.1550, LmjF36.3050, LmjF34.4100, LmjF07.1110, and LmjF07.0220), as well as several peptides for a number of likely contaminants including α- and β-tubulin, heat shock protein, mitochondrial and ribosomal proteins, translation factor eIF-3 subunit 8 and paraflagellar rod protein 1D.
Interestingly, no RNAP I-specific subunits co-precipitated with LmRPB6-TAP, even though RPB6 is shared between the three RNAP complexes in other organisms. This suggests that in L. major (and probably other trypanosomatids), the RPB6 isoform that we used (encoded by LmjF34.0890) is restricted to RNAP II and III, while the other isoform (LmRPB6z, encoded by LmjF25.0140) is restricted to RNAP I. Similarly, multiple peptides from only one isoform of LmRPB5 (encoded by LmjF18.1780) were seen in both the LmRPB2-TAP and LmRPB6-TAP complexes, and no peptides from the other isoform (encoded by LmjF18.0790) were observed.
4. Discussion
Gene organization and transcriptional processes in Leishmania and other trypanosomatids are unusual compared with other eukaryotes, suggesting that their transcriptional machinery may also differ from that in other organisms (Campbell et al., 2003; Clayton, 2002; Martinez-Calvillo et al., 2003). In support of this hypothesis, Blast analyses of the Tritryp genomes failed to identify most of the general transcription factors, and revealed at least two extensively divergent copies for each of the genes encoding RPB5, RPB6 and RPB10 (Ivens et al., 2005; Kelly et al., 2005; Nguyen et al., 2006). Moreover, the C-terminal domain of RPB1 (which is duplicated in T. brucei) lacks the characteristic heptapeptide repeats (Evers et al., 1989), and RPA2 presents a unique N-terminal extension domain, which may serve a parasite-specific function (Schimanski et al., 2003).
The TAP-tag protocol has been extensively used to isolate protein complexes in a variety of organisms. In T. brucei this procedure (with modifications in some cases) has been successfully used to analyze the exosome complex (Estevez et al., 2001), to identify members of the RNA editing complex (Panigrahi et al., 2003), to study the composition of RNAP I (Schimanski et al., 2003; Walgraffe et al., 2005; Nguyen et al., 2006) and RNAP II (Devaux et al., 2006; Das et al., 2006) complexes, to determine the composition of U1 small nuclear RNP (Palfi et al., 2005), and to identify proteins involved in synthesis of the spliced-leader RNA (Das et al., 2005; Schimanski et al., 2005). Similarly, in Leishmania tarentolae this protocol has been employed to analyze the editosome complex (Aphasizhev et al., 2003b). Here, we have used the TAP-tag procedure to further our knowledge about transcription in L. major by using two RNAP subunits, LmRPB2 and LmRPB6, as targets to isolate transcriptional complexes.
Only 10 of the 12 RNAP II subunits were originally identified by Blast searches in the Tritryps (Ivens et al., 2005) but the remaining two were identified by subsequent analyses (Kelly et al., 2005; Nguyen et al., 2006). In the experiments presented here, we have identified nine subunits (RPB1, RPB2, RPB3, RPB5, RPB6, RPB7, RPB8, RPB10 and RPB11) that co-purify with LmRPB2-TAP and/or LmRPB6-TAP (Table 1). This confirms two recent reports that also identified these subunits (except RPB10) in the T. brucei RNAP II complex isolated by TAP-tagging TbRPB9 (Devaux et al., 2006) or TbRPB4 (Das et al., 2006). The original Blast analyses of the Tritryp genomes revealed only 11 of the 17 RNAP III subunits (Ivens et al., 2005), with most of the RNAP III-specific subunits missing. The experiments presented here using LmRPB6-TAP confirmed the presence of six subunits (RPC1, RPC2, RPC6, RPB5, RPAC1and RPAC2), and identified putative orthologues of three previously unidentified subunits: RPC3 (LmjF27.2600), RPC5 (LmjF13.1370) and RPC9 (LmjF03.0790). Interestingly, the L. major genome contains a truncated copy (LmjF02.0680) of RPC3 that is not present in T. brucei or T. cruzi, and appears to have arisen during a recombination between the ends of chromosomes 2 and 27, which resulted in duplication of several other genes. While it is likely that this copy represents a pseudogene, our present data cannot exclude the possibility that it is present in RNAP III complexes. RPC3, RPC6 and RPC7 form a stable sub-complex which interacts with RPC1 (Flores et al., 1999) but we failed to detect RPC7, which, together with RPC4, have not yet been found in the Tritryps databases. More detailed bioinformatic analyses indicate that LmjF35.4170 is likely to encode the RPC4 orthologue on the basis of weak sequence conservation (12% identity, 31% similarity to human RPC4), especially within the most conserved, C-terminal, region of the protein (Supplementary Fig. S6). We detected a single peptide which corresponded to sequence from this protein in one experiment (see Table 1) but the peptide was not tryptic, so the identification must be regarded as uncertain.
In most other eukaryotes, five subunits (RPB5, RPB6, RPB8, RPB10 and RPB12) are shared between all three RNAPs (Geiduschek and Kassavetis, 2001; Hu et al., 2002). However, the TriTryp genomes contain two distinct paralogues of RPB5, RPB6 and RPB10 (Ivens et al., 2005; Kelly et al., 2005; Nguyen et al., 2006), and previous reports in T. brucei indicate that at least the RPB5 and RPB6 isoforms are segregated between RNAP I and RNAP II (Walgraffe et al., 2005; Nguyen et al., 2006; Devaux et al., 2006; Das et al., 2006). Our TAP-tag purifications with LmRPB6 as bait confirm these results and indicate that the RPB6 and RPB5 isoforms are found within both the RNAP II and RNAP III complexes, while the RPB6z and RPB5z isoforms are found only in RNAP I. Similarly, our experiments using TAP-tagged LmRPB2 indicate that the RPB10 isoform is associated with the RNAP II complex, while only the RPB10z isoform is associated with RNAP I in T. brucei (Nguyen et al., 2006). Thus, the RPB5, RPB6 and RPB10 subunits all appear to segregate between the RNAP I and RNAP II/III complexes in Tritryps and it appears that in all three cases, the duplication occurred very early in eukaryotic evolutionary history. Giardia intestinalis also has two isoforms of RPB5 (Seshadri et al., 2003), which show a similar level of sequence divergence. Thus, it is interesting to speculate that the different paralogues may represent ancient remnants of entirely separate RNAP complexes present in a primitive eukaryote ancestor.
In other eukaryotes, RPB6 plays an essential role in the assembly and stability of RNAP I, II and III, through specific interactions with the corresponding largest subunit (Minakhin et al., 2001; Tan et al., 2003) RPB6 has also been linked to transcription elongation, as it directly interacts with the transcription elongation factor TFIIS (Ishiguro et al., 2000). Phosphorylation of a serine residue at position 2 of RPB6 by casein kinase II (CKII) is apparently involved in the regulation of the function of this subunit (Kayukawa et al., 1999). Interestingly, this serine residue is present in the Tritryp RPB6 isoform associated with RNAP II and III, but it is absent in the RPB6z isoform (Supplementary Fig. S2A). It is also interesting to note that a mutation in a highly conserved residue of CR1 in RPB6 from yeast had a very different effect on the function of the three RNAPs, with the activities of RNAP II and III severely impaired, whereas the activity of RNAP I was only slightly altered (Tan et al., 2003). This indicates that the function of the common subunits may change, based on the RNAP context.
Our TAP-tag experiments revealed a number of proteins which apparently co-purified with the RNAP II and III complexes. While many of these appear to be due to non-specific contamination with tubulin, heat shock proteins, mitochondria and translation apparatus, several may represent more specific interactions. Thirteen proteins were found in both the LmRPB2-TAP and LmRPB6-TAP experiments, while a number were found only in individual experiments (Table 2). Many of these proteins represent those associated with RNA: e.g., RNA splicing factor PTSR-1 (LmjF07.0870), two RNA helicases (LmjF32.0400 and LmjF35.0370) and at least two RNA binding proteins (LmjF32.0750 and LmjF32.0150). Several others are likely to be associated with the DNA template: e.g. histones H2B, H3 and H4; a putative endonuclease (LmjF09.0050); and an ATPase associated with chromosome segregation (LmjF10.0160). The T. brucei orthologue of one of these helicases (LmjF32.0400) was also associated with the RNAP II complex (Das et al., 2006). Two other helicases (LmjF29.2090 and LmjF26.1560) may be associated with either RNA or DNA and may have a role in unwinding the DNA during transcription initiation and/or elongation. The association of some of these proteins with the T. brucei RNAP II complex has been described previously (Devaux et al., 2006). However, perhaps the most interesting result is the identification of an orthologue for the large subunit of the FACT complex (LmjF29.0020). FACT is required in higher eukaryotes for RNAP II transcription, where it facilitates elongation by destabilizing nucleosomal structure by removing one H2A-H2B dimmer during passage by the polymerase and then re-recruiting core histones to the DNA (Belotserkovskaya et al., 2003). Blastp searches of the L. major genome also identified a putative orthologue (encoded by LmjF32.0120) of the other FACT subunit, the structure-specific recognition protein 1 (SSRP1), which is a high mobility group (HMG)-like protein, although we did not find this protein associated with the RNAP II complex in these experiments. The conservation of these and other elongation-specific transcription factors in the Tritryp genomes suggests that transcription elongation may be more similar to other eukaryotes than is transcription initiation.
We believe the experiments described in this study represent the first comprehensive experimental characterization of the RNAP III complex from the trypanosomatids and confirm the composition of the RNAP II complex previously characterized in T. brucei (Devaux et al., 2006; Das et al., 2006). These results, together with similar studies for T. brucei RNAP I (Schimanski et al., 2003; Walgraffe et al., 2005; Nguyen et al., 2006) significantly expand our knowledge of the unique transcriptional machinery in these primitive parasitic protozoa.
Supplementary Material
Fig. 4.
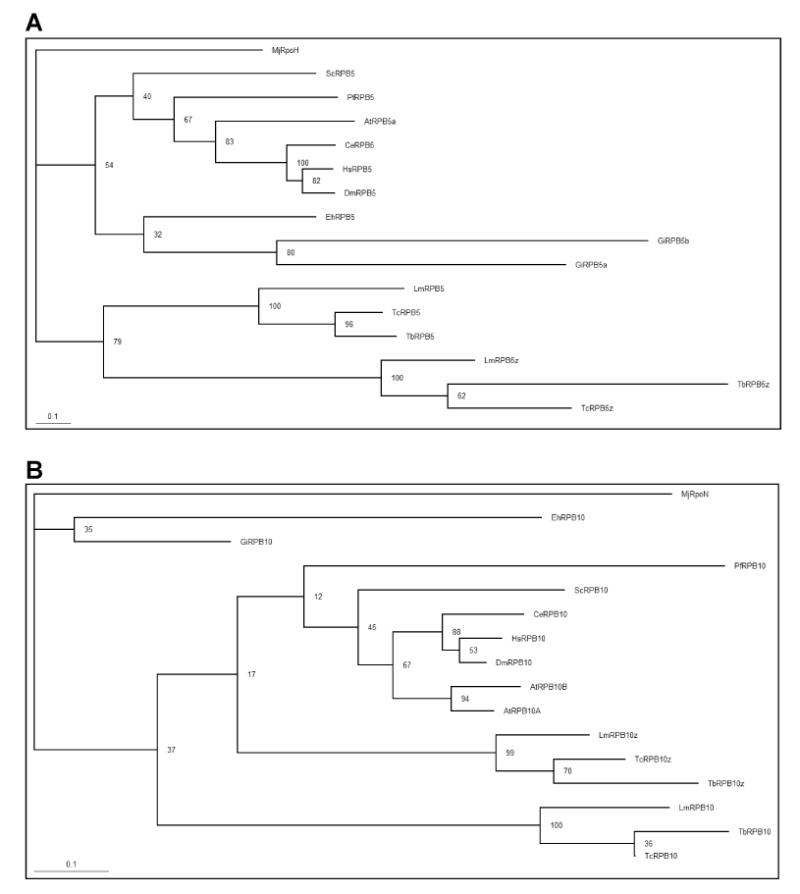
Phylogenetic analysis of RPB5 and RPB10 orthologues. Phylogenetic trees were constructed from the RPB5 (A) and RPB10 (B) sequence alignments shown in Supplementary Figs. 7 and 8, respectively, and rooted with the corresponding Methanococcus jannaschii sequences.
Acknowledgments
We thank Achim Schnaufer for help with the TAP-tag purifications, Aswini Panigrahi for help with the preparation of samples for mass spectrometry as well as analysis, and Yuko Ogata for assistance with analysis of the LC-MS/MS data. We also acknowledge Larry Simpson his kind gift of plasmid pREL1Lt-TAPmod. This work was supported by PHS grant 5 R01 AI053667 to P.J.M., and a postdoctoral fellowship from the International Training and Research in Emerging Infectious Diseases (ITREID) program to S.M.-C.
Footnotes
Note: Supplementary data associated with this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acker J, Wintzerith M, Vigneron M, Kedinger C. A 14.4 KDa acidic subunit of human RNA polymerase II with a putative leucine-zipper. DNA Seq. 1994;4:329–331. doi: 10.3109/10425179409020860. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003a;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R, Aphasizheva I, Simpson L. A tale of two TUTases. Proc Natl Acad Sci U S A. 2003b;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Campbell DA, Thomas S, Sturm N. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231–1240. doi: 10.1016/j.micinf.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Li H, Liu T, Bellofatto V. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol Biochem Parasitol. 2006;150:201–210. doi: 10.1016/j.molbiopara.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Das A, Zhang Q, Palenchar JB, Chatterjee B, Cross GA, Bellofatto V. Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol Cell Biol. 2005;25:7314–7322. doi: 10.1128/MCB.25.16.7314-7322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux S, Lecordier L, Uzureau P, Walgraffe D, Dierick JF, Poelvoorde P, Pays E, Vanhamme L. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol Biochem Parasitol. 2006;148:60–68. doi: 10.1016/j.molbiopara.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Estevez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers R, Hammer A, Köck J, Jess W, Borst P, Mémet S, Cornelissen AWCA. Trypanosoma brucei contains two RNA polymerase II largest subunit genes with an altered C-terminal domain. Cell. 1989;56:585–597. doi: 10.1016/0092-8674(89)90581-3. [DOI] [PubMed] [Google Scholar]
- Flores A, Briand JF, Gadal O, Andrau JC, Rubbi L, Van M V, Boschiero C, Goussot M, Marck C, Carles C, Thuriaux P, Sentenac A, Werner M. A protein-protein interaction map of yeast RNA polymerase III. Proc Natl Acad Sci U S A. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- Grondal EJM, Evers R, Kosubek K, Cornelissen AWCA. Characterization of the RNA polymerases of Trypanosoma brucei: Trypanosomal mRNAs are composed of transcripts derived from both RNA polymerase II and III. EMBO J. 1989;8:3383–3389. doi: 10.1002/j.1460-2075.1989.tb08502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucl Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Sun Y, Yuan CC, Kobayashi R, Myers MP, Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro A, Nogi Y, Hisatake K, Muramatsu M, Ishihama A. The Rpb6 subunit of fission yeast RNA polymerase II is a contact target of the transcription elongation factor TFIIS. Mol Cell Biol. 2000;20:1263–1270. doi: 10.1128/mcb.20.4.1263-1270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins C, Cadag E, Ciarloni L, Clayton C, Coulson RMR, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelinia G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Mueller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O'Neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schäfer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell J, Stuart KD, Barrell B, Myler PJ. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayukawa K, Makino Y, Yogosawa S, Tamura T. A serine residue in the N-terminal acidic region of rat RPB6, one of the common subunits of RNA polymerases, is exclusively phosphorylated by casein kinase II in vitro. Gene. 1999;234:139–147. doi: 10.1016/s0378-1119(99)00164-x. [DOI] [PubMed] [Google Scholar]
- Keller A, Purvine S, Nesvizhskii AI, Stolyar S, Goodlett DR, Kolker E. Experimental protein mixture for validating tandem mass spectral analysis. OMICS. 2002;6:207–212. doi: 10.1089/153623102760092805. [DOI] [PubMed] [Google Scholar]
- Kelly S, Wickstead B, Gull K. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem Soc Trans. 2005;33:1435–1437. doi: 10.1042/BST0331435. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- Martinez-Calvillo S, Nguyen D, Stuart K, Myler PJ. Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell. 2004;3:506–517. doi: 10.1128/EC.3.2.506-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Calvillo S, Stuart K, Myler PJ. Ploidy changes associated with disruption of two adjacent genes on Leishmania major chromosome 1. Int J Parasitol. 2005;35:419–429. doi: 10.1016/j.ijpara.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Martinez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart KD, Myler PJ. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11:1291–1299. doi: 10.1016/s1097-2765(03)00143-6. [DOI] [PubMed] [Google Scholar]
- Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci U S A. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nguyen TN, Schimanski B, Zahn A, Klumpp B, Günzl A. Purification of an eight subunit RNA polymerase I complex in Trypanosoma brucei. Mol Biochem Parasitol. 2006;149:27–37. doi: 10.1016/j.molbiopara.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Palfi Z, Schimanski B, Günzl A, Lucke S, Bindereif A. U1 small nuclear RNP from Trypanosoma brucei: a minimal U1 snRNA with unusual protein components. Nucl Acids Res. 2005;33:2493–2503. doi: 10.1093/nar/gki548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M, Nelson RG, Watkins KP, Agabian N. Trypanosome mRNAs share a common 5′ spliced leader sequence. Cell. 1984;38:309–316. doi: 10.1016/0092-8674(84)90552-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Sadhukhan PK, Chakraborty AK, Dasgupta A, Majumder HK. Chromatographic separation of DNA dependent RNA polymerases and molecular properties of RNA polymerase II from a Leishmania Spp. Mol Cell Biochem. 1997;171:105–114. doi: 10.1023/a:1006875631531. [DOI] [PubMed] [Google Scholar]
- Schimanski B, Klumpp B, Laufer G, Marhofer RJ, Selzer PM, Günzl A. The second largest subunit of Trypanosoma brucei's multifunctional RNA polymerase I has a unique N-terminal extension domain. Mol Biochem Parasitol. 2003;126:193–200. doi: 10.1016/s0166-6851(02)00273-6. [DOI] [PubMed] [Google Scholar]
- Schimanski B, Nguyen TN, Günzl A. Characterization of a multisubunit transcription factor complex essential for spliced-leader RNA gene transcription in Trypanosoma brucei. Mol Cell Biol. 2005;25:7303–7313. doi: 10.1128/MCB.25.16.7303-7313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri V, McArthur AG, Sogin ML, Adam RD. Giardia lamblia RNA polymerase II: amanitin-resistant transcription. J Biol Chem. 2003;278:27804–27810. doi: 10.1074/jbc.M303316200. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Sweetser D, Nonet M, Young RA. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Prysak MH, Woychik NA. Loss of the Rpb4/Rpb7 subcomplex in a mutant form of the Rpb6 subunit shared by RNA polymerases I, II and III. Mol Cell Biol. 2003;23:3329–3338. doi: 10.1128/MCB.23.9.3329-3338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walgraffe D, Devaux S, Lecordier L, Dierick JF, Dieu M, Van Den AJ, Pays E, Vanhamme L. Characterization of subunits of the RNA polymerase I complex in Trypanosoma brucei. Mol Biochem Parasitol. 2005;139:249–260. doi: 10.1016/j.molbiopara.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Yan S, Martinez-Calvillo S, Schnaufer A, Sunkin S, Myler PJ, Stuart K. A low background inducible promoter system in Leishmania donovani. Mol Biochem Parasitol. 2002;119:217–223. doi: 10.1016/s0166-6851(01)00418-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.