Abstract
The post-translational attachment of ubiquitin or ubiquitin-like modifiers (ULMs) to proteins regulates many cellular processes including the generation of innate and adaptive immune responses to pathogens. Vice versa, pathogens counteract immune defense by inhibiting or redirecting the ubiquitination machinery of the host. A common immune evasion strategy is for viruses to target host immunoproteins for proteasomal or lysosomal degradation by employing viral or host ubiquitin ligases. By degrading key host adaptor and signaling molecules, viruses thus disable multiple immune response pathways including the production of and response to interferons as well as other innate host defense mechanisms. Recent work further revealed that viruses inhibit the ligation of ubiquitin or ULMs or remove ubiquitin from host cell proteins. Thus, viruses succeed in either stabilizing negative regulators of innate immune signaling or thwart host cell proteins that are activated by ubiquitin or ULM-modification.
Introduction
Ubiquitin is a 76 amino acid protein that is conjugated to lysines of target proteins with the help of three main enzymes: The ubiquitin activating enzyme E1, one of dozens of E2 conjugating enzymes and a E3 substrate specific ubiquitin ligase, of which hundreds are known. Recent studies of viral ubiquitin ligases further revealed that ubiquitin can be conjugated to cysteines, serines, and threonines in the absence of lysines on the target protein [1, 2••]. The fate of target proteins depends on the number of attached ubiquitins, the mode of poly-ubiquitination at ubiquitin lysines 48, 63, or others, and the intracellular site at which ubiquitination occurs. Generally, ubiquitination of cytoplasmic and nuclear proteins leads to their degradation by the proteasome whereas ubiquitination of the cytoplasmic domains of transmembrane proteins results in their sorting to lysosomes via the multivesicular body pathway. An important exception to this general rule is the ubiquitin-controlled extraction of transmembrane proteins from the endoplasmic reticulum (ER), a quality control procedure during ER protein folding. In addition to playing a dominant role in controlling protein turnover, ubiquitination, particularly mono-ubiquitination, can also regulate protein function and protein/protein interaction. This is partially controlled by removal of ubiquitin from target proteins by ubiquitin hydrolases. Thus, similar to the control of phosphorylation by kinases and phosphatases, ubiquitin ligases and de-ubiquitination enzymes modulate substrate function by transient ubiquitination. In addition to ubiquitin, a number of ubiquitin-like modifiers (ULMs) similarly alter target substrates with various outcomes. Some of these ULMs are ubiquitin homologs (SUMO, ISG15) whereas others are unrelated by sequence (ATG6), but perform parallel roles. Each of these ULMs comes with designated E1, E2, and E3 enzyme-like proteins. In this review we will highlight the recent developments and trends in this very active field of investigation.
Essential role of the UPS for viral entry and replication
Viruses utilize the host ubiquitin pathway at each stage of their life cycle including entry, genome replication, and egress [3, 4]. This is illustrated by recent reports that diverse viral families are unable to enter cells or replicate if the ubiquitin proteasome system is disabled by proteasome inhibitors, a treatment that also depletes free ubiquitin. Such treatment trapped viruses in the endosomes and dense lysosomes, but did not affect initial endocytosis [5]. By contrast, proteasomal inhibitors blocked endocytosis of influenza virus due to the blockade of ubiquitination of epsin 1, a cargo specific adaptor for clathrin [6]. For herpes simplex virus it was shown that UPS activity was required at a post-penetration step to transport the incoming capsid to the nucleus [7]. Thus, several unrelated viral families depend on the UPS system even before the onset of viral replication. In poxvirus-infected cells, two groups reported that inhibitors of the proteasome or of E1 enzymes delayed expression of early viral genes and blocked the formation of virus replication factories resulting in complete inhibition of intermediate and late gene expression [8, 9•]. The UPS system is also required for the replication of coxsackie virus 3B since proteasome inhibition, ubiquitin knockdown, or increasing deubiquitinase activities all prevented CV3B replication [10]. Similarly, replication of human respiratory syncytial virus was decreased in the presence of proteasome inhibitors [11]. Although it has been speculated that proteasome inhibitors in clinical use might have anti-viral activity [8], it has yet to be demonstrated that these compounds are able to inhibit viral replication in vivo. Taken together, these studies highlight the importance of the UPS for viral infection.
Ubiquitin-mediated viral evasion of interferon-induction
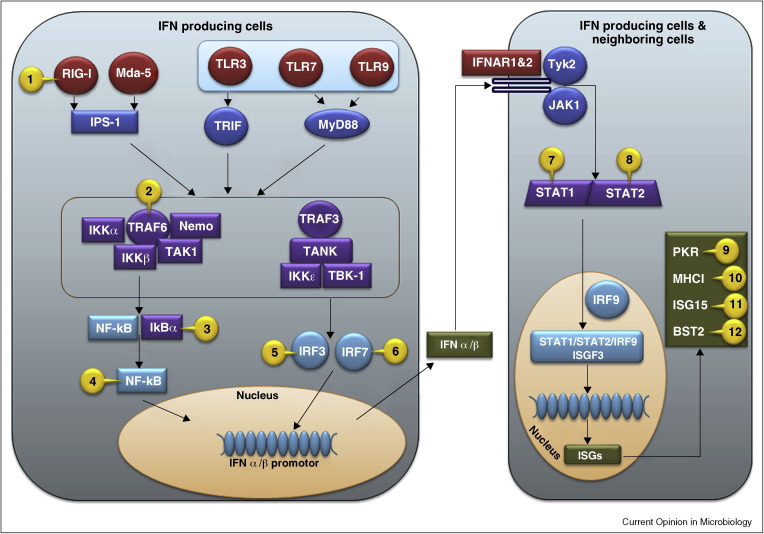
One reason the UPS is essential for viral replication is that many viruses use or inhibit the UPS to modulate the innate immune response, particularly the production and function of type I interferons (IFN), which include multiple IFN α subtypes and IFNβ. While the anti-viral activities of IFN have been known for over 50 years, the molecular details of how virus infection actually triggers IFN synthesis have only emerged in the past decade (for a recent review see [12]). As the roles and regulation of receptors, adaptors, signal transducers, and transcription factors in IFN induction come to light, corresponding viral counter-mechanisms are uncovered (reviewed in [13]) including many that involve the ubiquitin pathway [14]. The current model of virus-mediated induction of IFNβ and the IFN mediated responses with viral interventions is illustrated in Figure 1 .
Figure 1.
Examples of viral protein mediated intervention of host pathogen stimulated IFN stimulation pathways using UPS. Host anti-viral signals are initiated when cells sense the presence of pathogens via pattern-recognition receptors (PRRs) such as extracellular toll like receptors (TLRs), or intracellular RIG-I like helicases (RLHs) or DNA binding proteins. PRRs recognize molecular signatures of pathogens, the single or double stranded RNA or DNA. TLR3 activates Toll–interleukin (IL)-1-resistance (TIR) domain-containing adaptor inducing IFN-b (TRIF), whereas TLR7 and 9 activate myeloid differentiation factor 88 (MyD88). Melanoma differentiation-associated gene (mda-5) and retinoic acid-inducible gene-1 (RIG-I) activate tumor necrosis factor (TNF) receptor associated factor 3 (TRAF3) via Cardif/VISA/MAVS/IPS-1. Activated TRAF3 interacts with TRAF family member associated NFκB activator (TANK), TANK binding kinase 1 (TBK-1), and the related IκB Kinase ɛ (IKKɛ) to phosphorylate and translocate IRFs in the nucleus. In a parallel pathway, NFκb activation is initiated by poly-ubiquitination of TRAF6 and receptor interacting protein-1 (RIP1). These adaptors recruit transforming growth factor (TGF) β—activating enzyme 1 (TAK1), NFκB essential modifier (NEMO), and IκB Kinases (IKK) into a multi protein complex that phosphorylates inhibitor of NFκB (IκB) resulting in its ubiquitination and degradation by proteasomes. The degradation of IκB reveals the nuclear localization signal of NFκB that then translocates to the nucleus. In the nucleus, activated IRFs and NFκB assemble at the IFNβ promoter together with ATF-2/c-jun and other co-factors to stimulate IFN transcription. IFNs are secreted and induce an anti-viral state in neighboring cells by binding to the receptors IFNAR 1 & 2 and activating the tyrosine kinases JAK1 and Tyk2 that in turn activate signal transducer and activator of transcription (STATs). Phosphorylated STAT1 and STAT2 heterodimerize and translocate to the nucleus, where they associate with IRF-9 to form the hetrotrimeric transcription factor ISGF3. Binding of ISGF3 to the IFN stimulated response element (ISRE) induces transcription of IFN stimulated genes (ISGs). Viral intervention of these pathway proteins are marked in the figure as numbers and listed below in the virus (protein)[ref] format: 1. Influenza A virus (NS1) [22••]; 2. Measles virus (P) [27]; 3. African swine fever virus (A238L) [25], Rotavirus (NSP1) [28•]; 4.Murid herpesvirus-4 (ORF73) [26•], Poxvirus (CP77) [23]; 5. classical swine fever virus [15], Bovine viral diarrhea virus [16], Rotavirus [18, 19], HIV-1 (Vpr, Vif) [20]; 6. Ebola virus (VP35) [21•]; 7. Para influenza simian virus 5 [29, 30]; 8. Respiratory syncytial virus (NS1) [31]; 9. Hepatitis C virus [70]; 10. Human cytomegalovirus (US2, US11) [3], Kaposi's Sarcoma associated herpesvirus [KSHV] (K3, K5) [1, 2••]; 11. Influenza virus (NS1A) [42••, 46, 47, 48, 49], vaccinia virus (E3) [51•], Coronavirus (PLP) [53]; 12. HIV-1 (Vpu) [57, 61, 64, 65, 66], KSHV (K5) [63].
Viral inhibition of IRF3 activation
Examples for ubiquitin-mediated degradation of components of the signaling pathway leading to IFN synthesis are pestiviruses such as Classical swine fever virus (CSFV) and Bovine viral diarrhea virus (BVDV) [15, 16, 17] as well as rotaviruses [18, 19]. These viruses induce proteasome-dependent degradation of IRF3. HIV-I also mediates IRF3 degradation via Vpr-directed and Vif-directed ubiquitination of the protein [20]. Similarly, Ebola virus VP35 protein has been shown to inhibit IFN synthesis in immune cells by promoting SUMOylation of IRF7 by the cellular E3 ligase PIAS1 [21•]. IFN expression can also be impaired by blocking the signaling pathways leading from virus detection to transcription factor activation. This is well exemplified by the inhibition of RIG-I activity by the NS1 protein of influenza A virus. Gack et al. recently showed that this protein interacts with and blocks the activity of TRIM25, an E3 ligase required for ubiquitin-dependent interaction between RIG-I and IPS1 [22••]. NS1 was shown to prevent TRIM25-dependent ubiquitination of RIG-I and thus activation of IRF3-dependent IFN secretion thereby allowing the virus to evade its anti-viral effects. Moreover, in the absence of this phenotype mutant virus was highly attenuated thereby demonstrating its importance for virus replication and transmission.
Viral inhibition of NFκB activation
To prevent NFκB activation viral proteins either directly bind to NFκB to inhibit its translocation to the nucleus or they mediate degradation of NFκB. The poxviral protein CP77 prevents NFκB activation by binding with its N-terminal six ankyrin repeat region to the p65 subunit of NFκB [23]. Another poxvirus protein that contains pyrin domain containing protein M013 also has been shown to bind to RelA/p65 and prevent localization of NFκB to the nucleus [24]. African swine fever virus even encodes a homolog of human IκB, A238L that is resistant to phosphorylation and thus irreversibly binds and inactivates NFkB [25]. A direct degradation of p65/RelA was reported for the murid herpesvirus-4 (MuHV-4) latency associated protein ORF73. MuHV-4 ORF73 has an unconventional suppressor of cytokine signaling (SOCS)-box motif that associates with the host ubiquitin-ligase complex ElonginC/Cullin5/SOCS to mediate poly-ubiquitination and subsequent proteasomal degradation of p65/RelA [26•]. Functional deletion of SOCS-box motif in ORF73 ablated NFκB degradation and resulted in suppressed viral expansion in germinal center B cells and prevented persistent infection in mice indicating that suppression of NFkB activity is essential for viral persistence. Viruses also utilize host machinery that negatively regulates NFkB activation to turn off the innate immune response. For instance, the host de-ubiquitin (DUB) enzyme A20 terminates TLR-dependent NFκB activation by removing ubiquitins from ATF6. A20 is prematurely upregulated by measles virus P protein thus preventing NFκB activation [27]. An unusual targeting of the ubiquitin complex that mediates IκB degradation was observed for the rotavirus NSP1 protein. NSP1 mediates the ubiquitination and degradation of the F-Box substrate recognition protein, β-transducin repeat containing protein (β-TrCP) that binds to and degrades IκB through the ubiquitin-ligase complex Skp-1/Cul1/F-Box (SCF) [28•]. However, the same SCFβ-TrCP complex is utilized by HIV-1 Vpu to target host anti-viral effectors CD4 and bone marrow stromal antigen-2 (BST2). This demonstrates a case in which different viruses utilize the same host ubiquitin ligase in different contexts to target host proteins to their advantage.
Viral use of the ubiquitin system to inhibit IFN-dependent signal transduction
In addition to preventing the induction of IFN, viruses also inhibit the signal transduction pathway triggered upon binding of IFN to its receptor [13]. A key event in this signal cascade is the activation and nuclear translocation of STAT proteins that are frequent targets of viral counter-mechanisms. For example, parainfluenza simian virus 5 is known to degrade STAT1 via a proteasome-dependent mechanism [29] that involves co-opting a host cell E3 ligase [30]. Respiratory syncytial virus was found to degrade STAT2 through formation of an E3 ligase complex that includes both host proteins (Cul2, Rbx) as well as the viral NS1 protein [31]. More recently, Ashour et al. have shown that the Dengue virus NS5 protein mediates ubiquitination and proteasome-dependent degradation of STAT2 although the host cell molecules required for this are unknown [32]. Interestingly, this effect is exhibited only by the form of NS5 that is proteolytically processed by the viral polyprotein [32]. In addition to STAT1 and STAT2, STAT3 is also targeted by viral proteins. Mumps virus V protein targets both STAT1 and STAT3 but a mutant of this protein E95D that lost its ability to bind to and degrade STAT3 could still ubiquitinate and degrade STAT1 [33]. Virus with this mutation increased cell death resulting in a large-plaque phenotype.
Viral use of the ubiquitin system to prevent the function of IFN-induced genes
In the event that viruses fail to completely shut-down the induction of IFN or IFN-dependent signaling they face a multitude of anti-viral IFN-induced proteins (ISGs) [34]. Well-studied anti-viral ISGs include PKR, OAS, RNAseL, and Mx. While the pre-treatment of cells with IFN generally renders them resistant to viral infection, important ISGs that are induced during viral infection can be successfully counteracted by viruses. Here, we will focus on two ISGs whose function and viral counter-mechanisms are currently being elucidated: ISG15 and BST2/Tetherin.
ISG15
ISG15 is a ubiquitin-like modifier that is one of the most highly induced ISGs. This di-ubiquitin-like protein is conjugated to proteins (ISGylation), a process that utilizes UbE1L as E1 enzyme, UbcH6, UbcH8 or UbcM8 as the E2 enzyme and Herc5, HHARI, and Efp as E3 ubiquitin ligases that are also IFN-induced [35, 36]. This process is reversible via the action of the ubiquitin-specific protease UBP43. Although many ISGylated targets have been identified [37], the impact of ISGylation has yet to be established in individual cases. Recent examples include demonstration that IRF3 is positively regulated by ISG15 [38, 39] whereas RIG-I is negatively regulated [40]. However, a broadly anti-viral function of ISG15 has been established in ISG15 and UbE1L null mice that exhibit increased susceptibility to influenza A and B, HSV-1, MHV68, and Sindbis virus [41, 42••]. How ISG15 mediates anti-viral functions requires elucidation. Given the plethora of ISGylated proteins it is possible that anti-viral effects are virus-specific. One ISGylation anti-viral mechanism is inhibition of ubiquitin-ligase activity of Nedd4, which is required for budding of Ebola, vesicular stomatitis, and rabies viruses [43]. ISG15 and UbE1L over expression also inhibits ubiquitination of Tsg101 and Gag, a process essential for HIV budding [44].
Yet not all viruses are affected by ISGylation [45] and several viruses have developed countermeasures against ISGylation. The NS1 protein of influenza B virus binds to ISG15 in a species-specific manner [46, 47, 48] and thus inhibits protein ISGylation. By contrast, the NS1A protein of influenza A is ISGylated that disrupts its nuclear localization [49]. Sumoylation of NS1 was also reported [50]. Inhibition of ISGylation has also been reported for Vaccinia Virus (VACV) [51•]. Interestingly, this was dependent on the VACV E3L protein that was known previously as an inhibitor of PKR. E3L-deleted virus was growth-restricted in ISG15+, but not in ISG15− cells and mice. Viruses are also known to deconjugate ISG15. Nairoviruses and arteriviruses encode OTU domain proteases that hydrolyze both ubiquitin and ISG15 from cellular target proteins [52•]. Similarly, the coronavirus PLP protein acts as a de-ubiquitinating and de-ISGylating enzyme [53]. These viral counterstrategies probably pinpoint those ISGylation events most detrimental to a given viral species.
BST2/Tetherin
Bone marrow stromal antigen-2 (BST2/Tetherin) is an IFN-induced [54], glycosylated type II transmembrane protein with a unique topology since it also contains a glycosylphosphatidylinisotol (GPI) anchor at the C-terminus thus localizing it to the periphery of lipid rafts, forming a fence like structure [55]. We initially identified this protein in a proteomics screen as a new target of the viral E3 ligase K5 of Kaposi's sarcoma virus [56] and, more recently, in a similar screen as dominant target for the HIV Vpu protein [57]. KSHV-K5 is a viral homolog of the membrane-associated RING-CH (MARCH) family of transmembrane ubiquitin ligases [58] that share the ability to ubiquitinate the intracellular domains of selected target proteins [59]. By comparison, HIV-1 Vpu does not possess ubiquitin-ligase activity on its own but interacts with the cellular F-Box protein β-TRcP, a subunit of the cellular SCF-complex. The finding that BST2/Tetherin is targeted by different viral immune evasion proteins raised this protein from obscurity, particularly since recent work revealed that BST-2 is responsible for tethering mature HIV-1 particles to the cell membrane in the absence of Vpu [60••, 61]. As recently reviewed elsewhere [62], this seminal work stimulated extensive research by a number of groups revealing that this IFN-induced protein prevents the egress of several unrelated viral families, including gamma-2 herpesviruses (KSHV), lentiviruses (EIAV, FIV) retroviruses (SIV, RSV, MPMV, HTLV-1, PFV), Filoviruses (Ebola, MV), and arena viruses (Lassa). While the exact details of this retention require identification, it seems that unlike other ISGs, BST2/Tetherin acts at a very late stage of infection. Many viruses counteract BST-2 function using different molecular mechanisms. In addition to KSHV-K5 and HIV-1 Vpu, Ebolavirus envelope protein, and the nef protein of SIV and HIV-2 have been implicated in inhibiting BST2/Tetherin [62]. Since these evasion mechanisms are often highly species-specific, they probably contribute to host restriction. The molecular mechanism of BST2 downregulation by most viral proteins is not known, but some details emerged for KSHV-K5 and HIV-1 Vpu. In the case of KSHV-K5, cytosolic lysines of BST2 are directly ubiquitinated and this is required for lysosomal targeting [63]. By contrast, lysine-deleted BST2 is still targeted to lysosomes by HIV-1 Vpu in a βTRcP-dependent manner, suggesting that ubiquitination is indirectly involved in this process [57, 64]. In addition, Vpu can initiate the proteasomal destruction of BST2, particularly in situations when BST2 resides in the ER owing to overexpression [65, 66]. Thus, ubiquitination is central to counteracting the anti-viral activity of BST2.
Conclusions
Ubiquitin-enabled interference of viruses with the innate immune response emerges as a central immune evasion mechanism in almost every viral species studied. Since viruses specifically target those innate immune responses that are particularly detrimental to a given viral species, studying the manipulation of ubiquitination has revealed novel host defense mechanisms. The identification of novel targets for viral ubiquitin ligases has been accelerated by proteomics approaches suggesting that it will be possible to use viral ubiquitin ligases to identify novel important key elements of host defense pathways [56, 67, 68•, 69]. Studying ubiquitination events in virally infected cells thus holds great promise to unravel important modulators of the intricate relationship between host and pathogen.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
References
- 1.Cadwell K., Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 2••.Wang X., Herr R.A., Chua W.J., Lybarger L., Wiertz E.J., Hansen T.H. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol. 2007;177:613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 1 and 2 demonstrate that non-lysine residues can be targeted for ubiquitination by E3 ligases.
- 3.Isaacson M.K., Ploegh H.L. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe. 2009;5:559–570. doi: 10.1016/j.chom.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Randow F., Lehner P.J. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11:527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 5.Yu G.-Y., Lai M.M.C. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J Virol. 2005;79:644–648. doi: 10.1128/JVI.79.1.644-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C., Zhuang X. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc Natl Acad Sci USA. 2008;105:11790–11795. doi: 10.1073/pnas.0803711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delboy M.G., Roller D.G., Nicola A.V. Cellular proteasome activity facilitates herpes simplex virus entry at a postpenetration step. J Virol. 2008;82:3381–3390. doi: 10.1128/JVI.02296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satheshkumar P.S., Anton L.C., Sanz P., Moss B. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J Virol. 2009;83:2469–2479. doi: 10.1128/JVI.01986-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Teale A., Campbell S., Van Buuren N., Magee W.C., Watmough K., Couturier B., Shipclark R., Barry M. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J Virol. 2009;83:2099–2108. doi: 10.1128/JVI.01753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 8 and 9 reveal a central role for the UPS in the initial stages of poxviral infection. Both show that proteasomal inhibition or ubiquitin pathway inhibitors block viral replication.
- 10.Si X., Gao G., Wong J., Wang Y., Zhang J., Luo H. Ubiquitination is required for effective replication of coxsackievirus B3. PLoS One. 2008;3:e2585. doi: 10.1371/journal.pone.0002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lupfer C., Pastey M.K. Decreased replication of human respiratory syncytial virus treated with the proteasome inhibitor MG-132. Virus Res. 2010;149:36–41. doi: 10.1016/j.virusres.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi O., Akira S. Innate immunity to virus infection. Immunol Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale M., Jr., Sen G.C. Viral evasion of the interferon system. J Interferon Cytokine Res. 2009;29:475–476. doi: 10.1089/jir.2009.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhoj V.G., Chen Z.J. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 15.Bauhofer O., Summerfield A., Sakoda Y., Tratschin J.D., Hofmann M.A., Ruggli N. Classical swine fever virus Npro interacts with interferon regulatory factor 3 and induces its proteasomal degradation. J Virol. 2007;81:3087–3096. doi: 10.1128/JVI.02032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Rijnbrand R., Jangra R.K., Devaraj S.G., Qu L., Ma Y., Lemon S.M., Li K. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology. 2007;366:277–292. doi: 10.1016/j.virol.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szymanski M.R., Fiebach A.R., Tratschin J.D., Gut M., Ramanujam V.M., Gottipati K., Patel P., Ye M., Ruggli N., Choi K.H. Zinc binding in pestivirus N(pro) is required for interferon regulatory factor 3 interaction and degradation. J Mol Biol. 2009;391:438–449. doi: 10.1016/j.jmb.2009.06.040. [DOI] [PubMed] [Google Scholar]
- 18.Barro M., Patton J.T. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci USA. 2005;102:4114–4119. doi: 10.1073/pnas.0408376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graff J.W., Mitzel D.N., Weisend C.M., Flenniken M.L., Hardy M.E. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J Virol. 2002;76:9545–9550. doi: 10.1128/JVI.76.18.9545-9550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okumura A., Alce T., Lubyova B., Ezelle H., Strebel K., Pitha P.M. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology. 2008;373:85–97. doi: 10.1016/j.virol.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Chang T.H., Kubota T., Matsuoka M., Jones S., Bradfute S.B., Bray M., Ozato K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes how a viral protein increases the SUMOylation of IRF7 by interacting with the host E3 ligase PIAS1.
- 22••.Gack M.U., Albrecht R.A., Urano T., Inn K.S., Huang I.C., Carnero E., Farzan M., Inoue S., Jung J.U., Garcia-Sastre A. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; A novel mechanism of viral interference with innate immune signaling is described that interferes with the RIG-I ubiquitination by TRIM25.
- 23.Chang S.-J., Hsiao J.-C., Sonnberg S., Chiang C.-T., Yang M.-H., Tzou D.-L., Mercer A.A., Chang W. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-kB activation by tumor necrosis factor alpha but is independent of its host range function. J Virol. 2009;83:4140–4152. doi: 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman M.M., Mohamed M.R., Kim M., Smallwood S., McFadden G. Co-regulation of NF-kappaB and inflammasome-mediated inflammatory responses by myxoma virus pyrin domain-containing protein M013. PLoS Pathog. 2009;5:e1000635. doi: 10.1371/journal.ppat.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tait S.W., Reid E.B., Greaves D.R., Wileman T.E., Powell P.P. Mechanism of inactivation of NF-kappa B by a viral homologue of I kappa b alpha. Signal-induced release of i kappa b alpha results in binding of the viral homologue to NF-kappa B. J Biol Chem. 2000;275:34656–34664. doi: 10.1074/jbc.M000320200. [DOI] [PubMed] [Google Scholar]
- 26•.Rodrigues L., Filipe J., Seldon M.P., Fonseca L., Anrather J., Soares M.P., Simas J.P. Termination of NF-kappaB activity through a gammaherpesvirus protein that assembles an EC5S ubiquitin-ligase. EMBO J. 2009;28:1283–1295. doi: 10.1038/emboj.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates ubiquitination and degradation of p65/RelA by a viral protein.
- 27.Yokota S., Okabayashi T., Yokosawa N., Fujii N. Measles virus P protein suppresses Toll-like receptor signal through up-regulation of ubiquitin-modifying enzyme A20. FASEB J. 2008;22:74–83. doi: 10.1096/fj.07-8976com. [DOI] [PubMed] [Google Scholar]
- 28•.Graff J.W., Ettayebi K., Hardy M.E. Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism. PLoS Pathog. 2009;5:e1000280. doi: 10.1371/journal.ppat.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]; In contrast to the previously described utilization of this ubiquitin-ligase subunit by viruses to target host cell immunoproteins, this study demonstrates that rotavirus targets the beta-TrCP complex for degradation thus preventing NFkB activation.
- 29.Didcock L., Young D.F., Goodbourn S., Randall R.E. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulane C.M., Horvath C.M. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology. 2002;304:160–166. doi: 10.1006/viro.2002.1773. [DOI] [PubMed] [Google Scholar]
- 31.Elliott J., Lynch O.T., Suessmuth Y., Qian P., Boyd C.R., Burrows J.F., Buick R., Stevenson N.J., Touzelet O., Gadina M. Respiratory syncytial virus NS1 protein degrades STAT2 by using the Elongin-Cullin E3 ligase. J Virol. 2007;81:3428–3436. doi: 10.1128/JVI.02303-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashour J., Laurent-Rolle M., Shi P.Y., Garcia-Sastre A. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol. 2009;83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puri M., Lemon K., Duprex W.P., Rima B.K., Horvath C.M. A point mutation, E95D, in the mumps virus V protein disengages STAT3 targeting from STAT1 targeting. J Virol. 2009;83:6347–6356. doi: 10.1128/JVI.00596-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon Y.J., Yoo H.M., Chung C.H. ISG15 and immune diseases. Biochim Biophys Acta. 2010;1802:485–496. doi: 10.1016/j.bbadis.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harty R.N., Pitha P.M., Okumura A. Antiviral activity of innate immune protein ISG15. J Innate Immun. 2009;1:397–404. doi: 10.1159/000226245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C., Denison C., Huibregtse J.M., Gygi S., Krug R.M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci USA. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi H.X., Yang K., Liu X., Liu X.Y., Wei B., Shan Y.F., Zhu L.H., Wang C. Positive Regulation of IRF3 Activation by Herc5 via ISG15 Modification. Mol Cell Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu G., Reinert J.T., Pitha-Rowe I., Okumura A., Kellum M., Knobeloch K.P., Hassel B., Pitha P.M. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol Biol (Noisy-le-grand) 2006;52:29–41. [PubMed] [Google Scholar]
- 40.Kim M.J., Hwang S.Y., Imaizumi T., Yoo J.Y. Negative feedback regulation of RIG-I-mediated antiviral signaling by interferon-induced ISG15 conjugation. J Virol. 2008;82:1474–1483. doi: 10.1128/JVI.01650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giannakopoulos N.V., Arutyunova E., Lai C., Lenschow D.J., Haas A.L., Virgin H.W. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J Virol. 2009;83:1602–1610. doi: 10.1128/JVI.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Lenschow D.J., Lai C., Frias-Staheli N., Giannakopoulos N.V., Lutz A., Wolff T., Osiak A., Levine B., Schmidt R.E., Garcia-Sastre A. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci USA. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first in vivo data showing that ISG15 plays a crucial role in the host response to viral infection.
- 43.Malakhova O.A., Zhang D.E. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem. 2008;283:8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okumura A., Lu G., Pitha-Rowe I., Pitha P.M. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci USA. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osiak A., Utermöhlen O., Niendorf S., Horak I., Knobeloch K.-P. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol Cell Biol. 2005;25:6338–6345. doi: 10.1128/MCB.25.15.6338-6345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sridharan H., Zhao C., Krug R.M. Species specificity of the NS1 protein of influenza B virus: NS1 binds only human and non-human primate ubiquitin-like ISG15 proteins. J Biol Chem. 2010;285:7852–7856. doi: 10.1074/jbc.C109.095703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Versteeg G.A., Hale B.G., van Boheemen S., Wolff T., Lenschow D.J., Garcia-Sastre A. Species-specific antagonism of host ISGylation by the influenza B virus NS1 protein. J Virol. 2010;84:5423–5430. doi: 10.1128/JVI.02395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan W., Krug R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao C., Hsiang T.Y., Kuo R.L., Krug R.M. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc Natl Acad Sci USA. 2010;107:2253–2258. doi: 10.1073/pnas.0909144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal S., Rosas J.M., Rosas-Acosta G. Identification of the non-structural influenza A viral protein NS1A as a bona fide target of the Small Ubiquitin-like MOdifier by the use of dicistronic expression constructs. J Virol Methods. 2010;163:498–504. doi: 10.1016/j.jviromet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Guerra S., Caceres A., Knobeloch K.P., Horak I., Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]; An anti-poxviral function of ISG15s counteracted by the viral E3 protein, a protein also known to inhibit PKR.
- 52•.Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J., Richt J.A., Rowland R.R., Schmaljohn C.S., Lenschow D.J. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; A unique viral strategy to inhibit Ub-dependent and ISG15-dependent antiviral pathways is described. Two unrelated viral families use host OTU proteases to deconjugate ubiquitin and ISG15.
- 53.Clementz M.A., Chen Z., Banach B.S., Wang Y., Sun L., Ratia K., Baez-Santos Y.M., Wang J., Takayama J., Ghosh A.K. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol. 2010;84:4619–4629. doi: 10.1128/JVI.02406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blasius A.L., Giurisato E., Cella M., Schreiber R.D., Shaw A.S., Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- 55.Kupzig S., Korolchuk V., Rollason R., Sugden A., Wilde A., Banting G. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic. 2003;4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 56.Bartee E., McCormack A., Fruh K. Quantitative membrane proteomics reveals new cellular targets of viral immune modulators. PLoS Pathog. 2006;2:e107. doi: 10.1371/journal.ppat.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Douglas J.L., Viswanathan K., McCarroll M.N., Gustin J.K., Fruh K., Moses A.V. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J Virol. 2009;83:7931–7947. doi: 10.1128/JVI.00242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartee E., Mansouri M., Hovey Nerenberg B.T., Gouveia K., Fruh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol. 2004;78:1109–1120. doi: 10.1128/JVI.78.3.1109-1120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nathan J.A., Lehner P.J. The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp Cell Res. 2009;315:1593–1600. doi: 10.1016/j.yexcr.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 60••.Neil S.J., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]; References 60 and 61 independently demonstrate that, in the absnece of vpu, BST2 tethers HIV virions to cells thus inhibiting viral spread.
- 61.Van Damme N., Goff D., Katsura C., Jorgenson R.L., Mitchell R., Johnson M.C., Stephens E.B., Guatelli J. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Douglas J.L., Gustin J.K., Viswanathan K., Mansouri M., Moses A.V., Früh K. The Great Escape: viral strategies to counter BST-2/Tetherin. PLoS Pathog. 2010;6:e1000913. doi: 10.1371/journal.ppat.1000913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mansouri M., Viswanathan K., Douglas J.L., Hines J., Gustin J., Moses A.V., Fruh K. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol. 2009;83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell R.S., Katsura C., Skasko M.A., Fitzpatrick K., Lau D., Ruiz A., Stephens E.B., Margottin-Goguet F., Benarous R., Guatelli J.C. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5:e1000450. doi: 10.1371/journal.ppat.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goffinet C., Allespach I., Homann S., Tervo H.-M., Habermann A., Rupp D., Oberbremer L., Kern C., Tibroni N., Welsch S. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 66.Mangeat B., Gers-Huber G., Lehmann M., Zufferey M., Luban J., Piguet V. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dallaire F., Blanchette P., Branton P.E. A proteomic approach to identify candidate substrates of human adenovirus E4orf6-E1B55K and other viral cullin-based E3 ubiquitin ligases. J Virol. 2009;83:12172–12184. doi: 10.1128/JVI.01169-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68•.Dallaire F., Blanchette P., Groitl P., Dobner T., Branton P.E. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J Virol. 2009;83:5329–5338. doi: 10.1128/JVI.00089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reference [67] describes high throughput screening of the host proteome using mass spectrometry to identify new targets of cullin-based viral ubiquitin ligases. In reference 68 this method is used to identify a new target of a viral E3 ligase.
- 69.Hor S., Ziv T., Admon A., Lehner P.J. Stable isotope labeling by amino acids in cell culture and differential plasma membrane proteome quantitation identify new substrates for the MARCH9 transmembrane E3 ligase. Mol Cell Proteomics. 2009;8:1959–1971. doi: 10.1074/mcp.M900174-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garaigorta U., Chisari F.V. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe. 2009;6:513–522. doi: 10.1016/j.chom.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]