Summary
The yeast DEAD-box protein Mss116p is a general RNA chaperone that functions in mitochondrial group I and II intron splicing, translational activation, and RNA end processing. Here, we determined high-resolution X-ray crystal structures of Mss116p complexed with an RNA oligonucleotide and ATP analogs AMP-PNP, ADP-BeF3−, or ADP-AlF4−. The structures show the entire helicase core acting together with a functionally important C-terminal extension. In all structures, the helicase core is in a closed conformation with a wedge α-helix bending RNA 3′ of the central bound nucleotides, as in previous DEAD-box protein structures. Additionally, the C-terminal extension bends RNA 5′ of the central nucleotides, resulting in RNA crimping. Despite reported functional differences, we observe few structural changes in ternary complexes with different ATP analogs. The structures constrain models of DEAD-box protein function and reveal a new strand separation mechanism in which a protein uses two wedges to act as a molecular crimper.
Introduction
DEAD-box proteins are a large, ubiquitous family of ATP-dependent RNA helicases that mediate RNA and RNP structural rearrangements in a variety of cellular processes, including RNA splicing, translation, ribosome biogenesis, RNA degradation, and RNA transport (Linder, 2006). Members of helicase superfamily II, all DEAD-box proteins contain a helicase core consisting of two tandem RecA-like domains (domains 1 and 2) connected by a short linker (Jankowsky and Fairman, 2007). The helicase core contains the eponymous sequence motif II (D-E-A-D) and ten other conserved motifs, which function in ATP or RNA binding or form interdomain contacts. Many DEAD-box proteins also contain N- and/or C-terminal extensions, which differ between proteins and in some cases target the proteins to specific substrates via RNA-protein and/or protein-protein interactions (Linder, 2006; Silverman et al., 2003).
DEAD-box proteins differ from processive RNA helicases in unwinding RNA duplexes by local strand separation rather than by translocation through the duplex (Yang et al., 2007; Yang and Jankowsky, 2006). Studies of several DEAD-box proteins indicate that ATP and RNA bind cooperatively and stabilize the two flexibly attached core domains in a closed conformation (Lorsch and Herschlag, 1998a; Polach and Uhlenbeck, 2002; Theissen et al., 2008). An X-ray crystal structure of a closed conformation was first determined for the helicase core of the Drosophila DEAD-box protein Vasa complexed with single-stranded RNA (ssRNA) and the non-hydrolyzable ATP analog AMP-PNP (Sengoku et al., 2006). The structure showed the two core domains brought together with the eleven conserved DEAD-box protein motifs at the interface between the domains. The central nucleotide residues of the RNA bind in a cleft on one side of the interface, while AMP-PNP binds in a cleft on the opposite side. Importantly, the RNA is bound via its phosphate backbone and is bent by a wedge α-helix containing motif Ib, suggesting physical mechanisms for non-specific RNA binding and strand separation. Crystal structures of the human DEAD-box protein eIF4AIII within the exon-junction complex (EJC) and the human DEAD-box protein DDX19 (Dbp5p in yeast) showed virtually identical closed conformations for the core and the same modes of RNA binding and bending, suggesting that these are conserved features of DEAD-box proteins (Andersen et al., 2006; Bono et al., 2006; Collins et al., 2009; von Moeller et al., 2009).
Recent studies showed that ATP binding but not hydrolysis is required for RNA unwinding by DEAD-box proteins (Chen et al., 2008; Liu et al., 2008). This finding presumably reflects that ATP binding favors a conformation of the helicase core that has a high affinity for ssRNA, while the core alone or with bound ADP has a low affinity for ssRNA (Cordin et al., 2004; Lorsch and Herschlag, 1998b). ATP hydrolysis leads to the release of the bound RNA strand, allowing enzyme turnover. Further, several DEAD-box proteins, including Mss116p studied here, were shown to unwind RNA duplexes in the presence of the non-hydrolyzable ATP ground state analog ADP-beryllium fluoride (ADP-BeFx), but not AMP-PNP, another ground state analog, nor the transition state analog ADP-aluminum fluoride (ADP-AlFx; Liu et al., 2008). These findings raised the possibility that different ground and transition state ATP analogs might favor the formation of different DEAD-box protein conformations with different RNA-unwinding activities.
The closely related DEAD-box proteins CYT-19 from Neurospora crassa and Mss116p from Saccharomyces cerevisiae are amenable to facile genetic and biochemical analysis and have emerged as important model systems for studying DEAD-box protein function. These proteins act as general RNA chaperones that promote the efficient splicing of more than thirteen different mitochondrial (mt) group I and group II introns, translational activation, and RNA end processing (Huang et al., 2005; Mohr et al., 2002). As general RNA chaperones, CYT-19 and Mss116p bind RNA and RNP substrates non-specifically and disrupt or rearrange stable structures that are rate-limiting for RNA folding (Del Campo et al., 2009). The two proteins are largely interchangeable with the expression of CYT-19 ameliorating all the RNA splicing and translation defects in yeast mss116Δ strains and the purified proteins stimulating splicing of many of the same group I and II introns in vitro (Del Campo et al., 2009; Halls et al., 2007; Huang et al., 2005; Mohr et al., 2006; Mohr et al., 2002; Solem et al., 2006). The S. cerevisiae DEAD-box protein Ded1p and the E. coli DEAD-box protein SrmB can also substitute for Mss116p in vivo and in vitro to different extents that correlate with their RNA-unwinding activities (Del Campo et al., 2009). Studies of CYT-19 and Mss116p have provided insight into the mechanism of action of DEAD-box proteins (Chen et al., 2008; Liu et al., 2008; Yang et al., 2007) and how they function as RNA chaperones on physiological substrates (Bhaskaran and Russell, 2007; Del Campo et al., 2009; Del Campo et al., 2007; Halls et al., 2007; Mohr et al., 2002; Tijerina et al., 2006).
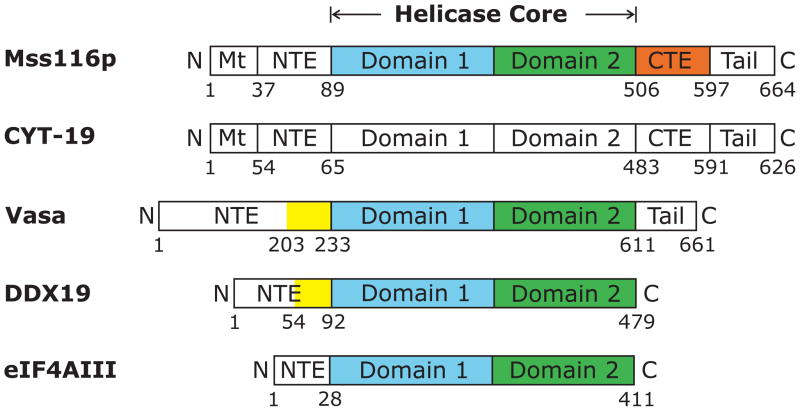
In Mss116p and CYT-19, the helicase core is preceded by a short N-terminal extension (NTE) and followed by an α-helical C-terminal extension (CTE) and a basic tail (Figure 1; Mohr et al., 2008). The helicase cores of Mss116p and CYT-19 are very similar to those of other DEAD-box proteins, including Vasa, DDX19, and eIF4AIII, whose structures were determined previously, but the N- and C-terminal extensions differ (Figures 1 and S1). The α-helical CTE found in Mss116p and CYT-19 is conserved in two subfamilies of yeast and fungal mt DEAD-box proteins related to Mss116p and CYT-19, respectively, and a similar α-helical CTE is also found in another DEAD-box protein family that includes S. cerevisiae Has1p (DDX18 in human), which functions in ribosome biogenesis (Mohr et al., 2008). Truncations or point mutations within the CTE of Mss116p and CYT-19 inactivate RNA-dependent ATPase activity and decrease RNA-binding affinity, suggesting that this region may stabilize the structure of the helicase core and contribute to RNA binding (Mohr et al., 2008). The basic tail, which follows the CTE, is predicted to be unstructured and is thought to provide an additional non-specific RNA-binding site that helps tether the helicase core to large RNA substrates for multiple rounds of RNA unwinding (Grohman et al., 2007; Mohr et al., 2008). Deletion of the basic tail in Mss116p and CYT-19 moderately decreases RNA-binding affinity and commensurately decreases group I and II intron splicing activity, and in Mss116p these effects were shown to occur without inhibiting RNA-dependent ATPase activity (Grohman et al., 2007; Mohr et al., 2008). The basic tail has analogs in other DEAD-box proteins, including the well-studied DEAD-box proteins Ded1p from yeast and SrmB from E. coli, while the Drosophila Vasa protein has a hydrophilic tail that is acidic rather than basic (Mohr et al., 2008).
Figure 1.
Schematics showing domains in the DEAD-box proteins S. cerevisiae Mss116p, N. crassa CYT-19, D. melanogaster Vasa, human DDX19, and human eIF4AIII. All of the proteins contain closely related helicase cores comprised of two RecA-like domains (domains 1 and 2) joined by a flexible linker. Mt, mitochondrial targeting sequence; NTE, N-terminal extension; CTE, C-terminal extension; Tail, hydrophilic tail based on hydropathy plots; the hydrophilic tail is basic in Mss116p and CYT-19 and acidic in Vasa (Mohr et al., 2008). The colored regions show portions of the NTE (yellow), domain 1 (blue), domain 2 (green), and CTE (orange) that have been visualized in the crystal structures of Mss116p (PDB ID 3i5x), Vasa (2db3), DDX19 (3fht), and eIF4AIII (2hyi). The domains are not drawn to scale.
Here, we determined high-resolution X-ray crystal structures of ternary complexes of Mss116p bound to ssRNA and AMP-PNP, ADP-BeF3−, or ADP-AlF4−. The structures provide insight into the function of the CTE and its interaction with the helicase core, constrain models of RNA unwinding by DEAD-box proteins, and reveal a new mode of local strand separation in which two wedge regions of a protein act together to crimp RNA.
Results
Structure Determination
We tested crystallization of full-length Mss116p and several C-terminal truncations that remove different lengths of the basic tail (Mohr et al., 2008) and obtained the best single crystals for Mss116p/Δ598-664. This protein begins with an N-terminal GlySer derived from the purification tag linked to Mss116p residue 37, which corresponds to the N-terminus of mature Mss116p after cleavage of its mt targeting peptide, and it contains the entire NTE, the helicase core, and the entire α-helical CTE up to the start of the basic tail at residue 597 (Figure 1). Although removal of the basic tail is expected to somewhat decrease RNA-binding affinity, Mss116p/Δ598-664 functions indistinguishably from full-length Mss116p in vivo (Huang, 2004), and like full-length Mss116p, it unwinds a model duplex in the presence of ADP-BeFx but not AMP-PNP (A. Putnam, E. Jankowsky, M.D., and A.M.L. unpublished data). The crystallization of Mss116p/Δ598-664 required the presence of both an RNA oligonucleotide and an ATP analog and was accomplished as described in Experimental Procedures and in greater detail elsewhere (Del Campo and Lambowitz, 2009).
We determined the structures of Mss116p/Δ598-664 bound to the RNA oligonucleotide U10 and three different ATP analogs: AMP-PNP, ADP-BeF3−, and ADP-AlF4−. The structures, hereafter referred to as the AMP-PNP, ADP-BeF3−, and ADP-AlF4− complexes, were determined to resolutions ranging from 1.9 to 2.1 Å, with R factors at or below 20% (Table 1). A fourth structure of Mss116p bound to AMP-PNP and U10 with a 5-bromouridine (Br-U10; referred to as the Br-U10 complex) enabled us to determine the register of the RNA on Mss116p (Figure S2). The structure of the native AMP-PNP complex was solved by single isomorphous replacement with anomalous scattering (SIRAS), using a SeMet-labeled AMP-PNP complex (Table 1), and the structures of the other three complexes were solved by refining the protein model against the other datasets. All of the complexes crystallized in the space group P21212 with similar unit cell dimensions and a single ternary complex in the asymmetric unit. In all of the structures, we were able to model 509 of the 563 amino acid residues (residues 88 to 596), the ATP analog, a single Mg2+ ion at the ATPase active site, and the entire path of U10 RNA. The latter was modeled by using information from all four complex structures (Figure S3). Most of the poorly ordered regions are at the ends of the protein and RNA (Figure S4). The NTE (residues 37-87) was disordered and could not be modeled. Because the complexes are very similar to each other except for a few solvent exposed side chains (root mean square deviation (rmsd) ≤ 0.15 Å over all 509 Cα atoms), we refer below to the highest resolution structure of the AMP-PNP complex unless indicated otherwise.
Table 1.
Crystallographic data and refinement statistics.
| Complex | AMP-PNP (SeMet) | AMP-PNP (native) | Br-U10/AMP-PNP | ADP-BeF3− (native) | ADP-AlF4− (native) |
|---|---|---|---|---|---|
|
Data collection | |||||
| Unit cella: a, b, c (Å) | 88.17, 126.48, 55.54 | 88.54, 126.52, 55.52 | 89.19, 126.35, 55.99 | 88.15, 126.02, 55.60 | 88.83, 126.11, 55.77 |
| Wavelength (Å) | 0.97931 | 0.91842 | 0.91842 | 0.97857 | 0.97857 |
| Total reflections | 585879 | 761109 | 270938 | 272033 | 571444 |
| Unique reflections | 24769 | 48438 | 21524 | 36177 | 44781 |
| Resolutionb (Å) | 30.0-2.4 (2.44-2.40) | 35.0-1.90 (1.94-1.90) | 35.0-2.5 (2.56-2.50) | 35.0-2.1 (2.15-2.10) | 35.0-1.95 (2.00-1.95) |
| Completeness (%) | 99.1 (91.9) | 96.4 (82.7) | 94.5 (75.5) | 98.1 (87.2) | 96.4 (71.2) |
| Overall I/σI) | 57.6 (10.1) | 30.6 (4.9) | 25.1 (3.5) | 21.8 (3.5) | 29.8 (2.7) |
| Rsymc (%) | 8.5 (22.8) | 7.7 (38.9) | 9.8 (42.6) | 9.3 (39.8) | 8.2 (41.9) |
|
Refinement | |||||
| Resolution | 34.0–1.90 | 35.0-2.5 | 33.4–2.1 | 33.6–1.95 | |
| No. of reflections | 45711 | 20426 | 34336 | 42491 | |
| Rwork (%) | 19.1 | 20.4 | 18.6 | 17.8 | |
| Rfreed (%) | 22.5 | 24.2 | 22.1 | 20.9 | |
| No. atoms | |||||
| Protein | 4018 | 3979 | 4000 | 4005 | |
| RNA | 190 | 191 | 190 | 190 | |
| Ligands | 32 | 32 | 32 | 33 | |
| Water | 289 | 85 | 218 | 298 | |
| Rmsd bonds (Å) | 0.008 | 0.008 | 0.007 | 0.007 | |
| Rmsd angles (°) | 1.149 | 1.073 | 1.056 | 1.031 | |
| Ramachandran favorede (%) | 98.8 | 99.4 | 99.4 | 99.4 | |
| Ramachandran allowed (%) | 1.2 | 0.6 | 0.6 | 0.6 | |
| PDB ID | 3i5x | 3i5y | 3i61 | 3i62 | |
All complexes crystallized in space group P21212.
The numbers in parentheses refer to the highest resolution shell.
Rsym = Σhkl Σi |Ihkl,i − 〈Ihkl〉|/Σhkl Σ 〈Ihkl〉
Rfree was calculated with 5% of reflections that were excluded from refinement.
Analysis by MolProbity (Davis et al., 2007).
The C-terminal Extension Is Part of Domain 2
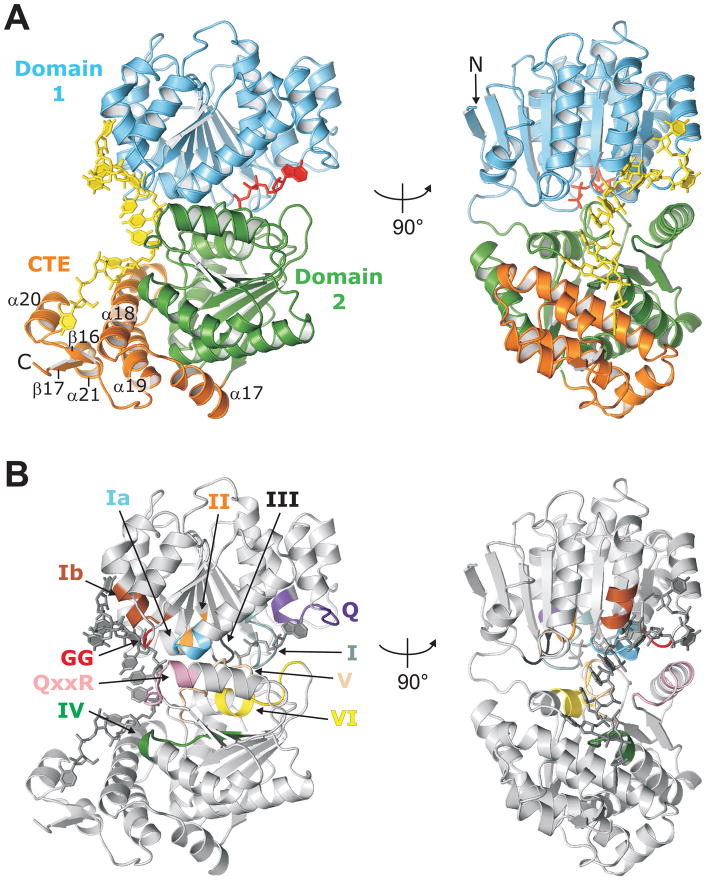
The structure determined for Mss116p/Δ598-664 includes both helicase core domains and the entire CTE (Figure 2). As in other DEAD-box proteins, the two helicase core domains have RecA-like folds, with the N-terminal domain (domain 1; residues 88-334) being an eight-stranded parallel β-sheet surrounded by eleven α-helices, and the C-terminal domain (domain 2; residues 335-505) being a seven-stranded parallel β-sheet surrounded by five α-helices. The CTE (residues 506-596) is a four-helix bundle (α18, 19, 20, and 21) with a flanking α-helix (α17) and a two-stranded parallel β-sheet (β16 and 17; Figures 2A and S1). Previously, hydropathy plots with a 20-amino acid residue sliding window indicated that Mss116p’s hydrophilic tail began at residue 580 (Mohr et al., 2008). The structure, however, shows that residues 580 to 594 form a structured part of the CTE (α21 and β17) on the side opposite the bound RNA (Figures 2A and S1). Thus, the putatively unstructured tail likely begins after residue 594. As stated above, we define the basic tail as starting at residue 597, which is disordered in the structure.
Figure 2.
Structure of Mss116p/Δ598-664 complexed with ssRNA and AMP-PNP. (A) Cartoon of Mss116p/Δ598-664 showing two views of the structure with domains colored as in Figure 1. The U10 RNA (yellow) and AMP-PNP (red) are shown in stick representation. (B) Location of DEAD-box protein motifs in the Mss116p structure. The views are the same as in (A), the motifs are colored (Q, purple; I, teal; Ia, light blue; GG, red; Ib, brown; II, orange; III, black; IV, green; QxxR, pink; V, peach; VI, yellow), and the RNA and AMP-PNP are dark gray.
As in other DEAD-box protein structures, domains 1 and 2 are in a closed conformation with the eleven conserved DEAD-box protein motifs at or near the interface between the two domains (Figure 2B). The central nucleotides of the U10 RNA are bound in a cleft that is perpendicular to the interface, and AMP-PNP is bound in a cleft formed by the interface on the opposite side. A search of the Protein Data Bank (PDB) with the Dali server (Holm et al., 2008) indicated that this arrangement is most similar to the structures of the DEAD-box proteins Drosophila Vasa (Sengoku et al., 2006), human eIF4AIII (Andersen et al., 2006; Bono et al., 2006), and human DDX19 (Collins et al., 2009; von Moeller et al., 2009), with a Z-score of ≥ 36 and a rmsd of ~2.0 Å over ~370 Cα atoms for each structure.
The CTE appears to be an addition to domain 2 rather than an independently folding domain. The extended α-helical region predicted previously for the CTE (Mohr et al., 2008) is seen to be a series of three α-helices (α17–19) that pack beneath and against the RNA-binding side of domain 2 with the remainder of the CTE packing against two of the above helices (α18 and 19; Figure 2A). The Dali server found no other proteins currently in the PDB with a structure that strongly matches the CTE (weak hits were to four helix bundles). The interface between the CTE and domain 2 buries 1374 Å2 of solvent accessible surface area, with mostly hydrophobic packing contacts (Krissinel and Henrick, 2007). Thus, the CTE could stabilize the helicase core, as well as interact with the 5′ end of the bound RNA.
Consistent with a structural role, Mss116p is expressed poorly and precipitates out of solution when the CTE is fully deleted (Mss116p/Δ508-664; Mohr et al., 2008 and unpublished data). Further, circular dichroism (CD) spectroscopy shows that C-terminal truncations within the CTE or just downstream of α19 significantly decrease the thermal stability of Mss116p (Figure S5; compare Mss116p/∆551-664 and Mss116p/∆569-664 to Mss116p and Mss116p/∆598-664). Notably, although Mss116p/∆569-664, which retains α19, and Mss116p/∆551-664, which disrupts α19, have similar CD spectra and thermal stabilities (Figure S5, the former has relatively high activity in vitro and is almost fully functional in vivo, while the latter is almost completely inactive in vitro and in vivo (Mohr et al., 2008). These findings suggest that α19 may play a critical role beyond overall structural stability (see Discussion).
Structures with Different Bound ATP Analogs Show No Changes Outside the ATP-Binding Site
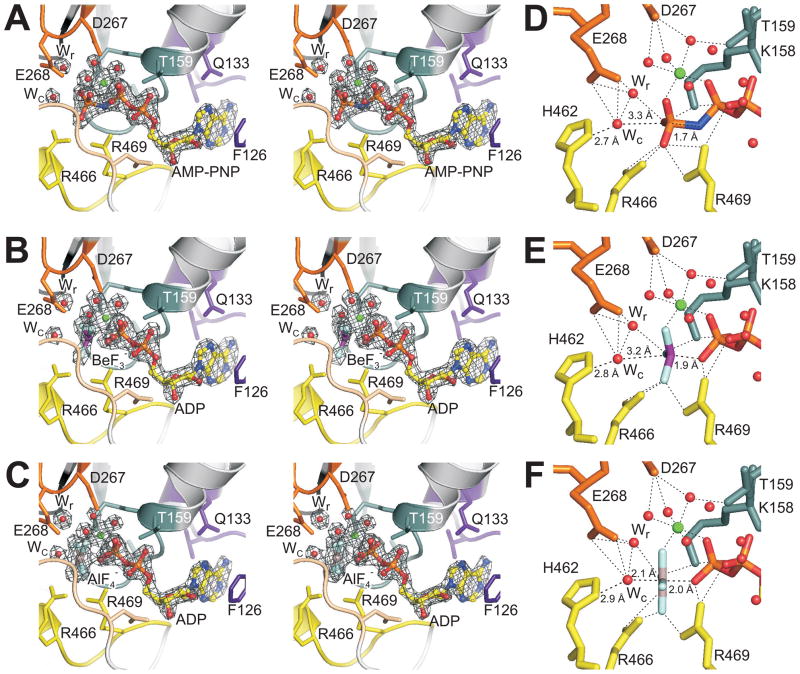
The finding that Mss116p could unwind an RNA duplex in the presence of the ATP analog ADP-BeFx but not AMP-PNP or ADP-AlFx raised the possibility that the structure of the ADP-BeF3− complex might differ in some way relevant to RNA binding (Liu et al., 2008). We find, however, that the structures with all three ATP analogs are virtually identical (rmsd of ≤ 0.15 Å over all 509 Cα atoms), with no discernible differences in how the RNA is bound and only small changes at the ATP-binding site. In the AMP-PNP complex, we observe clear electron density for AMP-PNP (Figure 3A). In the ADP-BeF3− complex, there is clear electron density for ADP and a trigonal group with tetrahedral geometry, consistent with BeF3− (Figure 3B; Chen et al., 2007). In the ADP-AlF4− complex, we see ADP and a square planar group, consistent with AlF4− (Figure 3C; Chen et al., 2007).
Figure 3.
Structure of the ATP-binding site of Mss116p/Δ598-664 with different bound ATP analogs. (A–C) Stereoviews of the ATP-binding site showing sigmaA-weighted m|Fo|− D|Fc| electron density contoured at 5 σ (gray) for an octahedrally coordinated Mg2+ ion, the putative catalytic (Wc) and relay (Wr) water molecules, and (A) AMP-PNP, (B) ADP-BeF3−, or (C) ADP-AlF4−. The ATP analog, Mg2+, and waters were removed from each model prior to map calculation. Mss116p is shown as a cartoon with side chains of residues that contact the ATP analog shown as sticks. (D–F) Details of coordination for (D) AMP-PNP, (E) ADP-BeF3−, and (F) ADP-AlF4−. In all panels, motif residues are colored as in Figure 2B, and ATP analog atoms are colored as follows: C (yellow), N (blue), O (red), P (orange), Be (purple), Al (gray), F (light blue), Mg (green). The Mg2+ ion and waters are shown as spheres.
In all three complexes, we can distinguish a Mg2+ ion coordinated in octahedral geometry to six ligands: the non-bridging β- and γ-phosphoryl oxygens of AMP-PNP (or the fluorine corresponding to this γ-phosphoryl oxygen position for ADP-BeF3− and ADP-AlF4−), and four water molecules. Additionally, two water molecules occupy the putative catalytic (Wc) and relay (Wr) positions described for the F1-ATPase (Dittrich and Schulten, 2005). In the F1-ATPase, ATP hydrolysis is thought to occur by in-line nucleophilic attack of Wc on the γ-phosphorous from the opposite side of the bridging phosphoryl oxygen, leading to a transition state with a planar γ-phosphate and two axial oxygens. This nucleophilic attack results in the release of Pi with inverted tetrahedral geometry. The nearby Wr is thought to be involved in a proton relay mechanism that increases the nucleophilicity of Wc (Dittrich and Schulten, 2005). Waters at positions equivalent to Wc and Wr have been identified in the Vasa and eIF4AIII structures (Nielsen et al., 2009) and can also been seen in the DDX19 structure (PDB ID 3fht; von Moeller et al., 2009).
The environments of the bound AMP-PNP and ADP-BeF3- are essentially the same, which, despite their differences in promoting RNA unwinding (Liu et al., 2008), is unsurprising given that both mimic ATP in the ground state. The structures show that the BeF3− group has a location and geometry similar to the γ-phosphoryl group of AMP-PNP (Figures 3D and E). In both structures, the Wc is coordinated by E268 (motif II) and H462 (motif VI) and is poised for in-line nucleophilic attack 3.3 Å from the γ-phosphorous of AMP-PNP and 3.2 Å from the Be of BeF3−. All the other side chains lining the ATP-binding pocket, even those making contact with the β- and γ-phosphoryl groups, are in the same conformation with similar contacts in the two complexes. Diffraction data from five additional isomorphous crystals of the ADP-BeF3− complex yielded electron density consistent with the same model (data not shown).
The binding of ADP-AlF4− induces several small but significant differences in the ATP-binding site. First, the AlF4− group is planar, mimicking the expected geometry of the transition state for ATP hydrolysis. As a result, the Al of AlF4− has moved ~0.5 Å toward Wc (with a slight adjustment of the bridging β-phosphoryl group), and Wc and the side chain of H462 have moved ~0.5 Å toward the Al, resulting in Wc being 2.1 Å from the Al (Figure 3F). Except for these differences, the ATP-binding site is the same as in the AMP-PNP and ADP-BeF3− complexes. A similar transition state structure was seen for the ternary complex of eIF4AIII in the EJC with bound ADP-AlF3, except that the lysine making ionic contacts with the β- and γ-phosphoryl oxygens of AMP-PNP (equivalent to K158 in Mss116p) moved completely away from this position with bound ADP-AlF3 (Nielsen et al., 2009). We observe no movement of this lysine with ADP-AlF4− bound to Mss116p, possibly reflecting that the lysine is held in place by the negative charge of the AlF4− anion.
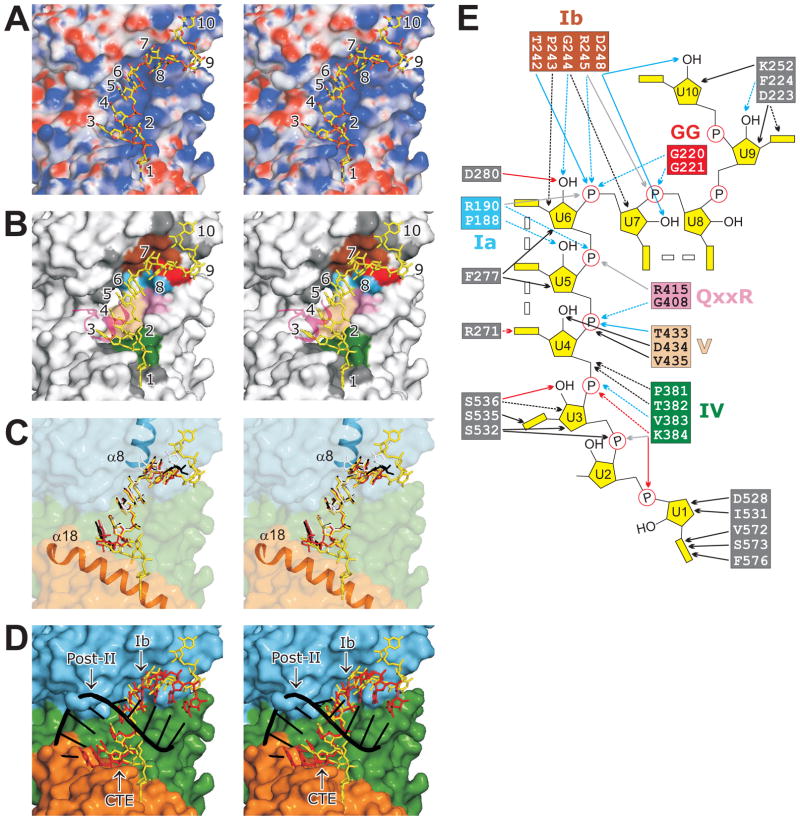
Mss116p Bends Single-Stranded RNA in Two Places
Mss116p binds U10 RNA in a cleft running perpendicular to the interface between domains 1 and 2. The cleft is basic, and the binding of the RNA is mainly through ionic contacts and hydrogen bonds (H-bonds) to the six phosphate groups of U3–U8 and H-bonds to the three 2′ OH groups of U6–U8 (Figures 4A and E). Most of the residues involved in RNA binding are from three conserved motifs of domain 1 (Ia, GG, and Ib), which contact U6–U8, and three conserved motifs of domain 2 (IV, QxxR, and V), which contact U3–U6 (Figures 4B, 4E, and S1). The binding of RNA via residues in these conserved motifs is essentially the same as in other DEAD-box protein structures (Andersen et al., 2006; Bono et al., 2006; Sengoku et al., 2006; Collins et al., 2009; von Moeller et al., 2009).
Figure 4.
RNA binding and bending by Mss116p. (A–D) Stereoviews of the RNA-binding cleft. (A) Molecular surface of Mss116p colored by electrostatic potential (blue is positive and red is negative) with the APBS plugin (Baker et al., 2001) in PyMOL. (B) Residues on the surface of Mss116p that contact U10 RNA. Residues are colored according to the conserved DEAD-box protein motif to which they belong or dark gray if not part of a conserved motif (see panel E). A portion of a symmetry related molecule (containing R584 and α20) that contacts the RNA is shown in transparent magenta. (C) Comparison of poly(U) RNAs bound to Mss116p (yellow), Vasa (red), eIF4AIII (white), and DDX19 (black). The RNA is shown after superposing the protein models of Vasa, eIF4AIII, and DDX19 onto Mss116p using PyMOL. The surface of Mss116p is colored by domains (see Figure 1) and is transparent to show the two α-helices (α8 and α18) involved in RNA bending. (D) Comparison of U10 RNA (yellow) bound to Mss116p (colored by domains) with an idealized A-form RNA duplex composed of a poly(U)10 strand (red) and a poly(A)10 strand (black). Regions that are involved in RNA bending (motif Ib and the CTE) or clash with the modeled poly(A) strand (Post-II) are labeled. (E) Cartoon showing contacts between Mss116p residues and U10 RNA. H-bonds, ionic contacts, hydrophobic contacts, and contacts through a water molecule are indicated by blue, light gray, black, and red arrows, respectively. Side- and main-chain interactions are indicated by solid and dashed lines, respectively. Double rectangles between RNA bases indicate base stacking. Residue colors are as in panel (B).
The bases of the centrally bound nucleotides U4, U5, and U6 stack with each other in near A-form geometry (Figures 4A and 4B). Between U6 and U7, the position of motif Ib in α8 of domain 1 causes a kink in the RNA that prevents U7 from stacking with U6 and continuing the A-form path (Figures 4C and 4E). This is the wedge α-helix, which was described in the structure of Vasa and other DEAD-box proteins and underlies current models of RNA-duplex unwinding by DEAD-box proteins (see Introduction). After the bend, U7 and U8 stack with each other, while U9 and U10 curl back onto the surface of Mss116p. The path of the RNA from U4 to U8 is very similar to that in Vasa, eIF4AIII, and DDX19 when the protein structures are superposed (Figure 4C).
Importantly, the Mss116p structures show that the 5′ end of the RNA (U1, U2, and U3) bends again due to the presence of the CTE. In the Vasa, eIF4AIII, and DDX19 structures, the base equivalent to U3 is also stacked with those of U4–U6, but in the Mss116p structure the ribose and base of U3 are rotated slightly and angled downward, making hydrophobic contacts with the surface of α18, while the phosphate of U3 makes an ionic contact to K384 (motif IV). After this slight bend, U2 and U1 are deflected to one side by the CTE. The ribose of U2 is angled upward and U1 is completely flipped over onto the CTE. The 5′ end of the RNA appears to assume the latter path in part because of bending by the CTE and in part because the CTE of a crystallographic symmetry mate packs into it (Figure 4B). While this crystal contact may influence the precise path of the 5′ end of the RNA in the structure, the location of the CTE is incompatible with continuation of an A-form helix, necessitating a second bend 5′ of the centrally bound nucleotides. This incompatibility is illustrated by superposing an ideal A-form U10 strand aligned with the stacked nucleotides of U4, U5, and U6 (Figure 4D). In this case, the 5′ end of the RNA exits the cleft differently but still runs into α18 necessitating the second bend. We conclude that the CTE introduces a second bend 5′ of the central bound nucleotides, which together with the first bend 3′ of the central bound nucleotides crimps the RNA strand.
Discussion
Here, we determined crystal structures of ternary complexes of Mss116p/Δ598-664 bound to ssRNA and three different ATP analogs, which mimic the ground and transition states for ATP hydrolysis. The structures, which include the entire helicase core and a functionally important α-helical CTE, provide insight into the function of the CTE, the potential for protein conformational changes associated with ATP binding and hydrolysis, and the mechanism of RNA unwinding. A key finding is that Mss116p utilizes two wedges, which act together to crimp RNA, a means of achieving local strand separation not seen in previous DEAD-box protein structures.
The CTE Stabilizes the Structure of the Helicase Core and Contributes to RNA Binding
The Mss116p/Δ598-664 structure shows that helicase core domains 1 and 2 are essentially the same as in other DEAD-box proteins, while the CTE is an additional compact module that is attached to and forms an extension of the RNA-binding side of domain 2. Thus, the CTE is positioned both to interact with the 5′ segment of the bound RNA strand and to stabilize the structure of domain 2. The CTE interacts extensively with domain 2 via packing interactions between the hydrophobic sides of α17, α18, and α19 and hydrophobic patches on the surface of domain 2, and C-terminal truncations that delete all or part of the CTE destabilize the protein, as judged by loss of activity and decreased thermal stability (see Results and below). Although the CTE can be fit readily onto domain 2 of other DEAD-box proteins (Figure S6), the hydrophobic patches on the surface of domain 2 with which it interacts are more extensive in Mss116p than in proteins, such as Vasa and DDX19, in which domain 2 may fold independently. This situation could reflect the evolution of Mss116p from an ancestral DEAD-box protein that acquired the CTE and then became dependent upon it for structural stability.
The basic tail (residues 597-664) is not present in Mss116p/Δ598-664, but the structure does show its point of attachment (residue 596), which is below the CTE on the RNA-binding side of domain 2. Thus, the basic tail is positioned to interact with distal upstream regions of the RNA, consistent with the hypothesis that it tethers the helicase core to larger RNA substrates for multiple rounds of RNA unwinding. The basic tail is predicted to be unstructured and is expected to interact non-specifically with the negatively charged phosphate backbone of RNA (Grohman et al., 2007; Mohr et al., 2008). Based on the Mss116p/Δ598-664 ternary complex structures, the 5′ segment of a longer bound RNA strand would emerge from the CTE at least two nucleotide residues upstream of U1, where it could immediately encounter the basic tail. However, a flexibly attached 68-residue tail could also potentially loop back to contact downstream regions of the bound RNA strand, help sequester the opposite RNA strand, or interact with and regulate the activity of the helicase core. Although it is possible that the CTE and basic tail have additional functions in targeting Mss116p to specific substrates via RNA-protein or protein-protein interactions, all RNA and RNP substrates tested thus far appear to interact with Mss116p non-specifically, presumably via phosphate-backbone interactions (Del Campo et al., 2009), as seen for the helicase core in the structures determined here.
Our results provide insight into how previously studied C-terminal truncations and mutations within the CTE and basic tail affect Mss116p activity. Mss116p/Δ508-664, with a C-terminal truncation that deletes the entire CTE, has decreased solubility and no detectable RNA-dependent ATPase activity (Mohr et al., 2008). The structure suggests that deletion of the CTE would destabilize the helicase core by exposing the hydrophobic surfaces of domain 2 that interact with the CTE to the solvent. Mss116p/Δ551-664 and Mss116p/Δ569-664, with shorter C-terminal truncations within or just downstream of α19 in the CTE, have CD spectra similar to that of the wild-type protein at 30°C, but decreased thermal stability compared to either the wild-type protein or Mss116p/Δ598-664, which lacks only the basic tail (Figure S5). These findings suggest that the short structured region containing β16, α20, α21, and β17 at the end of the CTE contributes to the stability of Mss116p.
Notably, despite having similar CD spectra and thermal stabilities, Mss116p/Δ569-664, which retains α19, has relatively high activity in vivo and in vitro, while Mss116p/Δ551-664, whose slightly longer C-terminal truncation removes most of α19, is almost completely inactive (Mohr et al., 2008). Thus, the disruption of α19 may affect the activity of Mss116p in some more specific manner than overall structural destabilization, such as impeding a required conformational change and/or inhibiting RNA binding. An effect on RNA binding could reflect that α19 helps position or support α18, which directly contacts the RNA, and is consistent with the finding that the apparent Kds for binding of group I and II introns by Mss116p/Δ551-664 are 61- to 304-fold higher than those for wild-type protein, compared to ~10-fold higher for Mss116p/Δ569-664, which retains α19 (Mohr et al., 2008). The previously described missense mutation Mss116p S557P in α19, which also results in loss of activity, and nearby pseudorevertants in α19, which restore activity (Huang, 2004; Mohr et al., 2008), also likely exert their effects by disrupting and restoring the structure of α19. Additionally, sequence alignments show that the G548V/L549S mutations in CYT-19, which inactivate the protein (Mohr et al., 2002), also fall within α19, supporting a conserved role for this helix in both Mss116p and CYT-19 (Figure S1).
Although an α-helical CTE similar to that of Mss116p is found in only some DEAD-box protein families, other DEAD-box proteins may use different N- or C-terminal extensions or partner proteins for similar functions involving structural stabilization or RNA unwinding. One example is the interaction between the DEAD-box protein eIF4A and a partner protein eIF4G, which contacts helicase core domains 1 and 2 on the side opposite the RNA-binding cleft and stimulates RNA-unwinding activity, presumably providing a means of regulating eIF4A’s activity (Schütz et al., 2008). A second example is the Barentsz protein (Btz or MLN51), which interacts with eIF4AIII in the EJC. The EJC structure shows that N- and C-terminal segments of Btz contact eIF4AIII domains 2 and 1, respectively, with Btz N-terminal residues P179-L193 contacting domain 2 in a position similar to α18 (I529-F546) of Mss116p’s CTE (Figure S6B). eIF4AIII by itself has low background ATPase and undetectable ssRNA binding and RNA-unwinding activities (Noble and Song, 2007), but, when activated by binding of Btz, eIF4AIII gains the ability to bind ssRNA along with RNA-stimulated ATPase and RNA-unwinding activities (Noble and Song, 2007). Further, the side chain of F188 makes a stacking contact with the base of U1 (U3 in Mss116p). Thus, Btz may function similarly to the CTE not only in stabilizing the structure of the helicase core but also in contributing to RNA crimping.
Mss116p Structures with Different Bound ATP Analogs Show Few Differences
Our structures show that the complexes of Mss116p/Δ598-664 with ssRNA and AMP-PNP, ADP-BeF3−, or ADP-AlF4− all crystallize in the same closed conformation. At the active site, the structures with the ground state analogs AMP-PNP and ADP-BeF3− are virtually identical, while the structure with the transition state analog ADP-AlF4− shows only small changes near the γ-phosphoryl group binding site (see Results). These small changes between the transition and ground state analogs do not translate into larger changes elsewhere in the protein. The lack of large conformational changes outside the ATP-binding site in ternary complexes with ssRNA and different ATP analogs agrees with the recent finding of six isomorphous structures of the dengue virus DExH-box protein NS3 bound to ssRNA or ssRNA plus ADP, AMP-PNP, ADP-VO4, or ADP-PO4 (Luo et al., 2008) and with structures of eIF4AIII in the EJC with bound AMP-PNP and ADP-AlF3 (Andersen et al., 2006; Bono et al., 2006; Nielsen et al., 2009). It remains possible that larger structural changes are prevented by crystal packing contacts and in solution ADP-BeFx induces a different DEAD-box protein conformation with larger changes that more efficiently carries out RNA unwinding. However, our results provide no indication of such a structure, nor is it necessary to postulate one to explain why ADP-BeFx but not other ATP analogs promotes RNA unwinding (see below).
Mss116p Is a Molecular Crimper
Because it is now clear that the ATP-bound state of DEAD-box proteins is competent for RNA-strand separation without ATP hydrolysis (Chen et al., 2008; Liu et al., 2008), we infer that the DEAD-box protein structures with bound ssRNA and ATP analogs may well reflect the final unwound state with the single strand separated from the duplex. The Mss116p structures show that the ssRNA in this state is bent on both sides of the centrally bound nucleotide residues. One bend is made by the motif Ib/wedge α-helix in domain 1, as seen previously in the structures of Vasa and other DEAD-box proteins, while the other bend is made by α18 in the CTE. Together these two wedges crimp the RNA strand in a manner that would separate it from a complementary strand in a duplex. Although the DEAD-box proteins Vasa, eIF4AIII, and DDX19 may accomplish RNA unwinding by inducing only a single bend with domain 1, it is also possible that they and other DEAD-box proteins similarly crimp RNA by producing two bends, either by using an ancillary domain not present in the crystallized constructs or an accessory protein, as discussed above for Btz acting together with eIF4AIII in the EJC.
Model for RNA Strand Separation by DEAD-Box Proteins
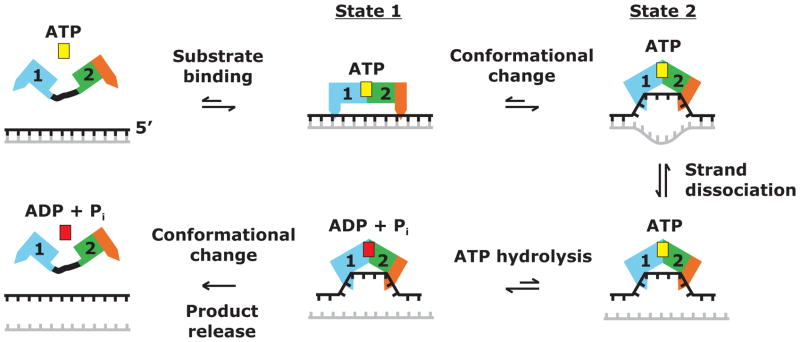
Considered together with previous data, the new structural information for Mss116p suggests a model of how DEAD-box proteins promote RNA-strand separation, which incorporates, refines, and extends previous models (Figure 5). Prior to the binding of ATP and RNA, the flexibly linked helicase core domains are in an open conformation, which can bind either ATP or RNA with no experimental evidence for a preferred order (Lorsh and Herschlag, 1998a; Theissen et al., 2008). The binding of both substrates induces a conformational change to a partially closed, pre-unwound state, denoted state 1, which can accommodate both strands of the RNA duplex (Nielsen et al., 2009). Following state 1, there is a further conformational change to the closed, unwound state 2, which we assume is reflected in the crystal structures. In state 2, the bound nucleotides of one strand of the RNA are bent or crimped by the protein and the duplex is disrupted.
Figure 5.
Model for the mechanism of RNA-strand separation by DEAD-box proteins. The DEAD-box protein with the helicase core in an open conformation binds ATP (yellow) and duplex RNA, leading to a partially closed pre-unwound state (state 1). State 1 then undergoes a conformational change to the closed unwound state (state 2) in which one RNA strand is bent or crimped. This conformational change is driven at least in part by the binding of one RNA strand in the RNA-binding cleft created by the interface of helicase core domains 1 and 2, and it actively unwinds the RNA duplex. ATP hydrolysis can occur during or after the conformational change from state 1 to state 2, with an additional conformational change required for the release of ADP + Pi (red) and the bound RNA strand. If the strands are incompletely separated or reassociate to reform the duplex, additional cycles of RNA binding and ATP hydrolysis may be needed for strand separation.
The ATP-dependent tight binding of a short (~6 nt) segment of the unwound RNA strand within the RNA-binding cleft of the helicase core presumably contributes to the driving force for the conformational change from state 1 to state 2. Reflecting this ATP-induced tight binding, the apparent Kd for the binding of a ssRNA oligonucleotide by Mss116p decreased from 28 nM in the absence of nucleotide to 2 nM in the presence of AMP-PNP, similar to findings for other DEAD-box proteins (Mohr et al., 2008 and references therein). All of the structures of DEAD-box proteins in a ternary complex with bound ssRNA, including the Mss116p structures here, are incompatible with binding an A-form duplex due to the conserved amino acid residues following motif II (post-II region), which would clash with the opposite strand (Figure 4D; Andersen et al., 2006). Thus, the conformational change from state 1 to state 2 is likely the key step that actively separates the two strands of a duplex.
ATP hydrolysis is not required for RNA unwinding and can be catalyzed by the helicase core during or after the conformational change from state 1 to state 2 (Nielsen et al., 2009; Theissen et al., 2008). The latter is best demonstrated by the finding that the related helicase core of dengue virus NS3 bound to ssRNA in state 2 can hydrolyze ATP soaked into the crystals (Luo et al., 2008). However, while ATP hydrolysis during the transition from state 1 to state 2 is not required for RNA unwinding, it may well enhance it, either by lowering the activation energy for the conformational change from state 1 to state 2 or by stabilizing the unwound state, possibly leading to the unwinding of additional nucleotide residues. Such enhancement may be why ATP gives higher single turnover rate constants for duplex unwinding than does ADP-BeFx (Liu et al., 2008).
Finally, another conformational change is necessary to release the bound RNA strand and the ATP-hydrolysis products, ADP and Pi, as shown by the finding that ATP-hydrolysis products are present in eIF4AIII in the EJC, where this conformational change is prevented by other proteins in the EJC (Nielsen et al., 2009). If the DEAD-box protein is tethered to an RNA substrate through another interaction (e.g., the basic tail in Mss116p), the process can restart close to the same start site.
Our finding of no large structural changes outside the ATPase active site for ternary complexes of Mss116p/Δ598-664 with different bound ATP analogs constrains models for DEAD-box protein structural transitions during RNA unwinding. Assuming that the DEAD-box protein structures with bound ATP analogs and ssRNA do in fact represent state 2, the structures show that all three ATP analogs are compatible with the formation of the same unwound state 2 when starting from ssRNA. The ability of Mss116p to promote RNA unwinding in the presence of ADP-BeFx but not AMP-PNP could then be explained in two ways, which are not mutually exclusive. The first possibility is that although all three ATP analogs can induce Mss116p to form state 2 with ssRNA, only ADP-BeFx can form state 1 with double-stranded RNA (dsRNA). If state 1 is an obligatory intermediate for formation of state 2 starting from dsRNA, then only ADP-BeFx would support unwinding. The second possibility is that all three ATP analogs can induce Mss116p to bind dsRNA in state 1 and ssRNA in state 2, to an extent sufficient to become trapped in a crystal lattice; however, in solution, only ATP or ADP-BeFx decreases the activation energy for the conformational change from state 1 to state 2 or stabilizes state 2 sufficiently to promote experimentally detectable RNA unwinding. Recent findings show that under RNA splicing conditions, Mss116p does in fact bind ssRNA oligonucleotides more tightly in the presence of ADP-BeFx than in the presence of AMP-PNP (F. Liu, E. Jankowsky, M.D. and A.M.L., unpublished data), consistent with the second possibility. Further testing of this model will require more detailed biochemical and structural analysis of the binding of DEAD-box proteins to dsRNA.
Experimental Procedures
Materials
The RNA oligonucleotide poly(U)10 (denoted U10) was synthesized by Integrated DNA Technologies, and 5′-UUU(5-BrU)UUUUUU-3′ (denoted Br-U10), with a 5-bromouridine for anomalous phasing, was synthesized by Dharmacon. SeMet and AlCl3 were from Acros Organics. AMP-PNP, ADP, BeCl2, and NaF were from Sigma-Aldrich. ADP-BeFx-Mg2+ and ADP-AlFx-Mg2+ were prepared by mixing BeCl2 or AlCl3 with equimolar amounts of ADP and MgCl2 and a 5-fold molar excess of NaF.
Protein Expression and Purification
The expression and purification of Mss116p/Δ598-664 for crystallization is described in detail elsewhere (Del Campo and Lambowitz, 2009). Briefly, a maltose binding protein-Mss116p/Δ598-664 fusion protein was expressed in E. coli strain Rosetta2 (EMD Biosciences), purified by amylose-affinity chromatography, cleaved with tobacco etch virus protease, and further purified by heparin-Sepharose chromatography and Superdex 200 gel filtration. SeMet-labeled Mss116p/Δ598-664 was expressed in Rosetta2 using PASM-5052 auto-inducing media (Studier, 2005) and purified like the native protein. Proteins used for crystallization were dialyzed into storage buffer (10 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1 mM DTT, 50 mM Arg + Glu, 50% glycerol) and concentrated to ~10 mg/ml, as measured by Bradford assay (Bio-Rad), using bovine serum albumin (Pierce Biotechnology) as a standard.
Crystallization
Ternary complexes were formed by incubating 90 μM Mss116p/Δ598-664, 180 μM ssRNA, 2 mM MgCl2, and 1 mM ATP analog for 10 min on the desktop. Sitting drops were assembled using 1 μl of complex, 0.8 μl of either 8% tacsimate, pH 7.0, 15% polyethylene glycol (PEG) 3350 (SeMet and Br-U10 complexes) or 0.2 M succinate pH 7.0, 15% PEG 3350 (AMP-PNP and ADP-AlF4− complexes), and 0.2 μl of a 1:100 dilution of crystal seed. Crystal seed for the AMP-PNP and ADP-AlF4− complexes was made as described (Del Campo and Lambowitz, 2009). The ADP-BeF3− complex did not require seeding and sitting drops were assembled using 1 μl of complex and 1 μl of 8% tacsimate pH 7.0, 15% PEG 3350, 2.5% ethylene glycol. For each sitting drop, the reservoir solution was 250 or 500 μl of a 1:1 mix of crystallization solution and Mss116p storage buffer (see above). The sitting drops were stored at 15° C. Crystals were removed from the drops and flash cooled immediately in liquid N2.
Structure Determination
For all crystals, synchrotron X-ray diffraction data were collected at LS-CAT beamlines 21-ID-D and -G at the Advanced Photon Source (APS), Argonne National Laboratory. Details of data collection and phasing are in Table 1. Diffraction intensities were indexed and scaled with HKL2000 (Otwinowski and Minor, 1997). SIRAS phasing was performed with autoSHARP (Vonrhein et al., 2007), using the SeMet AMP-PNP (8 Se sites) and native AMP-PNP complex datasets. ARP/wARP (Langer et al., 2008) built the initial protein model, and subsequent rounds of manual model building and refinement were done with Coot (Emsley and Cowtan, 2004) and Refmac (Murshudov et al., 1997). To build and refine models of the other complexes, the Rfree flags from the AMP-PNP complex dataset were transferred to the other datasets using tools from CCP4 (CCP4, 1994), and the protein model from the AMP-PNP dataset was then refined against the other datasets. Towards the end of refinement, TLS (translation, libration, screw) refinement in Refmac was done using four TLS groupings for Mss116p (residues 88-113; 114-334; 335-564; 565-596), a single grouping for the RNA, and a single grouping for the ATP analog. Model validation was performed using MolProbity (Davis et al., 2007). Structural figures were prepared using PyMOL (http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank Paul Paukstelis for assistance with data collection, data processing, refinement, helpful discussions, and comments on the manuscript. We thank Rick Russell, Eckhard Jankowsky, and Sabine Mohr for comments on the manuscript, and Eckhard Jankowsky for permission to cite unpublished data. We are grateful to Joe Brunzelle for help with data collection at the APS. Use of the APS was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). This work was supported by NIH grant GM037951 to A.M.L. M.D. was the recipient of NIH postdoctoral fellowship F32-GM76961.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Séraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen B, Doucleff M, Wemmer DE, De Carlo S, Huang HH, Nogales E, Hoover TR, Kondrashkina E, Guo L, Nixon BT. ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure. 2007;15:429–440. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci USA. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R, Karlberg T, Lehtiö L, Schütz P, van den Berg S, Dahlgren LG, Hammarström M, Weigelt J, Schüler H. The DExD/H-box RNA helicase DDX19 is regulated by an alpha-helical switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Tanner NK, Doére M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Lambowitz AM. Crystallization and preliminary X-ray diffraction of the DEAD-box protein Mss116p complexed with an RNA oligonucleotide and AMP-PNP. Acta Crystallogr F Struct Biol Cryst Commun. 2009;65:832–835. doi: 10.1107/S1744309109027225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz AM. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich M, Schulten K. Zooming in on ATP hydrolysis in F1. J Bioenerg Biomembr. 2005;37:441–444. doi: 10.1007/s10863-005-9487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: The C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and II intron splicing. Biochemical characterization of MSS116p, ATP-hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H-R. Ph.D. Thesis. University of Texas Southwestern Medical Center; Dallas: 2004. Functional studies of intron- and nuclear-encoded splicing factors in the mitochondria of Saccharomyces cerevisiae. [Google Scholar]
- Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998a;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998b;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A, et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nielsen KH, Chamieh H, Andersen CB, Fredslund F, Hamborg K, Le Hir H, Andersen GR. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS ONE. 2007;2:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci USA. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci USA. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.