Abstract
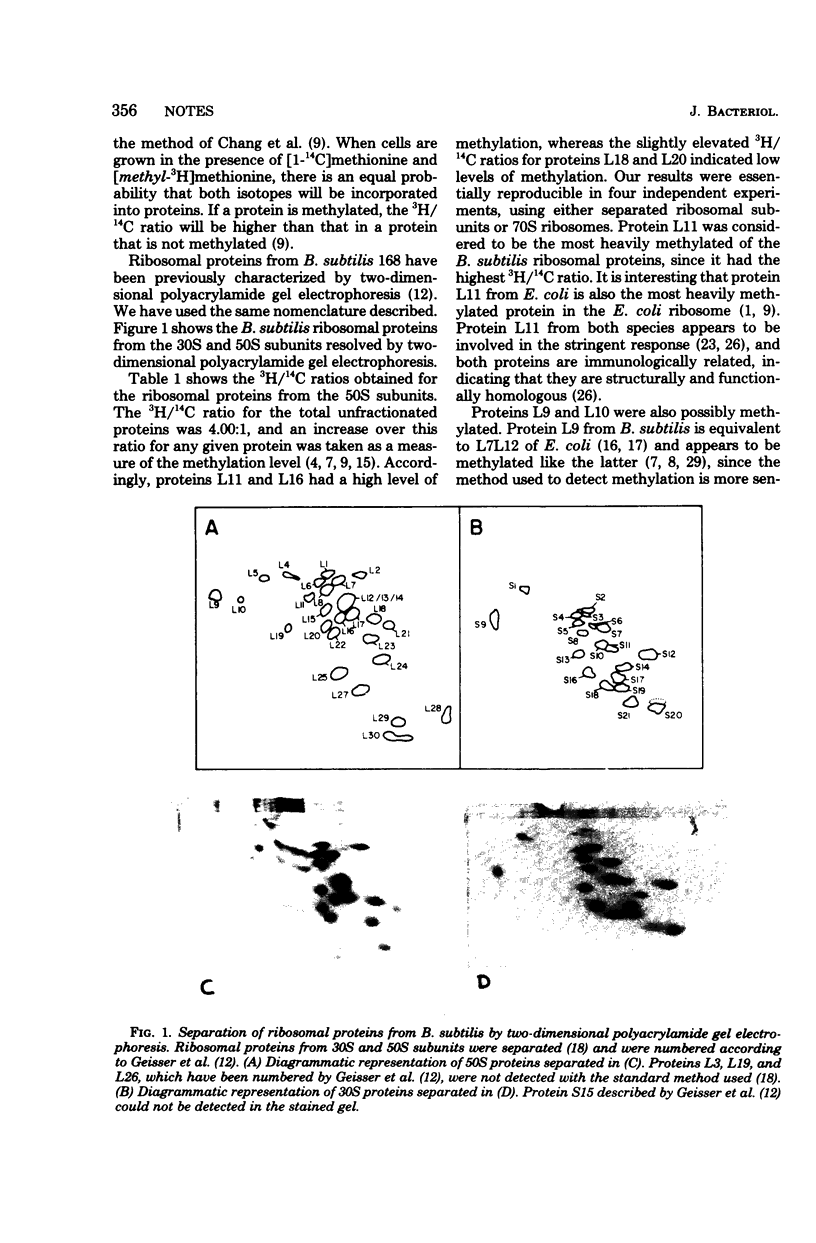
We measured the methylation of ribosomal proteins from the 30S and 50S subunits of Bacillus subtilis after growing the cells in the presence of [1-14C]methionine and [methyl-3H]methionine. Two-dimensional polyacrylamide gel electrophoretic analysis revealed a preferential methylation of the 50S ribosomal proteins. Proteins L11 and L16, and possibly L9, L10, L18, and L20, were methylated. On the other hand, only two possibly methylated proteins were found on the 30S subunit. A comparison of these results with those for Escherichia coli suggests a common methylation pattern for the bacterial ribosomal proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alix J. H., Hayes D., Nierhaus K. H. Properties of ribosomes and RNA synthesized by Escherichia coli grown in the presence of ethionine. V. Methylation dependence on the assembly of E. coli 50 S ribosomal subunits. J Mol Biol. 1979 Feb 5;127(4):375–395. doi: 10.1016/0022-2836(79)90228-6. [DOI] [PubMed] [Google Scholar]
- Alix J. H., Hayes D. Properties of ribosomes and RNA synthesized by Escherichia coli grown in the presence of ethionine. 3. Methylated proteins in 50 S ribosomes of E. coli EA2. J Mol Biol. 1974 Jun 15;86(1):139–159. doi: 10.1016/s0022-2836(74)80013-6. [DOI] [PubMed] [Google Scholar]
- Brosius J., Chen R. The primary structure of protein L16 located at the peptidyltransferase center of Escherichia coli ribosomes. FEBS Lett. 1976 Sep 15;68(1):105–109. doi: 10.1016/0014-5793(76)80415-2. [DOI] [PubMed] [Google Scholar]
- Cannon M., Cundliffe E. Methylation of basic proteins in ribosomes from wild-type and thiostrepton-resistant strains of Bacillus megaterium and their electrophoretic analysis. Eur J Biochem. 1979 Jul;97(2):541–545. doi: 10.1111/j.1432-1033.1979.tb13142.x. [DOI] [PubMed] [Google Scholar]
- Cannon M., Schindler D., Davies J. Methylation of proteins in 60S ribosomal subunits from Saccharomyces cerevisiae. FEBS Lett. 1977 Mar 15;75(1):187–191. doi: 10.1016/0014-5793(77)80083-5. [DOI] [PubMed] [Google Scholar]
- Chang C. N., Chang F. N. Methylation of ribosomal proteins in vitro. Nature. 1974 Oct 25;251(5477):731–733. doi: 10.1038/251731a0. [DOI] [PubMed] [Google Scholar]
- Chang C. N., Chang N. Methylation of the ribosomal proteins in Escherichia coli. Nature and stoichiometry of the methylated amino acids in 50S ribosomal proteins. Biochemistry. 1975 Feb 11;14(3):468–477. doi: 10.1021/bi00674a002. [DOI] [PubMed] [Google Scholar]
- Chang F. N., Chang C. N., Paik W. K. Methylation of ribosomal proteins in Escherichia coli. J Bacteriol. 1974 Nov;120(2):651–656. doi: 10.1128/jb.120.2.651-656.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. N., Navickas I. J., Chang C. N., Dancis B. M. Methylation of ribosomal proteins in HeLa cells. Arch Biochem Biophys. 1976 Feb;172(2):627–633. doi: 10.1016/0003-9861(76)90117-x. [DOI] [PubMed] [Google Scholar]
- Chang F. N. Temperature-dependent variation in the extent of methylation of ribosomal proteins L7 and L12 in Escherichia coli. J Bacteriol. 1978 Sep;135(3):1165–1166. doi: 10.1128/jb.135.3.1165-1166.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb D. G., Sarkar N., Pinzino C. J. The methylation of lysine residues in protein. J Biol Chem. 1966 Apr 25;241(8):1857–1862. [PubMed] [Google Scholar]
- Geisser M., Tischendorf G. W., Stöffler G. Comparative immunological and electrophoretic studies on ribosomal proteins of bacillaceae. Mol Gen Genet. 1973 Dec 20;127(2):129–145. doi: 10.1007/BF00333661. [DOI] [PubMed] [Google Scholar]
- Guha S., Szulmajster J. Isolation of 30S and 50S active ribosomal subunits of Bacillus subtilis, Marburg strain. J Bacteriol. 1975 Dec;124(3):1062–1066. doi: 10.1128/jb.124.3.1062-1066.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S. J., Kurland C. G., Voynow P., Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry. 1969 Jul;8(7):2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- Hernandez F., Cannon M. Methylation of proteins in 40 S ribosomal subunits from Saccharomyces cerevisiae. FEBS Lett. 1978 May 15;89(2):271–275. doi: 10.1016/0014-5793(78)80234-8. [DOI] [PubMed] [Google Scholar]
- Itoh T., Wittmann-Liebold B. The primary structure of Bacillus subtilis acidic ribosomal protein B-L9 and its comparison with Escherichia coli proteins L7/L12. FEBS Lett. 1978 Dec 15;96(2):392–394. doi: 10.1016/0014-5793(78)80445-1. [DOI] [PubMed] [Google Scholar]
- Jerez C. A., Mardones E., Amaro A. M. Alteration of the acidic ribosomal proteins from dormant spores of Bacillus subtilis. FEBS Lett. 1976 Sep 1;67(3):276–280. doi: 10.1016/0014-5793(76)80546-7. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt E., Wittmann H. G. Ribosomal proteins. VII. Two-dimensional polyacrylamide gel electrophoresis for fingerprinting of ribosomal proteins. Anal Biochem. 1970 Aug;36(2):401–412. doi: 10.1016/0003-2697(70)90376-3. [DOI] [PubMed] [Google Scholar]
- Kruiswijk T., Kunst A., Planta R. J., Mager W. H. Modification of yeast ribosomal proteins. Methylation. Biochem J. 1978 Oct 1;175(1):221–225. doi: 10.1042/bj1750221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault-Demare L., Chambliss G. H. Natural messenger ribonucleic acid-directed cell-free protein-synthesizing system of Bacillus subtilis. J Bacteriol. 1974 Dec;120(3):1300–1307. doi: 10.1128/jb.120.3.1300-1307.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik W. K., Kim S. Protein methylation. Science. 1971 Oct 8;174(4005):114–119. doi: 10.1126/science.174.4005.114. [DOI] [PubMed] [Google Scholar]
- Parker J., Watson R. J., Friesen J. D. A relaxed mutant with an altered ribosomal protein L11. Mol Gen Genet. 1976 Feb 27;144(1):111–114. doi: 10.1007/BF00277313. [DOI] [PubMed] [Google Scholar]
- Reporter M. Methylation of basic residues in structural proteins. Mech Ageing Dev. 1973 Mar;1(5):367–372. doi: 10.1016/0047-6374(73)90043-2. [DOI] [PubMed] [Google Scholar]
- Smith I., Paress P., Pestka S. Thiostrepton-resistant mutants exhibit relaxed synthesis of RNA. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5993–5997. doi: 10.1073/pnas.75.12.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhorst C., Möller W., Laursen R., Wittmann-Liebold B. The primary structure of an acidic protein from 50-S ribosomes of Escherichia coli which is involved in GTP hydrolysis dependent on elongation factors G and T. Eur J Biochem. 1973 Apr 2;34(1):138–152. doi: 10.1111/j.1432-1033.1973.tb02740.x. [DOI] [PubMed] [Google Scholar]
- Terhorst C., Wittmann-Liebold B., Möller W. 50-S ribosomal proteins. Peptide studies on two acidic proteins, A 1 and A 2 , isolated from 50-S ribosomes of Escherichia coli. Eur J Biochem. 1972 Jan 31;25(1):13–19. doi: 10.1111/j.1432-1033.1972.tb01661.x. [DOI] [PubMed] [Google Scholar]



