Introduction
Apoptosis is probably not the only form of cell death, but it is currently the only form for which the proteins that constitute the death pathway have been characterized at the structural, biochemical and genetic levels in great detail. Apoptosis is an ancient paradigm, going back to the earliest multicellular animals, but does not exist in plants, simple eukaryotes or microbes (Lamkanfi et al., 2002; Zmasek et al., 2007). Signal transduction in apoptosis is different to most well-described signal transduction pathways in that it does not require kinases or phosphatases, although these might well set a threshold for the ‘fitness’ of a cell to die. The two best-described apoptosis pathways in mammals, the intrinsic and the extrinsic pathways, converge on a common execution phase. This phase is driven by proteases known as caspases, which are typically subdivided into those that contain large prodomains and act at the apex of a proteolytic cascade (i.e. the initiator caspases-8, -9 and -10), and those that are activated by initiator caspases, through proteolytic cleavage, and in turn dismantle cells into easily engulfed apoptotic bodies (i.e. the effector caspases-3, -6 and -7) (Fuentes-Prior and Salvesen, 2004). It is essential to understand that caspases are signaling proteases, designed for specific protein cleavage and not for protein degradation. They make one or two cuts in their target proteins and do not destroy protein structure, but rather modify it, thus creating gain-of-function or loss-of-function events that drive forward the apoptotic phenotype (Timmer and Salvesen, 2007). A good example of gain of function by limited proteolysis is within the caspase cascade itself: the proteolytic action of the initiator caspases on the latent (zymogen) forms of the effector caspases. Importantly, the conversion of the death signal to the first proteolytic event in the intrinsic and extrinsic pathways has received a lot of attention, because control of the initiation phase is a major therapeutic target for both proliferative and degenerative diseases (Reed, 2002). This review summarizes the accumulated structural and biochemical evidence that defines the origin of the first proteolytic signal of the intrinsic
apoptosis pathway, that is, activation of the initiator caspase-9 by the cytosolic signaling platform known as the ‘apoptosome’.
In the extrinsic pathway, members of the tumor necrosis factor (TNF) receptor superfamily, including Fas (also known as CD95) and TNF-related apoptosis-inducing ligand (TRAIL) receptors 1 and 2 (TRAIL-R1/DR4 and TRAIL-R2/DR5), are clustered by their cognate ligands. This leads to recruitment and stabilization of a conformer of the adapter protein Fas-associated death domain (FADD) that is able to recruit procaspase-8 to form a death-inducing signaling complex (DISC) (Scott et al., 2009). The DISC is essentially a membrane-bound matrix that serves as a platform to increase the local concentration of procaspase-8, resulting in its activation by dimerization and intermolecular processing. Processed or active caspase-8 then cleaves and activates procaspases-3 and -7 (Fuentes-Prior and Salvesen, 2004).
The intrinsic pathway, largely conserved in worms and flies (Box 1), in mammals is triggered in response to distinct cellular stressors, including growth factor withdrawal, DNA damage and endoplasmic reticulum (ER) stress, and is transduced by members of the Bcl-2 family. Bcl-2 homology 3 (BH3)-only members of this family either directly activate the pro-apoptotic Bcl-2 family members BAX and BAK, or do so by antagonizing anti-apoptotic Bcl-2 family members (Brunelle and Letai, 2009). BAX and BAK are thought to homo-oligomerize to form pores in the outer mitochondrial membrane, through which pro-apoptotic proteins, including cytochrome c and second mitochondrial activator of caspases (Smac; also known as DIABLO), can pass from the intermembrane space into the cytoplasm. Cytochrome c, in particular, is a crucial mediator of the intrinsic pathway, as it activates yet another adaptor protein, known as apoptotic protease-activating factor-1 (Apaf-1), which homo-oligomerizes into a caspase-activating complex that sequentially recruits and activates the initiator caspase-9 and the effector caspases-3 and -7 (Bratton et al., 2001; Zou et al., 1997). The complex of Apaf-1 and caspase-9 is known as the apoptosome, and has a very different molecular mechanism to the DISC, although the apoptosome and the DISC share the common function of activating their respective apical caspases to initiate the apoptotic pathway (Riedl and Salvesen, 2007).
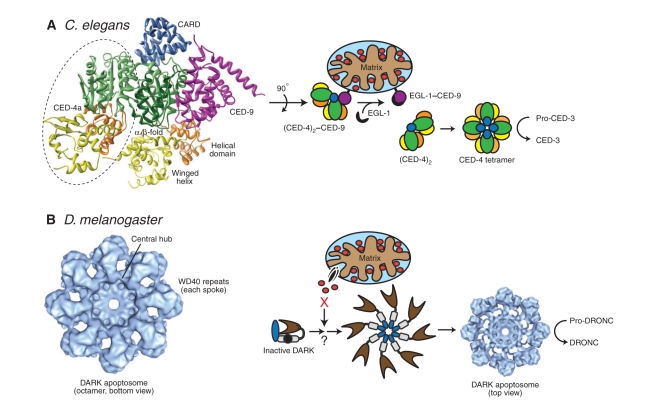
Box 1. Evolutionary conservation of apoptosome complexes
Apaf-1 and caspase-9 homologs are conserved in all multicellular animals, dating back to the simplest cnidarians such as sea anemones (Dunn et al., 2006). It seems that the intrinsic pathway might be a very primitive construct. Orthologs in C. elegans are very distant, but Drosophila melanogaster orthologs are closer to those in humans. Interestingly, the Apaf-1 homolog in C. elegans (CED-4) was originally thought to oligomerize into a tetrameric apoptosome, which in turn activates its cognate caspase, CED-3 (see panel A) (Yan et al., 2005). A more recent study, however, suggests that the worm apoptosome contains eight CED-4 proteins, organized as a tetramer of asymmetric CED-4 dimers (Qi et al., 2010).
The D. melanogaster Apaf-1 homolog, DARK, also oligomerizes into an octameric apoptosome that activates its initiator caspase, DRONC (panel B) (Yu et al., 2006), but, in contrast to human Apaf-1, DARK is refractory to cytochrome c activation, despite having WDRs (Dorstyn et al., 2004; Rodriguez et al., 1999). Consequently, DARK is either activated by an unknown ligand or has lost this regulatory feature altogether and undergoes a low level of continuous and constitutive activation (Muro et al., 2002). CED-4 does not contain WDRs and is held in check by its anti-apoptotic binding partner, CED-9. Following an apoptotic stimulus, CED-4 is displaced from CED-9 by the BH3-only homolog, EGL-1, which facilitates CED-4 oligomerization into the apoptosome (panel A) (Spector et al., 1997).
Despite these differences, both CED-4 and DARK use the same mechanism as Apaf-1. An N-terminal CARD domain, followed by a NOD/NB-ARC region with its characteristic AAA+ type ATPase domain, creates an oligomeric platform for activation of their cognate initiator caspases. They seem to be distinct, however, in their use of (d)ATP in apoptosome formation, because no ATPase activity is found in vitro for these divergent apoptosomes (Yan et al., 2005). This underlines the regulatory function of (d)ATP in apoptosome formation, setting them far apart from the traditional ATP-consuming, cycling ATPases.
Finally, although the stoichiometry of Apaf-1 and Apaf-1 homologs to initiator caspases has long been assumed to be 1:1 within their respective complexes, recent work with CED-4 suggests that the fly apoptosome possesses an internal cavity (or ‘hutch’) that accommodates and apparently activates only two CED-3 molecules (Qi et al., 2010). Thus, apoptosome assembly and the resulting stoichiometry of initiator caspase to activator probably play a crucial role in regulating its activity.
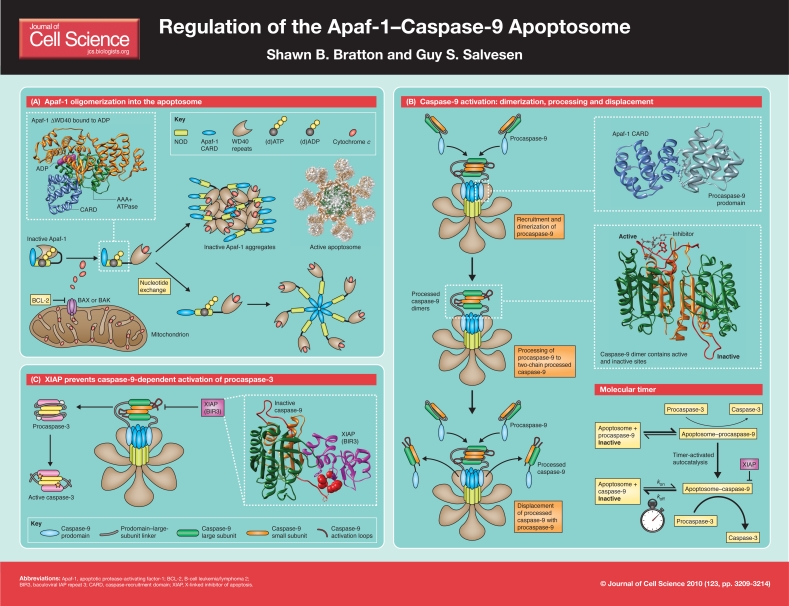
Apaf-1: structure, nucleotide hydrolysis and exchange, and oligomerization
Apaf-1 is a multidomain adapter protein containing an N-terminal caspase recruitment domain (CARD), followed by a nucleotide-binding and oligomerization domain (NOD, also known as NB-ARC) that is homologous to CED-4 in Caenorhabditis elegans, and a C-terminal regulatory Y-domain composed of 12–13 WD40 repeats (WDRs), which form seven- and six-blade β-propellers (Poster, panel A) (Box 1) (Riedl et al., 2005; Yu et al., 2005; Zou et al., 1997). Structural studies of the NOD indicate that it contains an AAA+ ATPase domain [with Walker A and B boxes that bind to (d)ATP and Mg2+], a winged-helix domain that contributes to nucleotide binding, and a superhelical domain (Riedl et al., 2005). Apaf-1 is normally present as a monomer in an inactive, locked conformation bound to dATP or ATP [hereafter referred to as (d)ATP] (Kim et al., 2005). However, upon cytochrome c binding to its WDRs (most probably between its β-propellers), Apaf-1 is thought to undergo a conformational change, driven by (d)ATP hydrolysis. In this semi-open conformation, Apaf-1 is susceptible to unproductive aggregation, but normally undergoes nucleotide exchange, accelerated in vitro by a complex composed of Hsp70, the tumor suppressor PHAPI and cellular apoptosis susceptibility (CAS) protein (Kim et al., 2005; Kim et al., 2008). Although the specific details remain unclear, following nucleotide exchange, the CARD and ancillary helical domains of Apaf-1 undergo additional (apparently radical) conformational changes to expose its AAA+ ATPase domain and allow multiple Apaf-1 proteins to oligomerize into a circular heptameric apoptosome complex (Poster, panel A) (Riedl and Salvesen, 2007; Yuan et al., 2010). To date, a high-resolution crystal structure of full-length Apaf-1 has not been attainable, much less a fully assembled complex. However, modeling of electron cryomicroscopic images at ~9.5 Å suggests the complex contains seven Apaf-1 molecules, wherein the CARDs form a ring situated directly above a central hub composed of the oligomerized NODs. When incubated with procaspase-9, the CARDs in both Apaf-1 and procaspase-9 form a flexibly tethered ‘disk’ that sits above the central hub, and each of the WDR domains is connected to the central hub through the superhelical domain. Overall, this gives the appearance of a wheel-like particle in which seven bent spokes radiate outward from the central hub and procaspase-9 enzymes (or at least their CARDs) sit atop the complex (Poster, panels A and B) (Yu et al., 2005; Yuan et al., 2010).
Caspase-9: structure, dimerization and activation, and cleavage
Conformational changes that occur following nucleotide exchange expose the CARD in Apaf-1, so that its oligomerization is coupled to recruitment of procaspase-9 to the resulting apoptosome. As previously noted, initiator caspases, like procaspase-9, possess long prodomains that facilitate their interaction with adapter proteins. The prodomain in caspase-9, in particular, contains a CARD that allows it to bind selectively to the CARD in Apaf-1 through homotypic interactions (Qin et al., 1999). Unlike other initiator caspases, including caspases-2, -8 and -10, the prodomain of caspase-9 is not removed during apoptosis; in fact, caspase-9 (in both its procaspase-9 and processed forms; discussed below) must remain bound to the apoptosome to retain substantial catalytic activity (Bratton et al., 2001; Rodriguez and Lazebnik, 1999; Stennicke et al., 1999). Although some studies suggest that this CARD–CARD interaction might induce a conformational change in monomeric caspase-9 that is necessary for its activation (Chao et al., 2005), most studies argue that instead the apoptosome serves primarily as a platform to increase the local concentration of procaspase-9 and facilitate its dimer-driven activation (Poster, panel B) (Boatright et al., 2003). In support of this ‘induced-proximity model’, incubation of a prodomain-less version of caspase-9 with kosmotropic salts induces caspase-9 dimerization and activation, as does replacing the prodomain in caspase-9 with a known dimerization domain, such as the GCN4 leucine zipper (Pop et al., 2006; Yin et al., 2006). Moreover, a chimeric caspase-8 protein, containing the prodomain of caspase-9, is activated by a truncated apoptosome, implying that a basic feature of all caspase-activating complexes is their ability to lower the threshold for initiator caspase dimerization (Pop et al., 2006). Whether the apoptosome induces additional conformational changes in caspase-9, which further enhance its activity following dimerization, remains an intensive area of investigation.
Interestingly, another notable difference between caspase-9 and the other initiator caspases (as well as effector caspases) is the length of its so-called activation loop, which connects its large and small subunits. In most caspases, this linker must be cleaved to allow the conformational changes necessary for active-site formation. In caspase-9, however, the linker is particularly long, so that, upon its recruitment and dimerization within the apoptosome, the linker is predicted to move and allow access to the active site in the absence of cleavage. In short, procaspase-9 has high catalytic activity when bound to the apoptosome (Renatus et al., 2001; Stennicke et al., 1999). The first proposed mechanism of caspase activation suggested that autocatalytic cleavage was a requirement for activity (Thornberry and Lazebnik, 1998), and the revelation that it is dispensable for caspase-9 thus raised a number of interesting questions, the most obvious of which is, if cleavage of caspase-9 is not required for its activation, then why does it occur? Recent studies suggest that procaspase-9 possesses higher affinity for the apoptosome, compared with the cleaved form of the protease, thereby facilitating a continuous cycle of procaspase-9 recruitment and activation, processing and release from the complex. Owing to its rapid autocatalytic cleavage within the apoptosome, however, procaspase-9 per se contributes little to the activation of procaspase-3. Thus, the apoptosome appears to function as a proteolytic-based ‘molecular timer’, in which the intracellular concentration of procaspase-9 sets the overall length of the timer, procaspase-9 autoprocessing activates the timer, and the rate at which a single processed caspase-9 molecule dissociates from the complex (and thus loses its capacity to activate procaspase-3) dictates how fast the timer ‘ticks’ over. Collectively, these results suggest that the purpose of procaspase-9 autoprocessing is not to activate caspase-9, but rather to initiate a molecular timer that regulates the duration of apoptosome activity (Poster, panel B) (Malladi et al., 2009).
Importantly, procaspase-9 processing also plays another regulatory role, in that it generates a neo-epitope that allows the processed protease to be directly inhibited by X-linked inhibitor of apoptosis (XIAP), a member of the IAP protein family. Biochemical and structural data indicate that the baculovirus IAP repeat 3 (BIR3) domain of XIAP binds to the neo-N-terminus of the small subunit of caspase-9, which arises following its processing, thereby forming a complex with caspase-9 that prevents the protease from undergoing dimerization (Poster, panel C) (Shiozaki et al., 2003; Srinivasula et al., 2001). If caspase-9 manages to process some caspase-3 or caspase-7 in this environment, XIAP can use its BIR2 domain (and adjacent residues) for binding and inhibiting these active effector caspases. Therefore, processing of procaspase-9 serves at least two roles: altering the inherent activity of the apoptosome and subjecting the apoptosome to further regulation by XIAP (Bratton et al., 2002; Malladi et al., 2009; Riedl et al., 2001; Scott et al., 2005).
Additional regulators of the apoptosome
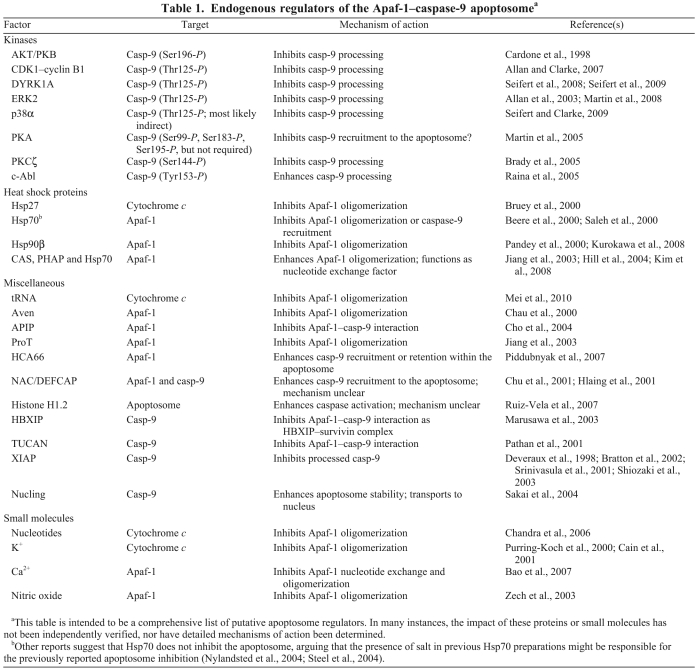
Given the crucial role of the apoptosome in mediating cell death, it is perhaps not surprising that a number of proteins and small molecules have been reported to regulate its function (Table 1). For example, several kinases phosphorylate caspase-9 at Thr125, Tyr153, Ser144 or Ser196 (although the latter two sites are not conserved in the mouse). ERK2, DYRK1A, CDK1–cyclin B1 and p38α phosphorylate caspase-9 at Thr125 and inhibit the cleavage of procaspase-9, whereas c-Abl reportedly phosphorylates caspase-9 at Tyr153, resulting in its activation (Allan and Clarke, 2007; Allan et al., 2003; Brady et al., 2005; Cardone et al., 1998; Martin et al., 2005; Martin et al., 2008; Raina et al., 2005; Seifert et al., 2008; Seifert and Clarke, 2009). Nevertheless, it is important to note that, in all cases, it is unclear how phosphorylation inhibits or activates caspase-9. Thr125 is located in the hinge region near the N-terminus of the large subunit and thus might inhibit prodomain exposure and caspase-9 recruitment to the apoptosome, but this has not been formally tested in an apoptosome reconstitution experiment.
Table 1.
Endogenous regulators of the Apaf-1–caspase-9 apoptosomea
Heat shock proteins (Hsps), including Hsp27, Hsp70 and Hsp90, reportedly also inhibit apoptosome formation and/or recruitment of caspase-9 to the complex by binding to cytochrome c or Apaf-1 (Beere et al., 2000; Bruey et al., 2000; Garrido et al., 1999; Kurokawa et al., 2008; Pandey et al., 2000; Saleh et al., 2000). The role of Hsp70 as a direct inhibitor of the apoptosome, however, is controversial, in part because of potential experimental artifacts and also because it apparently plays an important role in nucleotide (d)ATD–(d)ATP exchange (Hill et al., 2004; Jiang et al., 2003; Kim et al., 2008; Nylandsted et al., 2004; Steel et al., 2004). Recently, tRNA, various nucleotides (at physiological concentrations), and cations such as K+ and Ca2+ have been shown to inhibit Apaf-1 oligomerization by binding to cytochrome c or by inhibiting nucleotide exchange (Bao et al., 2007; Cain et al., 2001; Chandra et al., 2006; Mei et al., 2010; Purring-Koch and McLendon, 2000). Conversely, a number of additional proteins, including HCA66, NAC/DEFCAP, histone H1.2 and nucling, are thought to enhance apoptosome function (Chu et al., 2001; Hlaing et al., 2001; Piddubnyak et al., 2007; Ruiz-Vela and Korsmeyer, 2007; Sakai et al., 2004). In most cases, it remains unclear how this is achieved, but it will be interesting to assess their potential impact on the molecular timer of the apoptosome, as stabilization of processed caspase-9 within the apoptosome would be predicted to prolong its activity.
Future directions
Much has been learned about the apoptosome over the past decade but, equally, much remains unresolved. Low-resolution structures of the apoptosome exist, but the precise arrangement and stoichiometry of Apaf-1 and caspase-9 within the complex are unclear. Indeed, there is considerable discrepancy in some studies between the predicted size of the apoptosome and that observed by gel filtration analysis (Cain et al., 2000; Cain et al., 2001; Yu et al., 2005). The precise details regarding nucleotide exchange and the structural changes induced in its wake should also provide important details into the mechanisms of Apaf-1 oligomerization. Although caspase-9 activation almost certainly involves dimerization, the apoptosome could conceivably also induce additional conformational changes in caspase-9 that influence its activity. Once activated, it remains unclear why processing of caspase-9 reduces its affinity for the apoptosome – a prerequisite for the proteolytic-based molecular timer. The importance of the timer also awaits confirmation in vivo, but should be testable using non-cleavable caspase-9 knock-in mice, as they should be incapable of activating the timer. As already noted, caspase-9 undergoes post-translational modifications, and the apoptosome as a complex is regulated by a large number of proteins and small molecules, but the precise biochemical and structural details regarding how they inhibit or enhance apoptosome function remain almost entirely unknown. Finally, there is growing evidence that caspases are activated for non-apoptotic purposes, such as the induction of differentiation in numerous tissues (Yi and Yuan, 2009). Whether the apoptosome plays a role in this process is unknown, but if it did, how it could do so without killing the cell remains a puzzle. In short, this remarkable caspase-activating complex remains an important area of study in the field of apoptosis and beyond.
Acknowledgments
The authors gratefully acknowledge support from National Institutes of Health grants R01 CA129521 and P30 ES07784 (S.B.B.), as well as R01 CA69381 and R01 AG15402 (G.S.S.). Crystal structures of Apaf-1 ΔWDR [Protein Data Bank (PDB) code 1Z6T], caspase-9 prodomain bound to the Apaf-1 CARD (PDF code 3YGS), dimerized caspase-9 (PDB code 1JXQ), processed caspase-9 bound to the BIR3 domain of XIAP (PDB code 1NW9), and CED4 bound to CED9 (PDB code 2A5Y) were rendered using UCSF Chimera. Similarly, a space-filled model of the apoptosome was rendered using PDB code 3IYT and Electron Microscopy Data Bank EMD-5186. The authors also thank Christopher W. Akey for the 3D surface view of the fly apoptosome. Deposited in PMC for release after 12 months.
References
- Allan L. A., Clarke P. R. (2007). Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol. Cell 26, 301-310 [DOI] [PubMed] [Google Scholar]
- Allan L. A., Morrice N., Brady S., Magee G., Pathak S., Clarke P. R. (2003). Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat. Cell Biol. 5, 647-654 [DOI] [PubMed] [Google Scholar]
- Bao Q., Lu W., Rabinowitz J. D., Shi Y. (2007). Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Mol. Cell 25, 181-192 [DOI] [PubMed] [Google Scholar]
- Beere H. M., Wolf B. B., Cain K., Mosser D. D., Mahboubi A., Kuwana T., Tailor P., Morimoto R. I., Cohen G. M., Green D. R. (2000). Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2, 469-475 [DOI] [PubMed] [Google Scholar]
- Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pedersen I. M., Ricci J. E., Edris W. A., Sutherlin D. P., Green D. R., et al. (2003). A unified model for apical caspase activation. Mol. Cell 11, 529-541 [DOI] [PubMed] [Google Scholar]
- Brady S. C., Allan L. A., Clarke P. R. (2005). Regulation of caspase 9 through phosphorylation by protein kinase C zeta in response to hyperosmotic stress. Mol. Cell. Biol. 25, 10543-10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton S. B., Walker G., Srinivasula S., Sun X.-M., Butterworth M., Alnemri E. S., Cohen G. M. (2001). Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 20, 998-1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton S. B., Lewis J., Butterworth M., Duckett C. S., Cohen G. M. (2002). XIAP inhibition of caspase-3 preserves its association with the Apaf-1 apoptosome and prevents CD95- and Bax-induced apoptosis. Cell Death Differ. 9, 881-892 [DOI] [PubMed] [Google Scholar]
- Bruey J. M., Ducasse C., Bonniaud P., Ravagnan L., Susin S. A., Diaz-Latoud C., Gurbuxani S., Arrigo A. P., Kroemer G., Solary E., et al. (2000). Hsp27 negatively regulates cell death by interacting with cytochrome c. Nat. Cell Biol. 2, 645-652 [DOI] [PubMed] [Google Scholar]
- Brunelle J. K., Letai A. (2009). Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 122, 437-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain K., Bratton S. B., Langlais C., Walker G., Brown D. G., Sun X. M., Cohen G. M. (2000). Apaf-1 oligomerizes into biologically active ~700 kDa and inactive ~1.4 MDa apoptosome complexes. J. Biol. Chem. 275, 6067-6070 [DOI] [PubMed] [Google Scholar]
- Cain K., Langlais C., Sun X. M., Brown D. G., Cohen G. M. (2001). Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J. Biol. Chem. 276, 41985-41990 [DOI] [PubMed] [Google Scholar]
- Cardone M. H., Roy N., Stennicke H. R., Salvesen G. S., Franke T. F., Stanbridge E., Frisch S., Reed J. C. (1998). Regulation of cell death protease caspase-9 by phosphorylation. Science 282, 1318-1321 [DOI] [PubMed] [Google Scholar]
- Chandra D., Bratton S. B., Person M. D., Tian Y., Martin A. G., Ayres M., Fearnhead H. O., Gandhi V., Tang D. G. (2006). Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome C and inhibiting apoptosome. Cell 125, 1333-1346 [DOI] [PubMed] [Google Scholar]
- Chao Y., Shiozaki E. N., Srinivasula S. M., Rigotti D. J., Fairman R., Shi Y. (2005). Engineering a dimeric caspase-9: a re-evaluation of the induced proximity model for caspase activation. PLoS Biol. 3, 1079-1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau B. N., Cheng E. H., Kerr D. A., Hardwick J. M. (2000). Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol. Cell 6, 31-40 [PubMed] [Google Scholar]
- Cho D. H., Hong Y. M., Lee H. J., Woo H. N., Pyo J. O., Mak T. W., Jung Y. K. (2004). Induced inhibition of ischemic/hypoxic injury by APIP, a novel Apaf-1-interacting protein. J. Biol. Chem. 279, 39942-39950 [DOI] [PubMed] [Google Scholar]
- Chu Z. L., Pio F., Xie Z., Welsh K., Krajewska M., Krajewski S., Godzik A., Reed J. C. (2001). A novel enhancer of the Apaf1 apoptosome involved in cytochrome c-dependent caspase activation and apoptosis. J. Biol. Chem. 276, 9239-9245 [DOI] [PubMed] [Google Scholar]
- Deveraux Q. L., Takahashi R., Salvesen G. S., Reed J. C. (1997). X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388, 300-304 [DOI] [PubMed] [Google Scholar]
- Dorstyn L., Mills K., Lazebnik Y., Kumar S. (2004). The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J. Cell Biol. 167, 405-410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S. R., Phillips W. S., Spatafora J. W., Green D. R., Weis V. M. (2006). Highly conserved caspase and Bcl-2 homologues from the sea anemone Aiptasia pallida: lower metazoans as models for the study of apoptosis evolution. J. Mol. Evol. 63, 95-107 [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P., Salvesen G. S. (2004). The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 384, 201-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C., Bruey J. M., Fromentin A., Hammann A., Arrigo A. P., Solary E. (1999). HSP27 inhibits cytochrome c-dependent activation of procaspase-9. FASEB J. 13, 2061-2070 [DOI] [PubMed] [Google Scholar]
- Hill M. M., Adrain C., Duriez P. J., Creagh E. M., Martin S. J. (2004). Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 23, 2134-2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlaing T., Guo R. F., Dilley K. A., Loussia J. M., Morrish T. A., Shi M. M., Vincenz C., Ward P. A. (2001). Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of themammalian Ced-4 family of apoptosis proteins. J. Biol. Chem. 276, 9230-9238 [DOI] [PubMed] [Google Scholar]
- Jiang X., Kim H. E., Shu H., Zhao Y., Zhang H., Kofron J., Donnelly J., Burns D., Ng S. C., Rosenberg S., et al. (2003). Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science 299, 223-226 [DOI] [PubMed] [Google Scholar]
- Kim H. E., Du F., Fang M., Wang X. (2005). Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. USA 102, 17545-17550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. E., Jiang X., Du F., Wang X. (2008). PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol. Cell 30, 239-247 [DOI] [PubMed] [Google Scholar]
- Kurokawa M., Zhao C., Reya T., Kornbluth S. (2008). Inhibition of apoptosome formation by suppression of Hsp90beta phosphorylation in tyrosine kinase-induced leukemias. Mol. Cell. Biol. 28, 5494-5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., Declercq W., Kalai M., Saelens X., Vandenabeele P. (2002). Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 9, 358-361 [DOI] [PubMed] [Google Scholar]
- Malladi S., Challa-Malladi M., Fearnhead H. O., Bratton S. B. (2009). The Apaf-1•procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer. EMBO J. 28, 1916-1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. C., Allan L. A., Lickrish M., Sampson C., Morrice N., Clarke P. R. (2005). Protein kinase A regulates caspase-9 activation by Apaf-1 downstream of cytochrome c. J. Biol. Chem. 280, 15449-15455 [DOI] [PubMed] [Google Scholar]
- Martin M. C., Allan L. A., Mancini E. J., Clarke P. R. (2008). The docking interaction of caspase-9 with ERK2 provides a mechanism for the selective inhibitory phosphorylation of caspase-9 at threonine 125. J. Biol. Chem. 283, 3854-3865 [DOI] [PubMed] [Google Scholar]
- Marusawa H., Matsuzawa S., Welsh K., Zou H., Armstrong R., Tamm I., Reed J. C. (2003). HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 22, 2729-2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G., Yang X. (2010). tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell 37, 668-678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I., Hay B. A., Clem R. J. (2002). The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J. Biol. Chem. 277, 49644-49650 [DOI] [PubMed] [Google Scholar]
- Nylandsted J., Gyrd-Hansen M., Danielewicz A., Fehrenbacher N., Lademann U., Hoyer-Hansen M., Weber E., Multhoff G., Rohde M., Jaattela M. (2004). Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J. Exp. Med. 200, 425-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Saleh A., Nakazawa A., Kumar S., Srinivasula S. M., Kumar V., Weichselbaum R., Nalin C., Alnemri E. S., Kufe D., et al. (2000). Negative regulation of cytochrome c-mediated oligomerization of Apaf-1 and activation of procaspase-9 by heat shock protein 90. EMBO J. 19, 4310-4322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan N., Marusawa H., Krajewska M., Matsuzawa S., Kim H., Okada K., Torii S., Kitada S., Krajewski S., Welsh K., et al. (2001). TUCAN, an antiapoptotic caspase-associated recruitment domain family protein overexpressed in cancer. J. Biol. Chem. 276, 32220-32229 [DOI] [PubMed] [Google Scholar]
- Piddubnyak V., Rigou P., Michel L., Rain J. C., Geneste O., Wolkenstein P., Vidaud D., Hickman J. A., Mauviel A., Poyet J. L. (2007). Positive regulation of apoptosis by HCA66, a new Apaf-1 interacting protein, and its putative role in the physiopathology of NF1 microdeletion syndrome patients. Cell Death Differ. 14, 1222-1233 [DOI] [PubMed] [Google Scholar]
- Pop C., Timmer J., Sperandio S., Salvesen G. S. (2006). The apoptosome activates caspase-9 by dimerization. Mol. Cell 22, 269-275 [DOI] [PubMed] [Google Scholar]
- Purring-Koch C., McLendon G. (2000). Cytochrome c binding to Apaf-1: the effects of dATP and ionic strength. Proc. Natl. Acad. Sci. USA 97, 11928-11931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S., Pang Y., Hu Q., Liu Q., Li H., Zhou Y., He T., Liang Q., Liu Y., Yuan X., et al. (2010). Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141, 446-457 [DOI] [PubMed] [Google Scholar]
- Qin H., Srinivasula S. M., Wu G., Fernandes-Alnemri T., Alnemri E. S., Shi Y. (1999). Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature 399, 549-557 [DOI] [PubMed] [Google Scholar]
- Raina D., Pandey P., Ahmad R., Bharti A., Ren J., Kharbanda S., Weichselbaum R., Kufe D. (2005). c-Abl tyrosine kinase regulates caspase-9 autocleavage in the apoptotic response to DNA damage. J. Biol. Chem. 280, 11147-11151 [DOI] [PubMed] [Google Scholar]
- Reed J. C. (2002). Apoptosis-based therapies. Nat. Rev. Drug Discov. 1, 111-121 [DOI] [PubMed] [Google Scholar]
- Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. (2001). Dimer formation drives the activation of the cell death protease caspase-9. Proc. Natl. Acad. Sci. USA 98, 14250-14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl S. J., Salvesen G. S. (2007). The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell. Biol. 8, 405-413 [DOI] [PubMed] [Google Scholar]
- Riedl S. J., Renatus M., Schwarzenbacher R., Zhou Q., Sun C., Fesik S. W., Liddington R. C., Salvesen G. S. (2001). Structural basis for the inhibition of caspase-3 by XIAP. Cell 104, 791-800 [DOI] [PubMed] [Google Scholar]
- Riedl S. J., Li W., Chao Y., Schwarzenbacher R., Shi Y. (2005). Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434, 926-933 [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Oliver H., Zou H., Chen P., Wang X., Abrams J. M. (1999). Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat. Cell Biol. 1, 272-279 [DOI] [PubMed] [Google Scholar]
- Rodriguez J., Lazebnik Y. (1999). Caspase-9 and APAF-1 form an active holoenzyme. Genes Dev. 13, 3179-3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Vela A., Korsmeyer S. J. (2007). Proapoptotic histone H1.2 induces CASP-3 and -7 activation by forming a protein complex with CYT c, APAF-1 and CASP-9. FEBS Lett. 581, 3422-3428 [DOI] [PubMed] [Google Scholar]
- Sakai T., Liu L., Teng X., Mukai-Sakai R., Shimada H., Kaji R., Mitani T., Matsumoto M., Toida K., Ishimura K., et al. (2004). Nucling recruits Apaf-1/pro-caspase-9 complex for the induction of stress-induced apoptosis. J. Biol. Chem. 279, 41131-41140 [DOI] [PubMed] [Google Scholar]
- Saleh A., Srinivasula S. M., Balkir L., Robbins P. D., Alnemri E. S. (2000). Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat. Cell Biol. 2, 476-483 [DOI] [PubMed] [Google Scholar]
- Scott F. L., Denault J. B., Riedl S. J., Shin H., Renatus M., Salvesen G. S. (2005). XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 24, 645-655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott F. L., Stec B., Pop C., Dobaczewska M. K., Lee J. J., Monosov E., Robinson H., Salvesen G. S., Schwarzenbacher R., Riedl S. J. (2009). The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature 457, 1019-1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert A., Clarke P. R. (2009). p38alpha- and DYRK1A-dependent phosphorylation of caspase-9 at an inhibitory site in response to hyperosmotic stress. Cell Signal. 21, 1626-1633 [DOI] [PubMed] [Google Scholar]
- Seifert A., Allan L. A., Clarke P. R. (2008). DYRK1A phosphorylates caspase-9 at an inhibitory site and is potently inhibited in human cells by harmine. FEBS J. 275, 6268-6280 [DOI] [PubMed] [Google Scholar]
- Shiozaki E. N., Chai J., Rigotti D. J., Riedl S. J., Li P., Srinivasula S. M., Alnemri E. S., Fairman R., Shi Y. (2003). Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell 11, 519-527 [DOI] [PubMed] [Google Scholar]
- Spector M. S., Desnoyers S., Hoeppner D. J., Hengartner M. O. (1997). Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature 385, 653-656 [DOI] [PubMed] [Google Scholar]
- Srinivasula S. M., Hegde R., Saleh A., Datta P., Shiozaki E., Chai J., Lee R. A., Robbins P. D., Fernandes-Alnemri T., Shi Y., et al. (2001). A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 410, 112-116 [DOI] [PubMed] [Google Scholar]
- Steel R., Doherty J. P., Buzzard K., Clemons N., Hawkins C. J., Anderson R. L. (2004). Hsp72 inhibits apoptosis upstream of the mitochondria and not through interactions with Apaf-1. J. Biol. Chem. 279, 51490-51499 [DOI] [PubMed] [Google Scholar]
- Stennicke H. R., Deveraux Q. L., Humke E. W., Reed J. C., Dixit V. M., Salvesen G. S. (1999). Caspase-9 can be activated without proteolytic processing. J. Biol. Chem. 274, 8359-8362 [DOI] [PubMed] [Google Scholar]
- Thornberry N. A., Lazebnik Y. (1998). Caspases: enemies within. Science 281, 1312-1316 [DOI] [PubMed] [Google Scholar]
- Timmer J. C., Salvesen G. S. (2007). Caspase substrates. Cell Death Differ. 14, 66-72 [DOI] [PubMed] [Google Scholar]
- Yan N., Chai J., Lee E. S., Gu L., Liu Q., He J., Wu J. W., Kokel D., Li H., Hao Q., et al. (2005). Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature 437, 831-837 [DOI] [PubMed] [Google Scholar]
- Yi C. H., Yuan J. (2009). The Jekyll and Hyde functions of caspases. Dev. Cell 16, 21-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q., Park H. H., Chung J. Y., Lin S. C., Lo Y. C., da Graca L. S., Jiang X., Wu H. (2006). Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol. Cell 22, 259-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Acehan D., Menetret J. F., Booth C. R., Ludtke S. J., Riedl S. J., Shi Y., Wang X., Akey C. W. (2005). A structure of the human apoptosome at 12.8 Å resolution provides insights into this cell death platform. Structure 13, 1725-1735 [DOI] [PubMed] [Google Scholar]
- Yu X., Wang L., Acehan D., Wang X., Akey C. W. (2006). Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 355, 577-589 [DOI] [PubMed] [Google Scholar]
- Yuan S., Yu X., Topf M., Ludtke S. J., Wang X., Akey C. W. (2010). Structure of an apoptosome-procaspase-9 CARD complex. Structure 18, 571-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech B., Kohl R., von Knethen A., Brune B. (2003). Nitric oxide donors inhibit formation of the Apaf-1/caspase-9 apoptosome and activation of caspases. Biochem. J. 371, 1055-1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek C. M., Zhang Q., Ye Y., Godzik A. (2007). Surprising complexity of the ancestral apoptosis network. Genome Biol. 8, R226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H., Henzel W. J., Liu X., Lutschg A., Wang X. (1997). Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405-413 [DOI] [PubMed] [Google Scholar]