Abstract
Trypanosomes evade host immunity by exchanging variant surface glycoprotein (VSG) coats. VSG genes are transcribed from telomeric expression sites, which contain a diverse family of expression-site-associated genes (ESAGs). We have discovered that the mRNAs for one ESAG family, ESAG9, are strongly developmentally regulated, being enriched in stumpy forms, a life-cycle stage in the mammalian bloodstream that is important for the maintenance of chronic parasite infections and for tsetse transmission. ESAG9 gene sequences are highly diverse in the genome and encode proteins with weak similarity to the massively diverse MASP proteins in Trypanosoma cruzi. We demonstrate that ESAG9 proteins are modified by N-glycosylation and can be shed to the external milieu, this being dependent upon coexpression with at least one other family member. The expression profile and extracellular release of ESAG9 proteins represents a novel and unexpected aspect of the transmission biology of trypanosomes in their mammalian host. We suggest that these molecules might interact with the external environment, with possible implications for infection chronicity or parasite transmission.
Keywords: Trypanosoma brucei, Antigenic variation, ESAG, Transmission, Secretion
Introduction
African trypanosomes evade mammalian immune responses by an extreme capacity for antigenic variation (McCulloch, 2004). The parasite surface is completely covered by a dense monolayer of protein tethered to the cell surface by a glycophosphatidyl inositol anchor. This coat, composed of variant surface glycoprotein (VSG), is homogeneous such that only one antigenic variant is expressed at a time on any given parasite. Periodically, antibody responses to the predominant VSG cause complement-mediated lysis of the majority of parasites, followed by outgrowth of parasites that have switched to a distinct antigenic variant. This results in the undulating waves of parasitaemia that are characteristic of trypanosome infections.
A further contributor to the undulating profile of a trypanosome infection is the developmental regulation of proliferation status. Early in each wave of parasitaemia, trypanosomes multiply rapidly as morphologically slender forms. However, as parasite numbers increase, an unidentified parasite-derived signal (stumpy induction factor; SIF) stimulates growth arrest in G1 (Vassella et al., 1997). This quorum sensing (QS) mechanism induces the developmental switch to morphologically stumpy forms, which do not divide, thereby limiting the increase in parasite numbers and promoting the generation of a chronic infection (Lythgoe et al., 2007). Although stumpy forms are irreversibly committed to cell division arrest in the bloodstream, this does not represent a simple example of self-destructive cooperation, as seen in some bacterial populations (Ackermann et al., 2008; Gardner and Kummerli, 2008). Rather, the stumpy forms are adapted for life-cycle progression via their transmission to tsetse flies, the vector for African trypanosomiasis (Dean et al., 2009; Engstler and Boshart, 2004; Tasker et al., 2000). In particular, only stumpy forms in the bloodstream parasite population express members of a surface transporter family of proteins (PAD proteins) that allow detection of the citrate/cis-aconitate differentiation signal upon transmission to tsetse flies (Dean et al., 2009). This is assisted by developmentally regulated and thermoregulated protein trafficking to the parasite surface that is characteristic of stumpy forms (Engstler and Boshart, 2004). Also, stumpy forms are relatively resistant to proteolytic assault in the tsetse midgut and to other stresses that rapidly result in the killing of slender forms (Nolan et al., 2000; Sbicego et al., 1999). Combined with the enhanced resistance of stumpy forms to the developing antibody response in the mammalian host via hydrodynamic flow (Engstler et al., 2007), these characteristics enable trypanosome populations to achieve a balance between rapid establishment and maintenance of the parasitaemia, development of a sustained long-term infection and optimisation of their transmission potential.
Long-term laboratory passage of trypanosome lines without tsetse transmission produces monomorphism, i.e. the selection of parasites that have lost the capacity to generate stumpy forms in vivo and that are analogous to QS signal-blind mutants in bacteria (Diggle et al., 2007). These have provided useful experimental tools because they exhibit a reduced frequency of antigen switching (Robinson et al., 1999), enabling analysis of the underlying molecular regulation of antigenic variation. Studies of these lines have revealed that VSG genes are always expressed from telomeric expression sites, of which only one of about 25 possible sites is active at any one time (Borst, 2002). The active expression site is apparently selected through association with an expression site body at the nuclear envelope (Navarro and Gull, 2001). VSG expression sites have a conserved overall structure (Hertz-Fowler et al., 2008). Upstream of the VSG gene itself are a number of expression-site-associated genes (ESAGs) (Cully et al., 1985; Pays et al., 2001). These are co-transcribed with the VSG gene by RNA polymerase I (Gunzl et al., 2003), the promoter being positioned 45–60 kb upstream (Kooter et al., 1987; Pays et al., 1989). The function of ESAGs is largely unknown, although the products of ESAG6 and ESAG7 form a heterodimeric transferrin receptor (Bitter et al., 1998; Salmon et al., 1994; Schell et al., 1991), and ESAG4 encodes an adenylate cyclase activity (Pays et al., 1989). Also, bioinformatic analysis of ESAG5 protein suggests that it bears similarity to a human lipid binding/lipid transfer family of proteins (Barker et al., 2008). In addition, Trypanosoma brucei rhodesiense express an unusual ESAG, SRA, which confers resistance to an innate immune component of human serum, ApoL-1 (Vanhamme et al., 2003; Xong et al., 1998). Thus, where characterised, ESAGs are involved in host–parasite interactions unrelated to antigenic variation.
We have discovered that an unusual expression-site-associated gene family, ESAG9, is developmentally regulated, being specifically expressed only in stumpy forms. We demonstrate that several ESAG9 genes are upregulated during the transition from slender to stumpy forms in T. brucei, contrasting with the monoallelic expression characteristic of VSG genes. Surprisingly, we find that ESAG9 protein can be shed from slender form parasites when ectopically coexpressed with another ESAG9 copy. Confirming the biological relavence of this, ESAG9 proteins are also shed by wild-type stumpy cells generated in vivo. Our results demonstrate that ESAG9 proteins are stumpy stage-specific molecules implicated in trypanosome chronicity or transmission biology.
Results
Developmental expression of a VSG expression-site-associated gene
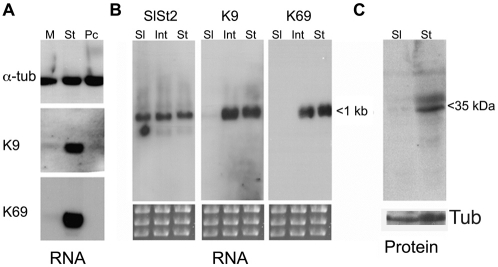
Previous analyses have demonstrated that tsetse-transmission competence requires the development of bloodstream stumpy forms (Dean et al., 2009; Robertson, 1912; Tasker et al., 2000; Wijers and Willett, 1960). To understand the molecular characteristics of stumpy forms we used cDNA subtraction selection to identify transcripts that differ in expression between isogenic monomorphic slender (T. brucei EATRO 2340; GUP 2965) and stumpy forms (T. brucei EATRO 2340; GUP2962), each expressing the same VSG gene (GUTat 7.2) from the same VSG expression site. This resulted in the selection for a number of stumpy-enriched transcripts, of which the most frequently isolated were two members of the ESAG9 family (Florent et al., 1991), represented by clones K9 and K69. To confirm the differential expression of these transcripts, northern blots of RNA derived from monomorphic slender (GUP 2965) and stumpy cells (GUP2962) and also from cultured procyclic forms (T. brucei s427) were hybridised with riboprobes for K9 and K69 and for the constitutively expressed transcript α-tubulin. Fig. 1A establishes that both K9 and K69 transcripts are highly enriched in the stumpy samples, each being expressed at a low level in monomorphic cells and absent in procyclic forms. Hybridisation of cultured procyclic forms generated 7 days after the in vitro differentiation of T. brucei EATRO 2340 GUP 2962 stumpy forms also generated very low expression, confirming that the observed expression profile did not represent strain specific differences between procyclic form parasites (data not shown). The upregulation of ESAG9 transcripts in stumpy form cells has also been recently confirmed by microarray analysis of different life-cycle stages (Jensen et al., 2009; Kabani et al., 2009).
Fig. 1.
Differential expression of ESAG9 transcripts. (A) Expression of clones K9 and K69 analysed against total RNA derived from monomorphic (M), or stumpy forms of pleomorphic T. brucei EATRO 2340 GUTat 7.2 (St) and procyclic forms (Pc). A constitutively expressed transcript, α-tubulin is shown as a control. (B) Expression of K9 and K69 in slender (Sl), intermediate (Int) and stumpy forms of pleomorphic T. brucei EATRO 2340. A control constitutively expressed transcript is also shown. (C) Equivalent whole cell protein samples of slender and stumpy forms of pleomorphic T. brucei EATRO 2340 probed with an anti-peptide antibody to ESAG9-K9. The protein migrates at approximately 35 kDa, contrasting with a predicted molecular mass of 27 kDa.
The precise biological relationship between laboratory-passaged monomorphic forms and pleomorphic slender forms is not clear (Matthews et al., 2004). Therefore, to assay the expression of K9 and K69 during the course of a pleomorphic trypanosome infection, RNA was harvested when the parasites were at a density of less than 2×107/ml and >99% slender in morphology. Parasites were also harvested at a density of 1×108/ml but before the appearance of significant numbers (i.e. <10%) of stumpy forms, this representing an ‘intermediate form’ population. Finally, parasites were harvested at 2×108–5×108/ml, when the population was >90% stumpy by morphology. Fig. 1B demonstrates that K9 and K69 were both almost undetectable in slender cells, but fully upregulated in intermediate and stumpy forms. Thus, the mRNAs for these ESAG9 family members are developmentally regulated in bloodstream trypanosomes, being induced early in the transition to stumpy forms prior to morphological transformation.
Genomic analysis of the ESAG9 gene family in T. brucei
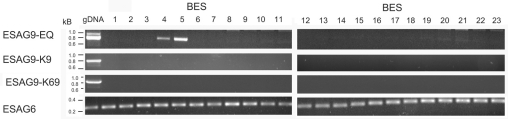
Two closely related ESAG9 genes have been previously detected in the equine parasite T. equiperdum (Florent et al., 1991). Although monomorphic, these parasites are believed to be a recently diverged subspecies of T. brucei (Claes et al., 2005; Lai et al., 2008). In T. equiperdum, one ESAG9 gene was expressed from within the BoTat 178 VSG expression site, whereas the other was transcribed from a non-expression site location in a VSG-independent manner (Florent et al., 1991). Analysis of the T. brucei TREU927/4 genome database revealed that the ESAG9-K9 gene is located in a chromosome internal position on chromosome 7 (Tb927.07.170) whereas the ESAG9-K69 gene is absent from the available genome dataset. Although VSG expression sites are underrepresented, the T. brucei genome database (available at http://www.geneDB.org) contains 19 ESAG9 genes, of which ten are annotated as pseudogenes. The annotated ESAG9 genes are each located at chromosome-internal and subtelomeric positions, and each is closely adjacent to other ESAGs, VSG genes or VSG pseudogenes. Hence, ESAG9 genes appear to co-associate with VSG gene expression sites, expression site components or expression site relics, frequently at subtelomeric positions (diagrammatic representations of the genomic location of several ESAG9 genes are shown in supplementary material Fig. S1). In order to evaluate whether ESAG9-K9 and ESAG9-K69 genes were also represented in VSG expression sites, a TAR library comprising the cloned telomeres of T. brucei EATRO 2340 (a kind gift of Gloria Rudenko, University of Oxford, Oxford, UK) (Young et al., 2008) was analysed for the presence of each gene copy using gene-specific PCR primers. This demonstrated that neither K9 or K69 was found associated with the cloned telomeric sequences (Fig. 2), although Tb927.5.4620 (labelled ESAG9-EQ on Fig. 2 because it is the gene most closely related to the T. equiperdum ESAG9 genes; see below) was found on two TAR clones. This matches the expression site location for the closest homologue of this gene in T. brucei s427 in BES211 (Hertz-Fowler et al., 2008) (supplementary material Fig. S1). Thus, ESAG9 sequences are occasionally found within VSG expression sites, but neither ESAG9-K9 or ESAG9-K69 were expressed from, nor located in, a VSG expression site in the parasite strain examined.
Fig. 2.
Analysis of T. brucei EATRO 2340 TAR clones for the presence of ESAG9 gene sequences. T. brucei EATRO 2340 genomic DNA (far left) and TAR cloned bloodstream form expression sites (1–23, a gift from Gloria Rudenko) were analysed by PCR using gene-specific primers for the presence of the following genes: Tb927.5.4620 (the gene most similar to the ESAG sequences in T. equiperdum), ESAG9-K9, ESAG9-K69 and ESAG6, which is present in every expression site that has been sequenced so far. ESAG6 was indeed found to be in all expression sites, Tb927.5.4620 was in two expression sites (4 and 5) and ESAG9-K9 and ESAG9-K69 were absent from all expression sites in T. brucei EATRO 2340.
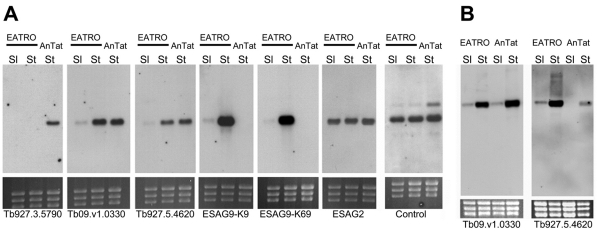
Given the genomic distribution of ESAG9 genes within and outside of VSG expression sites, we sought to analyse whether one, two or a repertoire of several ESAG9 genes could be expressed in T. brucei and whether the expression of each of these was stumpy-enriched. Thus, the expression of ESAG9-K9, ESAG9-K69 and three further ESAG9 genes identified in the T. brucei TREU 927/4 genome sequence (Tb927.3.5790, Tb927.5.4620 and Tb09.v1.0330) were analysed in monomorphic slender and pleomorphic stumpy forms of T. brucei GUTat 7.2, and in stumpy forms of a pleomorphic line of T. brucei expressing VSG AnTat1.1. Fig. 3A demonstrates that each line expressed more than one ESAG9 gene, with two of the genes tested being expressed in both T. brucei GUTat 7.2 and T. brucei AnTat1.1 (Tb09.v1. 0330; Tb927.5.4620), in each case this expression was stumpy-enriched (Fig. 3A,B). By contrast, Tb927.3.5790 was expressed in T. brucei AnTat 1.1 stumpy forms but not in T. brucei EATRO 2340 slender or stumpy forms, whereas ESAG9-K9 and ESAG9-K69 were expressed in T. brucei EATRO 2340 but not AnTat1.1. This reflected the presence or absence of each gene: analysis of the genomic DNA of each line by PCR demonstrated that Tb927.3.5790 was absent from T. brucei EATRO 2340 genomic DNA whereas ESAG9-K9 and ESAG9-K69 were absent from the genome of T. brucei AnTat1.1 (data not shown).
Fig. 3.
Expression of ESAG9 genes in T. brucei. (A) Expression profile of five ESAG9 genes on monomorphic slender (EATRO Sl) and pleomorphic stumpy forms of T. brucei EATRO 2340 GUTat 7.2 (EATRO St) and pleomorphic stumpy forms of T. b. brucei AnTat1.1 (AnTat St). Loading of each lane is indicated by the ethidium bromide-stained rRNAs. An ESAG2-specific riboprobe was also hybridised with the same RNA samples, as was a constitutively expressed control transcript (Tb11.02.3640). (B) Stumpy-enriched expression of two ESAG9 genes expressed in both T. brucei EATRO 2340 and T. brucei brucei AnTat1.1. RNA from pleomorphic slender (early parasitaemia) and stumpy (late parasitaemia) cells is shown for each line.
Where present, therefore, the expression of all ESAG9 genes tested was elevated in stumpy forms. Contrasting with this, a control transcript (Tb11.02.3640) was expressed equally in slender and stumpy forms of each line, as was another expression-site-associated gene, ESAG2 (Fig. 3A). Moreover, ESAG9 expression was not upregulated in bloodstream forms arrested in G2 by VSG-specific RNAi ablation (Sheader et al., 2005; Gloria Rudenko, personal communication), eliminating non-specific elevation of ESAG9 mRNA upon cell cycle arrest, albeit in a distinct cell-cycle stage to stumpy forms. We conclude that ESAG9 expression is developmentally elevated in a stumpy-specific manner, that multiple members of the ESAG9 family are upregulated upon the transition to stumpy forms, and that different trypanosome lines express distinct repertoires of these genes.
ESAG9 genes encode a diverse protein family with a conserved motif
Fourteen intact ESAG9 genes have been identified in the genomes of different trypanosome species and strains: T. brucei TREU927/4 (nine genes), T. brucei Lister 427 expression sites (one gene), T. brucei EATRO 2340 (two genes; ESAG9-K9 and ESAG9-K69, the latter also being present in TREU927/4), T. equiperdum (two genes) and T. b. gambiense (one gene). No closely related sequences were identified in the incomplete genomes of T. congolense or T. vivax. To search ESAG9 proteins for motifs that suggest function, we aligned the predicted protein sequences of all available intact ESAG9 genes (supplementary material Fig. S2). Many of the predicted ESAG9 proteins have relatively low identity (18–39%) to each other and to the T. equiperdum proteins, demonstrating that ESAG9s are a diverse protein family. However, one T. brucei TREU927/4 ESAG9 member (Tb927.5.4620) and the ESAG9 encoded in an expression site in T. brucei Lister 427 (Tb427.BES122.10) were highly homologous to each other (88% identity) and to the genes previously characterised in T. equiperdum (ESAG9u, ESAG9c). Tb927.5.4620 exhibited 67 and 84% identity with ESAG9u and ESAG9c of T. equiperdum, respectively, and Tb427.BES122.10 exhibited 69 and 85% identity, respectively. It appears therefore that ESAG9 proteins are very diverse as a family, and yet can be well conserved between different trypanosome species. PSI-BLAST searches did not reveal the presence of ESAG9-related proteins in any other species, although searches against other kinetoplastid genomes frequently recognised at low similarity (e-value = 4×10−5–0.028, with a typical value of 0.001) distinct T. cruzi MASP proteins, which are members of a large and diverse mucin glycoprotein superfamily of unknown function (Bartholomeu et al., 2009). The weak similarity was restricted to the central diverse core of MASP proteins, distinct from their conserved C-terminal region.
Although the ESAG9 proteins are diverse, a restricted region of conservation was observed near the N-terminus of each protein. Thus, approximately 10–20 amino acids after the predicted cleavage site of the N-terminal signal sequence, there exists the motif CX3WX8CX5G, which is highly conserved among the ESAG9 family. Searching available sequence databases for proteins that share this motif identified a related sequence conserved among members of the protein phosphatase PP2C family (supplementary material Fig. S3), these residues being required for Mn2+-binding and functional activity of these enzymes (Barford et al., 1998; Kusuda et al., 1998). Similarity to this conserved motif was also observed in members of the SRCR family of scavenger receptors. Outside of this region, however, no further extensive similarity to these or other proteins was detected.
Protein expression and ectopic protein expression of ESAG9-K9 and ESAG9-K69
To evaluate whether the observed differential mRNA profile of the ESAG9 genes was matched at the protein level, an antibody was raised against a peptide sequence specific for ESAG9-K9 (NH2-QVHDGEQRDLEGRGC), this being located at a position not conserved in other ESAG9 family members (supplementary material Fig. S2). When this antibody was reacted with protein samples from monomorphic slender and stumpy forms, a protein of ~35 kDa was detected specifically in stumpy parasites (Fig. 1C). This was slightly larger than the molecular mass predicted from the primary ESAG9-K9 amino acid sequence (27 kDa), suggesting possible posttranslational modification of the protein (see below).
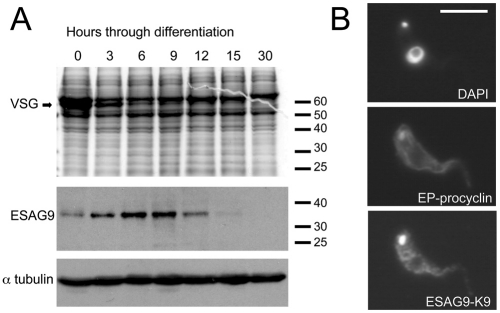
The expression profile of ESAG9-K9 was also examined during the synchronous differentiation of a homogenous population of stumpy form parasites to procyclic forms in vitro. Thus, stumpy cells were harvested from a rodent infection and differentiation of the cells to procyclic forms was initiated with cis-aconitate and protein samples prepared at 3 hourly intervals up to 15 hours, and then after 30 hours. Under these conditions, the VSG coat was maintained for 3–6 hours, but significantly decreased thereafter, matching the expected kinetics for this differentiation event (Matthews and Gull, 1994; Ziegelbauer and Overath, 1990) (Fig. 4A). By contrast, ESAG9-K9 was detected weakly in stumpy form cell extracts but enriched between 3–9 hours after the initiation of differentiation, and undetectable beyond 15 hours. Immunofluorescence assay of cells 6 hours after the initiation of differentiation confirmed ESAG9-K9 expression on differentiating cells, with the staining being dispersed in a punctate pattern with concentration around the nucleus and, in some instances, a signal from the flagellar pocket region or closely associated structures (Fig. 4B). The punctate staining profile was reminiscent of the trypanosome endoplasmic reticulum distribution, which was explored further using a transgenic cell line (see below). In contrast to ESAG9-K9, an anti-peptide antibody raised against ESAG9-K69 reacted very poorly with either stumpy cells or cells undergoing differentiation such that its expression and cellular localisation could not be determined.
Fig. 4.
Expression profile of ESAG9-K9 during differentiation to procyclic forms. (A) ESAG9 expression during stumpy–procyclic form differentiation. Stumpy forms of T. brucei EATRO 2340 were induced to differentiate to procyclic forms. Analysis of the Coomassie Blue stained gel shows that the VSG runs at approximately 50 kDa and is lost between 3 and 6 hours. The same protein samples were probed with anti-peptide antibody to ESAG9-K9 and trypanosome α-tubulin. (B) Cells harvested 6 hours after the initiation of differentiation and probed with antibody to ESAG9-K9 and EP procyclin (to demonstrate differentiation). A DAPI stained image of the same cell is also shown.
Expression analysis of ESAG9 proteins in transgenic bloodstream forms
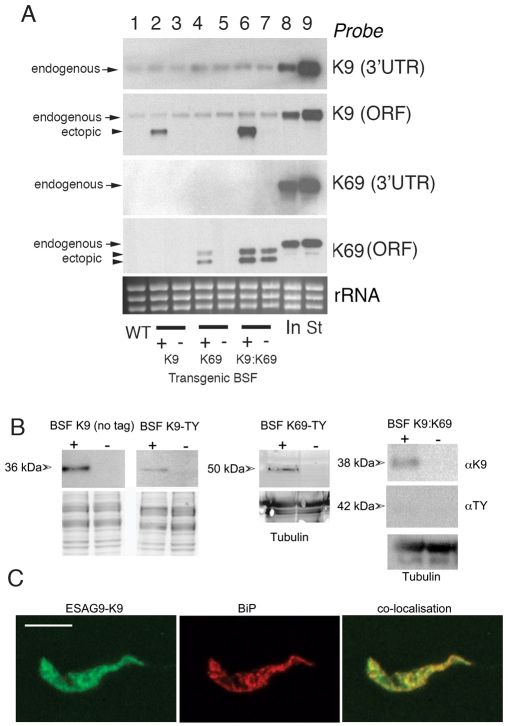
The transient or low level expression of ESAG9 proteins rendered them difficult to study in stumpy cells or parasites undergoing differentiation. Therefore, to assist characterisation of these proteins, we generated monomorphic bloodstream cells engineered to express either ESAG9-K9 or ESAG9-K69 under tetracycline regulation. To aid detection, but to avoid perturbation of potential N-terminal or C-terminal targeting signals, an internal epitope tag sequence, the ten amino acid Ty1 tag (Bastin et al., 1996), was incorporated into each gene at an equivalent position to replace the ten amino acids internal to either ESAG9-K9 (amino acids 64–73; generating cell line BSF ESAG9-K9-Ty) or ESAG9-K69 (amino acids 55–64; generating cell line BSF ESAG9-K69-Ty), this region having no clear conservation between distinct ESAG9 gene copies (supplementary material Fig. S2). An expression construct was also generated containing an untagged copy of ESAG9-K9, detectable with the K9-specific antibody, to eliminate any potential disruption of ESAG9 caused by the epitope tag sequence. Finally, to match the observed coexpression of these genes in T. brucei EATRO2340 stumpy forms, we generated a further cell line capable of the simultaneous inducible expression of both untagged ESAG9-K9 and Ty1-tagged ESAG9-K69 (generating cell line BSF K9:K69).
After selection and the induction of transgene expression with tetracycline, K9 and K69 transcripts were generated at levels equivalent to (K9), or less than (K69), endogenous levels of these transcripts in T. brucei EATRO 2340 stumpy forms (Fig. 5A). This indicated that overexpression artefacts were unlikely to contribute to any functional analyses. At the protein level, ectopically expressed ESAG9 proteins were detectable from cell lines expressing ESAG9-K9 (untagged and Ty1 tagged) and ESAG9-K69-Ty1 (Fig. 5B). In the double-expresser BSF K9:K69 cell line, ESAG9-K9 was expressed but ESAG9-K69 was barely detectable in the cell lysate because it is released from the cells (see later). The molecular mass at which ESAG9-K69 protein migrated varied between the BSF K69 cell line, where it ran at 50 kDa, and the BSF K9:K69 cell line, where the ESAG9-K69 ran at 42 kDa, both higher than the predicted molecular mass of 26 kDa for this protein. In cell line BSF-K9-Ty, ESAG9-K9 migrated at the same molecular mass as the endogenous protein in stumpy forms, indicating appropriate processing and possible posttranslational modification. Moreover, by immunofluorescence, the punctate expression observed in differentiating cells (Fig. 4D) was also detectable in this cell line with the Ty-tag specific antibody. This signal matched that of the ER marker BiP (a gift of Jay Bangs, University of Wisconsin, Madison, WI), and precise colocalisation of the two proteins was observed using confocal microscopy (Fig. 5D). Apparently, the ectopic expression of ESAG9-K9 in monomorphic slender cells faithfully reproduces the expression and location of this protein in pleomorphic stumpy and differentiating cells. The ectopic expression of the different ESAG9 proteins, however, had no effect on parasite growth in vitro (supplementary material Fig. S4) or in vivo (data not shown).
Fig. 5.
Secretion of ESAG9 proteins. (A) Northern blots of the mRNA levels of ESAG-K9 (no tag) and ESAG9-K69-Ty in transgenic T. brucei s427 monomorphic forms. In each case probes were used that detected either the endogenous ESAG9 transcript only (this being directed to the 3′UTR missing in the transgenes) or to the open reading frame (ORF; detecting both the endogenous and ectopically expressed copy). The gene for ESAG9-K9 is present in T. brucei s427 and the low level expression of this gene is detected in the monomorphic slender forms. By contrast, the gene for ESAG9-K69 is absent from the T. brucei s427 genome and no endogenous transcript is detected. The respective levels of ESAG9-K9 and ESAG9-K69 are also shown in early intermediate (In) and stumpy (St) forms of T. brucei EATRO 2340 (lanes 8, 9). Tetracycline-inducible expression of ESAG9-K9 (lanes 2, 3), ESAG9-K69 (lane 4,5) and both ESAG9-K9 and K69 is detected in the single and double expresser lines, though the ESAG-K69 transgene expression is leaky in the double expresser. The ESAG9-K69 generates two mRNAs believed to represent the use of alternative polyadenylation sites in the expression construct. (B) Ectopic expression of ESAG9-K9 (no tag, or Ty-tagged) and of ESAG9-K69, or both proteins, in monomorphic bloodstream forms of T. brucei. Protein samples were derived from cells harvested after 48 hours with (+) or without (−) tetracycline (to induce transgene expression) and probed for expression of each protein using either ESAG9-K9-specific antiserum (K9-no tag samples and BSF K9:K69) or the Ty1-specific antibody, BB2. Relative loading was verified by Coomassie Blue staining or reaction with antibody to α-tubulin. In the BSF K9:K69 cell line, ESAG9-K69 expression was barely detectable due to its extracellular release (see Fig. 7). (C) Colocalisation of Ty-tagged ESAG9-K9 in BSF K9-Ty cells with an ER marker, BIP. The localisation of ESAG9-K9 (left) (detected using the BB2 antibody) and BIP (centre) were analysed using confocal microscopy. In the overlaid image (right; yellow) extensive colocalisation was evident. Scale bar: 15 μm.
Initially, the transgenic cell lines were used to assess post-translational modifications of the ESAG9 proteins. In the first instance, we examined the potential for GPI anchor addition to ESAG9-K9, a modification predicted by the D-GPI and GPI-SOM, but not Big-PI, algorithms. This was evaluated by submitting the cells to hypotonic lysis under conditions where the parasite GPI-PLC induces the release of mature GPI-anchored proteins into the supernatant. As a control, we monitored the release of the GPI-anchored VSG 221 protein expressed on the transgenic cell line. Fig. 6A demonstrates that ESAG9-K9 remained cell-associated upon hypotonic lysis whereas the VSG control was very rapidly released into the soluble fraction, indicating that the ESAG9-K9 protein was either not GPI anchored in these cells or that GPI-PLC could not gain access to the protein. Hypotonic release of ESAG9-K9 from stumpy forms of T. brucei EATRO 2340 was also not observed, although in that case VSG release could not be verified due to the absence of a suitable VSG-specific antibody (data not shown).
Fig. 6.
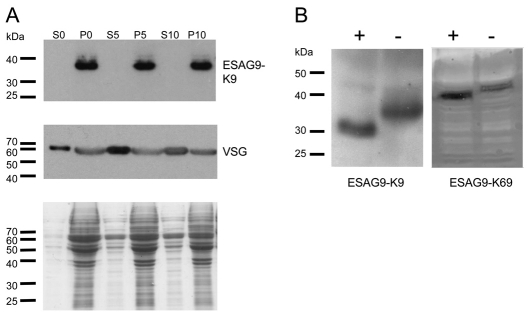
Post-translational modifications of ESAG9 proteins. (A) Hypotonic lysis of BSF K9 cells. Cells induced to ectopically express ESAG9-K9 were harvested and incubated in hypotonic lysis buffer for 0, 5 or 10 minutes and then centrifuged to separate the pellet (P) and supernatant (S) fractions. The samples were analysed by western blotting, which revealed that the VSG was released into the supernatant by the action of endogenous GPI-PLC enzyme even at the zero minutes, whereas ESAG9-K9 remained associated with the cell pellet. The Coomassie Blue-stained gel demonstrates the relative abundance of proteins in the S and P fractions. (B) N-Glycosylation of ESAG9 proteins. Monomorphic bloodstream form cells expressing either ESAG9-K9 or ESAG9-K69 were subjected to enzymatic digestion with PNGaseF (+), and controls were incubated for the same length of time in digestion buffer without the addition of enzyme (−). Western blotting with BB2 antibody indicated that ESAG9-K9 changed size by approximately 4 kDa as a result of enzyme treatment, and ESAG9-K69 by 2 kDa.
In addition to GPI addition, the potential for N-glycosylation was evaluated for ESAG9-K9 and ESAG9-K69, each of which were predicted to contain this modification at either three positions (ESAG9-K9) or two positions (ESAG9-K69). Thus, cell lysates from monomorphic bloodstream form cells ectopically expressing either ESAG9-K9 or ESAG9-K69 were subjected to treatment with peptide N-glycosidase F (PNGase F) enzyme, and the migration of the ectopically expressed protein examined (Fig. 6B). This demonstrated that ESAG9-K9 ran approximately 4 kDa lower after treatment, whereas ESAG9-K69 ran approximately 2 kDa lower. These size changes are compatible with the presence of either two or one glycan chains, respectively, assuming that these glycans are of a high-mannose type (because of the ER localisation), generating an approximate mass of 1700–1900 Da (Jones et al., 2005).
ESAG9 proteins are secreted
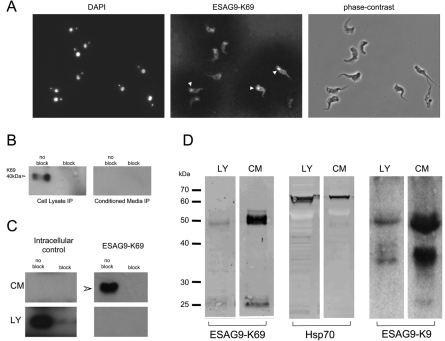
Examination of the cell line expressing ESAG9-K9 and ESAG9-K69 together (BSF K9:K69) revealed the expression of barely detectable ESAG9-K69 by western blotting using the Ty1-specific antibody BB2, despite the detectable (though weak) expression of ESAG9-K9 (Fig. 5B). However, immunofluorescence using the BB2 antibody revealed an occasional signal from the flagellar pocket of fixed cells and a clear corona of staining visible on the slide surrounding the cells (Fig. 7A). This suggested that ESAG9-K69 protein was being released from the parasites. To investigate this, conditioned media from the BSF-K69 and BSF K9:K69 cell line was subjected to immunoprecipitation using the BB2 antibody, in order to select the Ty1 epitope-tagged ESAG9-K69 protein. To ensure specificity, a blocking peptide control was also used that binds to the BB2 antibody and thereby prevents it from recognising the Ty tag. This immunoprecipitation analysis generated a clear signal from the BSF K69 line associated with the cell pellet, but not from the medium, demonstrating an absence of detectable protein release from this cell line (Fig. 7B). By contrast, however, the same analysis using the BSF K9:K69 cell line, expressing both ESAG9-K9 and ESAG9-K69, demonstrated that ESAG9-K69 protein was released to the cell medium, with no detectable cell-associated signal (Fig. 7C). To eliminate the possibility that the released protein was a consequence of cell lysis, a parallel immunoprecipitation was also carried out using an antibody specific for the cytoplasmic RNA binding protein, TbZFP3, again in the presence or absence of an anti-TbZFP3 blocking peptide to ensure specificity (intracellular control; Fig. 7C). In this case, TbZFP3 was exclusively selected from the cellular fraction, demonstrating cellular integrity under the selection conditions used.
Fig. 7.
Location of ESAG9-K69 protein in BSF K9:K69 cells engineered to express both ESAG9-K9 and ESAG9-K69 concurrently. (A) The ESAG9-K69 was detected using BB2 antibody and is visible in some cells as a signal from, or nearby, the flagellar pocket (white arrowheads) and also as a speckled staining on the slide itself. (B) Immunoprecipitation from cell lysate and conditioned media of monomorphic cells ectopically expressing ESAG9-K69. Immunoprecipitation was carried out with the BB2 antibody, which recognises the Ty epitope tag in the ESAG9-K69 sequence, in the presence (block) or absence (no block) of blocking peptide. Samples were analysed by western blotting using the BB2 antibody. There was a strong signal for ESAG9-K69 from the cell lysate in the absence of blocking peptide. (C) Immunoprecipitation from cell lysate (LY) and conditioned media (CM) of monomorphic cells ectopically expressing both ESAG9-K9 and ESAG9-K69. Immunoprecipitation was carried out with either BB2 antibody that recognises the Ty epitope tag in the ESAG9-K69 sequence, or anti-TbZFP3 antibody, which is an intracellular control, in the presence or absence of blocking peptides. Samples were analysed by western blotting using the respective antibodies. There was a strong signal for the intracellular control in the cell lysate in the absence of ZFP3 blocking peptide (no block), and no signal from the conditioned media. There was a strong signal for ESAG9-K69 from the conditioned media in the absence of blocking peptide and no signal from the cell lysate. Conditioned media from T. brucei EATRO 2340 stumpy form cells. (D) Stumpy form cells were harvested from a mouse 6 days post-innoculation and incubated in serum-free media. The conditioned media was then concentrated and analysed by western blotting with ESAG9-K69, Hsp70 (a control for cell integrity) and ESAG9-K9 antibodies. For all antibodies, there was a signal from both the cell lysate and concentrated conditioned media lanes. However, the signal from the CM lanes was much stronger for ESAG9-K69 and ESAG9-K9 than for Hsp70. Size markers are in kDa.
The ectopic expression of proteins in transgenic parasites has the potential to generate aberrant location or trafficking properties in the expressed protein. Therefore, we assessed the potential secretion of ESAG9-K69 in T. brucei EATRO 2340 cells using the anti-peptide antibody specific for that protein. Thus, stumpy cells were harvested from a mouse infection, purified by DE52 chromatography and incubated for 4 hours in serum-free medium, in order to permit effective accumulation of released proteins. Thereafter, the stumpy-conditioned medium was concentrated 1300-fold by using a 10 kDa Mr vivaspin column and the relative distribution of ESAG9-K69 between the conditioned medium and cellular fraction determined (Fig. 7D). As before, an antibody to an intracellular marker protein, in this case HSP70, was assessed simultaneously to indicate the extent of released protein attributable to cell lysis. Although some release of HSP70 was evident in the conditioned medium, as expected after incubation of recently harvested stumpy forms maintained under serum-free conditions, the ESAG-K69 was found to be preferentially concentrated in the medium fraction, with approximately tenfold enrichment with respect to the intracellular HSP70 control (i.e. 39.4 and 4.4%, respectively, release into the medium, accounting for the cell number equivalents in the assay). Reactivity of the same samples with antibody to ESAG9-K9 also revealed an enrichment of that protein in the extracellular fraction. Interestingly, however, much of this protein migrated at 36 and 50 kDa, the latter matching the migration of ESAG9-K69. These higher molecular mass forms might represent hetero- or homodimerisation of the expressed ESAG9 proteins or represent protein aggregation during medium concentration, a phenomenon sometimes observed with glycosylated proteins (data not shown).
We conclude the ESAG9-K9 and ESAG9-K69 can be secreted by trypanosomes. Moreover, analysis in transgenic bloodstream form lines demonstrated that the release of ESAG9-K69 was dependent upon the coexpression of at least one other ESAG9 protein, ESAG9-K9.
Consequences of ESAG9 secretion in vitro and ex vivo
The release of ESAG9 protein by stumpy forms suggested a potential role in either optimising transmission or in interactions with the mammalian immune system. Because stumpy forms express a large and diverse family of ESAG9 transcripts (Fig. 3), RNAi-mediated ablation of the expression of all family members was not feasible. Moreover, the inability of these monomorphic lines to establish in tsetse flies (our unpublished observations) restricted our ability to assay transmission in vivo. Hence, we exploited available in vitro and ex vivo assays to assess the consequences of ESAG9 secretion for parasite survival, and to monitor any effects on mammalian immune function.
One factor potentially important in parasite survival during transmission is the resistance of the differentiating parasite to complement in the blood meal, which in mosquitos remains active for 6 hours or more (Margos et al., 2001). In tsetse, this time coincides with the loss of the trypanosome VSG coat, which ordinarily protects the bloodstream trypanosomes from complement activated by the alternative pathway. Therefore, to assess whether ESAG9 protein could protect parasites from complement during tsetse infection, procyclic form trypanosomes were incubated with the conditioned medium from BSF K9:K69 cells in the presence of either Guinea Pig serum (GPS; as a source of complement) or heat-inactivated Guinea Pig or foetal calf serum (HI-GPS, HI-FCS), as controls. The viability of each population was then assessed over 24 hours in an Alamar Blue viability assay. Supplementary material Fig. S5 demonstrates that GPS effectively blocked procyclic form growth when compared to either HI-GPS or HI-FCS, as expected. However, no difference was observed in the presence of BSF K9:K69 conditioned medium. A titration of GPS also revealed no protective effect of ESAG9 protein in the cell culture medium (supplementary material Fig. S6), confirming that exogenous ESAG9 K9:K69 does not render procyclic form parasites resistant to complement-mediated lysis under the assay conditions used.
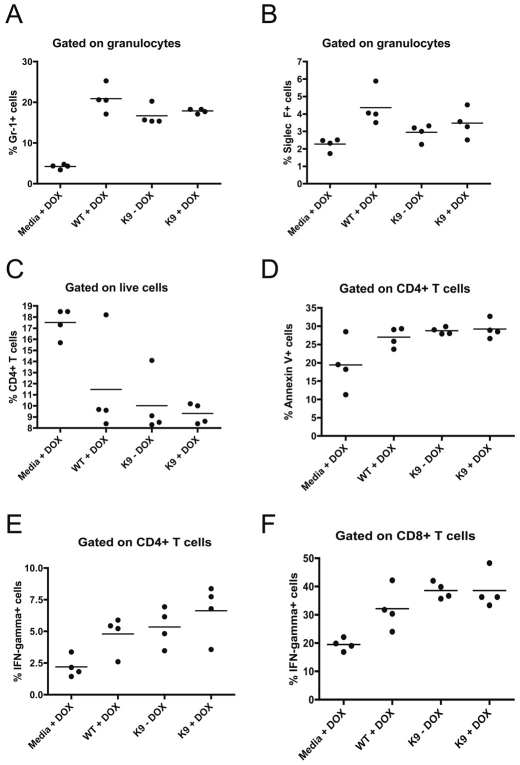
To assess the potential effects of ESAG9 in the mammalian host we used an ectopic expression approach in the monomorphic lines that do not normally express ESAG9. Hence, the BSF K9:K69 cell line was grown in mice administered either without, or with, doxycycline (DOX) in their drinking water to induce ectopic ESAG9 expression. Transgene expression was confirmed at the transcript level by northern blotting of mRNA derived from the in vivo grown parasites, this indicating tightly inducible and close to physiological levels of K9 expression and subphysiological, leaky expression of K69 when compared to T. brucei EATRO 2340 stumpy forms (supplementary material Fig. S7). As controls, mice were alternatively infected with the parental monomorphic cell line or were injected with HMI-9 medium, these mice being subsequently supplied with, or without, DOX in their drinking water. Initially, in the trypanosome-infected mice, the progression of the parasitaemia over approximately 5 days was assayed. This revealed no consistent difference in the overall virulence of the respective parasite lines. Thereafter, the immune responses generated in each group were assayed by ex vivo FACS analysis. Specifically, spleens were excised from four mice in each group at 4 days post-infection and assayed for a number of immunological responses known to be associated with trypanosome infection (Beschin et al., 1998; Millar et al., 1999; Sternberg and Mabbott, 1996). These included the prevalence of different classes of immune cells, the apoptosis of lymphocytes, and expression of interferon-γ (IFN-γ), interleukin-10 (IL-10) and Foxp3 by CD4+ and CD8+ T-cells. As expected, there were consistent differences between the naïve mice and the trypanosome-infected mice (encompassing WT +DOX, BSF K9:K69 +DOX and BSF K9:K69 −DOX). Thus, trypanosome infection generated an increase in the percentage of granulocytes that were eosinophils (Siglec F+) and neutrophils (Gr-1+), a decrease in the percentage of lymphocytes that were positive for CD4 and an increase in apoptosis of CD4+ T-cells. Moreover, an increase in the production of IFN-γ by both CD4+ and CD8+ T-cells was detected by intracellular cytokine staining (Fig. 8). Although the responses between trypanosome-infected and naïve mice reflected expectations, no statistically significant effects on the host immune response due to the expression of ESAG9 proteins were observed.
Fig. 8.
Ex vivo FACS analysis using the mouse model of trypanosomiasis. Mice were inoculated with transgenic (BSF K9.K69) parasites induced (+DOX) or not induced (−DOX) for the ectopic expression of ESAG9-K9 and ESAG9-K69. Controls were wild-type monomorphic parasites (WT) or naïve inoculations (Media). The following traits were assayed: (A) the percentage of live granulocytes that were Gr-1-positive (neutrophils); (B) the percentage of live granulocytes that were Siglec F-positive (eosinophils); (C) the percentage of live lymphocytes that were CD4+; (D) the percentage of CD4+ T-cells that were undergoing apoptosis (assayed by Annexin V); (E) the percentage of CD4+ T-cells that were expressing IFN-γ, as assayed by intracellular cytokine staining; and (F) the percentage of CD8+ T-cells that were expressing IFN-γ.
Discussion
ESAGs co-transcribed with the surface coat antigens of the African trypanosome are believed to contribute to the survival of parasites in the mammalian bloodstream. Here, we have discovered that one gene family, previously found contained within a VSG expression site, is exclusively expressed only in the transmissible bloodstream stage of the parasite stumpy forms. Further, we have discovered that many copies of the gene are expressed, at least at the RNA level, from outside of expression sites, with at least two copies, ESAG9-K9 and ESAG9-K69, being translated into protein.
Surprisingly, ESAG9 proteins can be released from bloodstream stumpy forms, and from monomorphic cells expressing both ESAG9-K9 and ESAG9-K69. This suggests that different ESAG9 proteins might cooperate to assist trafficking to the flagellar pocket of the parasite for extracellular release from stumpy forms. Perurbation of this, either through the expression of a single ESAG9 protein type in vitro, or during the normal differentiation process, might result in the accumulation of intracellular ESAG9 protein within the endoplasmic reticulum, this being visualised as transiently cell-associated protein during differentiation. Nonetheless, ESAG9 expression in bloodstream stumpy cells appears to be significantly extracellular, generating the potential for interaction with the mammalian immune system in chronic infections or providing function as a pre-adaptation for parasite transmission.
No other secreted proteins in the trypanosomatids have yet been characterised to this extent. A member of the cyclophilin family, CypA, is reported to be secreted by bloodstream form Trypanosoma congolense parasites (Pelle et al., 2002) but a function has not as yet been definitively ascribed to this protein. Also, Trypanosoma cruzi shed SAP proteins (Baida et al., 2006), although these are implicated in host cell invasion and so not relevant to the extracellular T. brucei. However, it is interesting to note that SAP proteins share some similarity with the T. cruzi MASP family, which, in turn, were weakly recognised in BLAST searches using ESAG9 sequences, but not other ESAGs. MASP proteins are extensively glycosylated surface proteins with no assigned function; however, they too have been proposed to be released from T. cruzi parasites and speculated to modulate host immune regulation (Bartholomeu et al., 2009).
Similar to the MASP protein family, the sequence of predicted ESAG9 proteins is remarkably diverse. Indeed, little sequence identity was recognised between ESAG9 proteins beyond the presence of an N-terminal signal sequence and potential GPI-anchor addition site at the C-terminus, which at least in the case of ESAG9-K9 appeared not be utilised. Nonetheless, a well-conserved motif was identified in the N-terminal region of the mature protein, represented by the sequence CX3WX8CX5G. This motif is similar to one half of the manganese binding motif present in protein phosphatase type 2 enzymes, and also to the cysteine-rich repeats in the scavenger receptor-cysteine-rich (SRCR) family of proteins, which are among the most evolutionarily ancient family of proteins of the immune system. We also note that the recently identified trypanosomatid protein, trypanosome-suppressive immunomodulating factor (TSIF), is a cysteine-rich protein, this being found to have inflammatory and immunosuppressive proteins in vitro and in ex vivo assays when expressed as a recombinant protein (Gomez-Rodriguez et al., 2009). Unfortunately, we have not succeeded in expressing recombinant ESAG9 protein; however, our expression of ESAG9 proteins in monomorphic cells under inducible regulation probably represents the most biologically appropriate assay for examining the function of ESAG9 proteins in vivo. Although this has not demonstrated a consistent effect on immune markers, our experiments do not preclude an immunomodulatory function for ESAG9 due to the limitations in what is experimentally tractable. For example, although we were able to express two copies of ESAG9 proteins in our transgenic lines, the mRNA expression profile in stumpy forms suggested that many copies of different ESAG9 proteins are expressed such that the overall ESAG9 expression levels, or protein repertoire, achieved in our experiments might have been insufficient. Furthermore, the necessity of using virulent monomorphic lines, which do not form stumpy cells and so do not ordinarily express ESAG9 proteins, meant that immune responses in chronic infections could not be assayed. These factors, combined with the inability to deplete the expression of the diverse members of the ESAG9 family in pleomorphic cells by RNAi, leaves the potential immunomodulatory function of these protein an open question.
Our experiments provide the first analysis of a diverse family of ESAGs expressed predominantly in the transmission stage of the trypanosome. Other ESAG proteins have recently been speculated to be involved in immune regulation (e.g. ESAG5) and stumpy forms have been shown to demonstrate novel mechanisms promoting their survival in the mammalian bloodstream (Engstler et al., 2007). The expression of ESAG9 proteins might assist the survival of those cells expressing the proteins, for example, at the peak of a wave of parasitaemia or during their early establishment in the fly, where both mammalian and tsetse immune factors might impact on them. A more intriguing prospect, however, is the potential for generating an effect on the immune system to enhance the viable chronicity of trypanosome infections in the field. In this case, individual parasites, irreversibly division-arrested and doomed to destruction in the blood, would contribute to sustaining a long-term infection, either through aiding the survival of the remaining proliferative parasites, or by ameliorating the potentially damaging effects of the anti-parasite immune responses on the host. Such a cooperative action by the parasites would provide an interesting evolutionary context to stumpy form biology over and above their role in parasite transmission.
Materials and Methods
Ethics statement
Animal experiments in this work were carried out in accordance with the local ethical approval requirements of the University of Edinburgh and the UK Home Office Animal (Scientific Procedures) Act (1986).
Trypanosomes
T. brucei EATRO 2340 monomorphic (GUP2965) or pleomorphic (GUP 2962) were used both being gifts of Mike Turner at the University of Glasgow, Scotland. T. brucei AnTat1.1 was a gift of Jay Bangs (University of Wisconsin). Each line was grown in female Balb/c mice. Monomorphic T. brucei Lister s427 were used in transgene expression analyses.
Construct generation and production of transgenic cell lines
ESAG9-K9 and ESAG9-K69 were each isolated from a PCR-select subtractive hybridisation screen between monomorphic slender and stumpy cells of T. brucei EATRO 2340. ESAG9-K9 and ESAG9-K69 were then PCR amplified from genomic DNA using primers that added a 5′HindIII and 3′BamHI site to the native genes and then cloned into the HindIII/BamHI sites of the trypanosome expression vector pHD451 (Biebinger et al., 1997). The internal Ty1 epitope tag sequence (Bastin et al., 1996) was incorporated by annealing separate amplicons of the 5′ or 3′ half of the genes with the epitope tag sequence being incorporated in to the 3′ or 5′ end of each half-product respectively. Annealing and amplification using ESAG9-K9 or ESAG9-K69 forward and reverse primers (incorporating a 5′HindIII site, or 3′ BglII site) generated the internally tagged genes, which were validated by DNA sequencing.
For parasite transfection, 10 μg of each construct was transfected in to bloodstream forms of T. brucei s427 engineered to express the tetracycline repressor protein, enabling inducible transgene expression. Transformants were selected using either 2 μg/ml hygromycin (ESAG9-K9) or 0.5 μg ml−1 puromycin (ESAG9-K69).
For parasite differentiation analyses, in vitro grown bloodstream forms in mid-log growth (4×106 ml−1), or stumpy cells harvested from rodents, were supplemented with cis-aconitate pH 7.0 at a concentration of 6 mM. Samples for immunofluorescence were harvested and analysed as described previously (Tasker et al., 2000) and the images processed using Adobe Photoshop 7.
Conditioned media was generated by growing cells in either HMI-9 media supplemented with 5% FCS or serum-free media. The parasites were removed by centrifugation and the resulting supernatant filtered through a 0.20 μm filter and concentrated by centrifugation through Vivaspin columns with a 10 kDa cut-off (Sartorius).
Protein, RNA analyses
Protein and RNA purification was carried out as described (Tasker et al., 2000). Genomic DNA was prepared as described (MedinaAcosta and Cross, 1993). Western blotting and northern blotting were each carried out as described (Tasker et al., 2000). Antibodies to ESAG9-K9 and ESAG9-K69 were raised to the relevant peptides and affinity purified against the immunogens (Eurogentec, Belgium).
N-Glycosylation analysis
Some 6×107 bloodstream form cells were harvested by centrifugation, lysed, and incubated in the presence or absence or PNGase-F according to the manufacturer's instructions (New England Biosciences).
Immunoprecipitation
Prior to the immunoprecipitation, the required volume of antibody was incubated for 30 minutes with 10 μg of the blocking peptide or with an equivalent volume of buffer. The antibody was then incubated with cell lysate for one hour at 4°C. An equivalent volume of Protein G beads (Sigma) was added and incubated for 1 hour at 4°C. The G beads were washed and the protein eluted by boiling in Laemmli buffer.
Ex vivo FACS analysis
Red blood cell lysed spleen cell samples were surface-stained with FITC-conjugated α-CD4 (BioLegend), phycoerythrin (PE)-conjugated α-CD8 (BD Pharmingen), Alexa Fluor 488-conjuated α-F4/80 (Caltag Laboratories), PE-conjugated α-Siglec F (BD Pharmingen), Biotin-conjugated α-Gr-1 in combination with Streptavidin APC (BD Pharmingen), PerCP-conjugated α-B220 (BD Pharmingen), APC-conjugated α-Annexin V (BD Pharmingen), APC-conjugated α-IL-10 (Pharmingen) and PE-conjugated α-FoxP3 (eBioscience). For intracellular cytokine staining, 5×106 spleen cells were incubated with 0.5 μg/ml phorbol myristate acetate (Sigma) and 1 μg/ml Ionomycin (Sigma) for 1 hour at 37°C, 5% CO2 before treatment with 10 μg/ml of the protein transport inhibitor Brefeldin A (Sigma) for a following 3 hours. Cells were then surface-stained with anti-CD4 and anti-CD8 prior to intracellular staining using e-Bioscience fixation and permeabilisation buffers and APC-conjugated anti-IFN-γ (BD Pharmingen). 100,000 cells from each sample were analysed using a FACScalibur machine (Beckton Dickinson) and the data analysed with FlowJo software (Tree Star).
Supplementary Material
Acknowledgments
We thank Pamela Davies and Deborah Hall for excellent technical assistance, and the Matthews laboratory and colleagues at IIIR, University of Edinburgh, for discussions and criticism. We thank Mike Lehane (Liverpool School of Tropical Medicine) for experiments with tsetse flies. We also thank Jay Bangs (University of Wisconsin), Dave Barry (Glasgow Biomedical Research Centre), Gloria Rudenko (University of Oxford) and Keith Gull (University of Oxford) for sharing reagents and unpublished data. Work was supported by a strategic award from the Wellcome Trust for the Centre for Immunity, Infection and Evolution. Deposited in PMC for immediate release. This article is freely accessible online from the date of publication.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/19/3401/DC1
References
- Ackermann M., Stecher B., Freed N. E., Songhet P., Hardt W. D., Doebeli M. (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987-990 [DOI] [PubMed] [Google Scholar]
- Baida R. C., Santos M. R., Carmo M. S., Yoshida N., Ferreira D., Ferreira A. T., El Sayed N. M., Andersson B., da Silveira J. F. (2006). Molecular characterization of serine-, alanine-, and proline-rich proteins of Trypanosoma cruzi and their possible role in host cell infection. Infect. Immunol. 74, 1537-1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D., Das A. K., Egloff M. P. (1998). The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27, 133-164 [DOI] [PubMed] [Google Scholar]
- Barker A. R., Wickstead B., Gluenz E., Gull K. (2008). Bioinformatic insights to the ESAG5 and GRESAG5 gene families in kinetoplastid parasites. Mol. Biochem. Parasitol. 162, 112-122 [DOI] [PubMed] [Google Scholar]
- Bartholomeu D. C., Cerqueira G. C., Leao A. C., daRocha W. D., Pais F. S., Macedo C., Djikeng A., Teixeira S. M., El-Sayed N. M. (2009). Genomic organization and expression profile of the mucin-associated surface protein (masp) family of the human pathogen Trypanosoma cruzi. Nucleic Acids Res. 37, 3407-3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P., Bagherzadeh Z., Matthews K. R., Gull K. (1996). A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77, 235-239 [DOI] [PubMed] [Google Scholar]
- Beschin A., Brys L., Magez S., Radwanska M., De Baetselier P. (1998). Trypanosoma brucei infection elicits nitric oxide-dependent and nitric oxide-independent suppressive mechanisms. J. Leukoc. Biol. 63, 429-439 [DOI] [PubMed] [Google Scholar]
- Biebinger S., Wirtz L. E., Lorenz P., Clayton C. (1997). Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol. 85, 99-112 [DOI] [PubMed] [Google Scholar]
- Bitter W., Gerrits H., Kieft R., Borst P. (1998). The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature 391, 499-502 [DOI] [PubMed] [Google Scholar]
- Borst P. (2002). Antigenic variation and allelic exclusion. Cell 109, 5-8 [DOI] [PubMed] [Google Scholar]
- Claes F., Buscher P., Touratier L., Goddeeris B. M. (2005). Trypanosoma equiperdum: master of disguise or historical mistake? Trends Parasitol. 21, 316-321 [DOI] [PubMed] [Google Scholar]
- Cully D. F., Ip H. S., Cross G. A. (1985). Coordinate transcription of variant surface glycoprotein genes and an expression site associated gene family in Trypanosoma brucei. Cell 42, 173-182 [DOI] [PubMed] [Google Scholar]
- Dean S. D., Marchetti R., Kirk K., Matthews K. (2009). A surface transporter family conveys the trypanosome differentiation signal. Nature 459, 213-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle S. P., Griffin A. S., Campbell G. S., West S. A. (2007). Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411-414 [DOI] [PubMed] [Google Scholar]
- Engstler M., Boshart M. (2004). Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 18, 2798-2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstler M., Pfohl P., Herminghaus S., Boshart M., Wiegerttjes G., Heddergott N., Overath P. (2007). Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 131, 505-515 [DOI] [PubMed] [Google Scholar]
- Florent I. C., Raibaud A., Eisen H. (1991). A family of genes related to a new expression site-associated gene in Trypanosoma equiperdum. Mol. Cell. Biol. 11, 2180-2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A., Kummerli R. (2008). Social evolution: this microbe will self-destruct. Curr. Biol. 18, R1021-R1023 [DOI] [PubMed] [Google Scholar]
- Gomez-Rodriguez J., Stijlemans B., De Muylder G., Korf H., Brys L., Berberof M., Darji A., Pays E., De Baetselier P., Beschin A. (2009). Identification of a parasitic immunomodulatory protein triggering the development of suppressive M1 macrophages during African trypanosomiasis. J. Infect. Dis. 200, 1849-1860 [DOI] [PubMed] [Google Scholar]
- Gunzl A., Bruderer T., Laufer G., Schimanski B., Tu L. C., Chung H. M., Lee P. T., Lee M. G. (2003). RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryotic Cell 2, 542-551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz-Fowler C., Figueiredo L. M., Quail M. A., Becker M., Jackson A., Bason N., Brooks K., Churcher C., Fahkro S., Goodhead I., et al. (2008). Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS ONE 3, e3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen B. C., Sivam D., Kifer C. T., Myler P. J., Parsons M. (2009). Widespread variation in transcript abundance within and across developmental stages of Trypanosoma brucei. BMC Genomics 10, 482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. C., Mehlert A., Guther M. L., Ferguson M. A. (2005). Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N-glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. J. Biol. Chem. 280, 35929-35942 [DOI] [PubMed] [Google Scholar]
- Kabani S., Fenn K., Ross A., Ivens A., Smith T. K., Ghazal P., Matthews K. (2009). Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 10, 427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooter J. M., van der Spek H. J., Wagter R., d'Oliveira C. E., van der Hoeven F., Johnson P. J., Borst P. (1987). The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T. brucei. Cell 51, 261-272 [DOI] [PubMed] [Google Scholar]
- Kusuda K., Kobayashi T., Ikeda S., Ohnishi M., Chida N., Yanagawa Y., Shineha R., Nishihira T., Satomi S., Hiraga A., et al. (1998). Mutational analysis of the domain structure of mouse protein phosphatase 2Cbeta. Biochem. J. 332, 243-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai D. H., Hashimi H., Lun Z. R., Ayala F. J., Lukes J. (2008). Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc. Natl. Acad. Sci. USA 105, 1999-2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe K. A., Morrison L. J., Read A. F., Barry J. D. (2007). Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc. Natl. Acad. Sci. USA 104, 8095-8100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margos G., Navarette S., Butcher G., Davies A., Willers C., Sinden R. E., Lachmann P. J. (2001). Interaction between host complement and mosquito-midgut-stage Plasmodium berghei. Infect. Immunol. 69, 5064-5071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. R., Gull K. (1994). Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J. Cell Biol. 125, 1147-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. R., Ellis J. R., Paterou A. (2004). Molecular regulation of the life cycle of African trypanosomes. Trends Parasitol. 20, 40-47 [DOI] [PubMed] [Google Scholar]
- McCulloch R. (2004). Antigenic variation in African trypanosomes: monitoring progress. Trends Parasitol. 20, 117-121 [DOI] [PubMed] [Google Scholar]
- MedinaAcosta E., Cross G. A. M. (1993). Rapid isolation of DNA from Trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol. Biochem. Parasitol. 59, 327-330 [DOI] [PubMed] [Google Scholar]
- Millar A. E., Sternberg J., McSharry C., Wei X. Q., Liew F. Y., Turner C. M. (1999). T-Cell responses during Trypanosoma brucei infections in mice deficient in inducible nitric oxide synthase. Infect. Immunol. 67, 3334-3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M., Gull K. (2001). A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414, 759-763 [DOI] [PubMed] [Google Scholar]
- Nolan D. P., Rolin S., Rodriguez J. R., Van Den Abbeele J., Pays E. (2000). Slender and stumpy bloodstream forms of Trypanosoma brucei display a differential response to extracellular acidic and proteolytic stress. Eur. J. Biochem. 267, 18-27 [DOI] [PubMed] [Google Scholar]
- Pays E., Tebabi D., Pays A., Coquelet H., Revelard P., Salmon D., Steinert M. (1989). The genes and transcripts of an antigen gene expression site from T. brucei. Cell 57, 835-845 [DOI] [PubMed] [Google Scholar]
- Pays E., Lips S., Nolan D., Vanhamme L., Perez-Morga D., Xong H. V., Chamekh M., Chimfwembe C. E., Van Den Abbeele J., Pays A., et al. (2001). The VSG expression sites of Trypanosoma brucei: multipurpose tools for the adaptation of the parasite to mammalian hosts Mol. Biochem. Parasitol. 114, 1-16 [DOI] [PubMed] [Google Scholar]
- Pelle R., McOdimba F., Chuma F., Wasawo D., Pearson T. W., Murphy N. B. (2002). The African trypanosome cyclophilin A homologue contains unusual conserved central and N-terminal domains and is developmentally regulated. Gene 290, 181-191 [DOI] [PubMed] [Google Scholar]
- Robertson M. (1912). Notes on the polymorphism of Trypanosoma gambiense in the blood and its relation to the exogenous cycvle in Glossina palpalis. Proc. R. Soc. Lond. B. Biol. Sci. 85, 241-539 [Google Scholar]
- Robinson N. P., Burman N., Melville S. E., Barry J. D. (1999). Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell. Biol. 19, 5839-5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon D., Geuskens M., Hanocq F., Hanocq Q.-J., Nolan D., Ruben L., Pays E. (1994). A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell 78, 75-86 [DOI] [PubMed] [Google Scholar]
- Sbicego S., Vassella E., Kurath U., Blum B., Roditi I. (1999). The use of transgenic Trypanosoma brucei to identify compounds inducing the differentiation of bloodstream forms to procyclic forms. Mol. Biochem. Parasitol. 104, 311-322 [DOI] [PubMed] [Google Scholar]
- Schell D., Evers R., Preis D., Ziegelbauer K., Kiefer H., Lottspeich F., Cornelissen A., Overath P. (1991). A transferrin-binding protein of Trypanosoma brucei is encoded by one of the genes in the variant surface glycoprotein gene expression site. EMBO J. 10, 1061-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheader K., Vaughan S., Minchin J., Hughes K., Gull K., Rudenko G. (2005). Variant surface glycoprotein RNA interference triggers a precytokinesis cell cycle arrest in African trypanosomes. Proc. Natl Acad. Sci. USA 102, 8716-8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg M. J., Mabbott N. A. (1996). Nitric oxide-mediated suppression of T cell responses during Trypanosoma brucei infection: soluble trypanosome products and interferon-gamma are synergistic inducers of nitric oxide synthase. Eur. J. Immunol. 26, 539-543 [DOI] [PubMed] [Google Scholar]
- Tasker M., Wilson J., Sarkar M., Hendriks E., Matthews K. (2000). A novel selection regime for differentiation defects demonstrates an essential role for the stumpy form in the life cycle of the African trypanosome. Mol. Biol. Cell 11, 1905-1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L., Paturiaux-Hanocq F., Poelvoorde P., Nolan D. P., Lins L., Van Den Abbeele J., Pays A., Tebabi P., Van Xong H., Jacquet A., et al. (2003). Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422, 83-87 [DOI] [PubMed] [Google Scholar]
- Vassella E., Reuner B., Yutzy B., Boshart M. (1997). Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J. Cell Sci. 110, 2661-2671 [DOI] [PubMed] [Google Scholar]
- Wijers D., Willett K. (1960). Factors that may influence the infection rate of Glossina Palpalis with Trypanosoma gambiense. II. – The number and morphology of the trypanosomes present in the blood of ther host at the time of the infective feed. Ann. Trop. Med. Parasitol. 54, 341-350 [PubMed] [Google Scholar]
- Xong H. V., Vanhamme L., Chamekh M., Chimfwembe C. E., Van Den Abbeele J., Pays A., Van Meirvenne N., Hamers R., De Baetselier P., Pays E. (1998). A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell 95, 839-846 [DOI] [PubMed] [Google Scholar]
- Young R., Taylor J. E., Kurioka A., Becker M., Louis E. J., Rudenko G. (2008). Isolation and analysis of the genetic diversity of repertoires of VSG expression site containing telomeres from Trypanosoma brucei gambiense, T. b. brucei and T. equiperdum. BMC Genomics 9, 385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer K., Overath P. (1990). Surface antigen change during differentiation of Trypanosoma brucei. Biochem. Soc. Trans. 18, 731-733 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.