Abstract
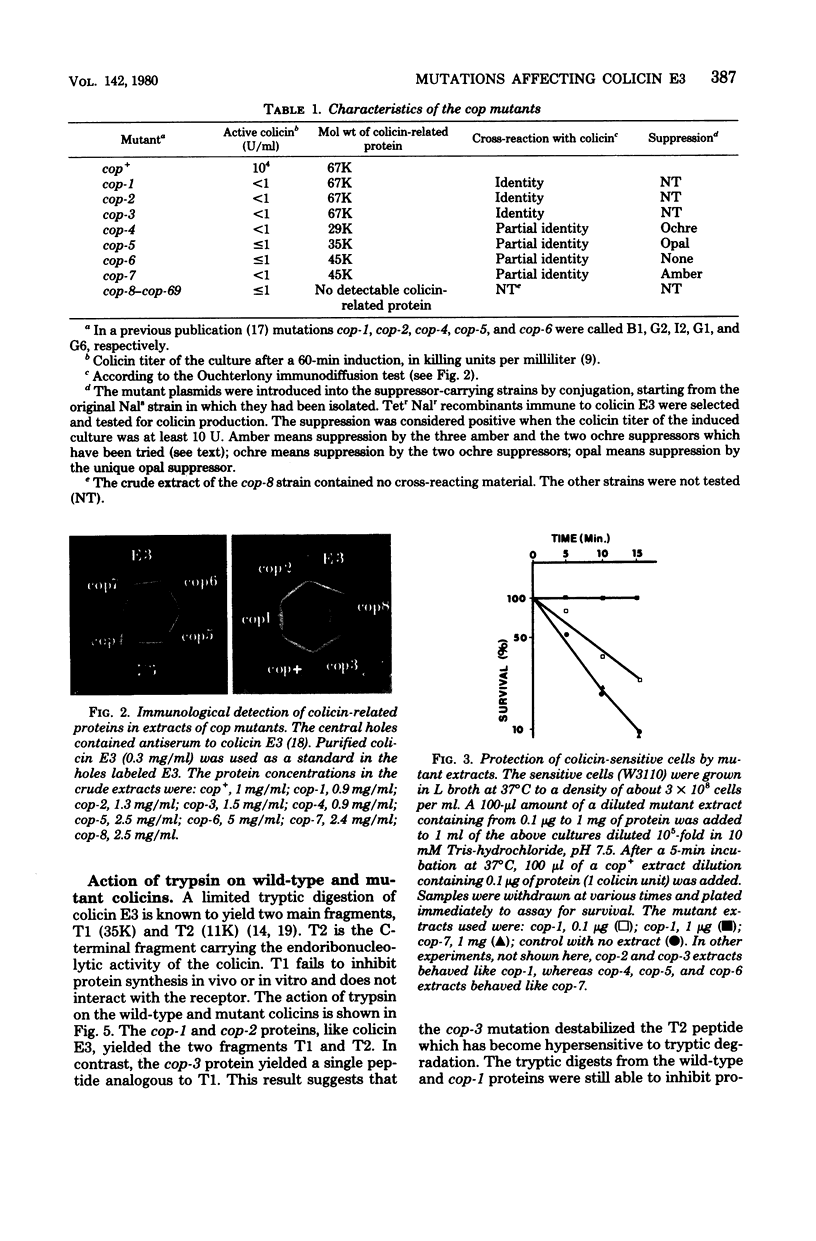
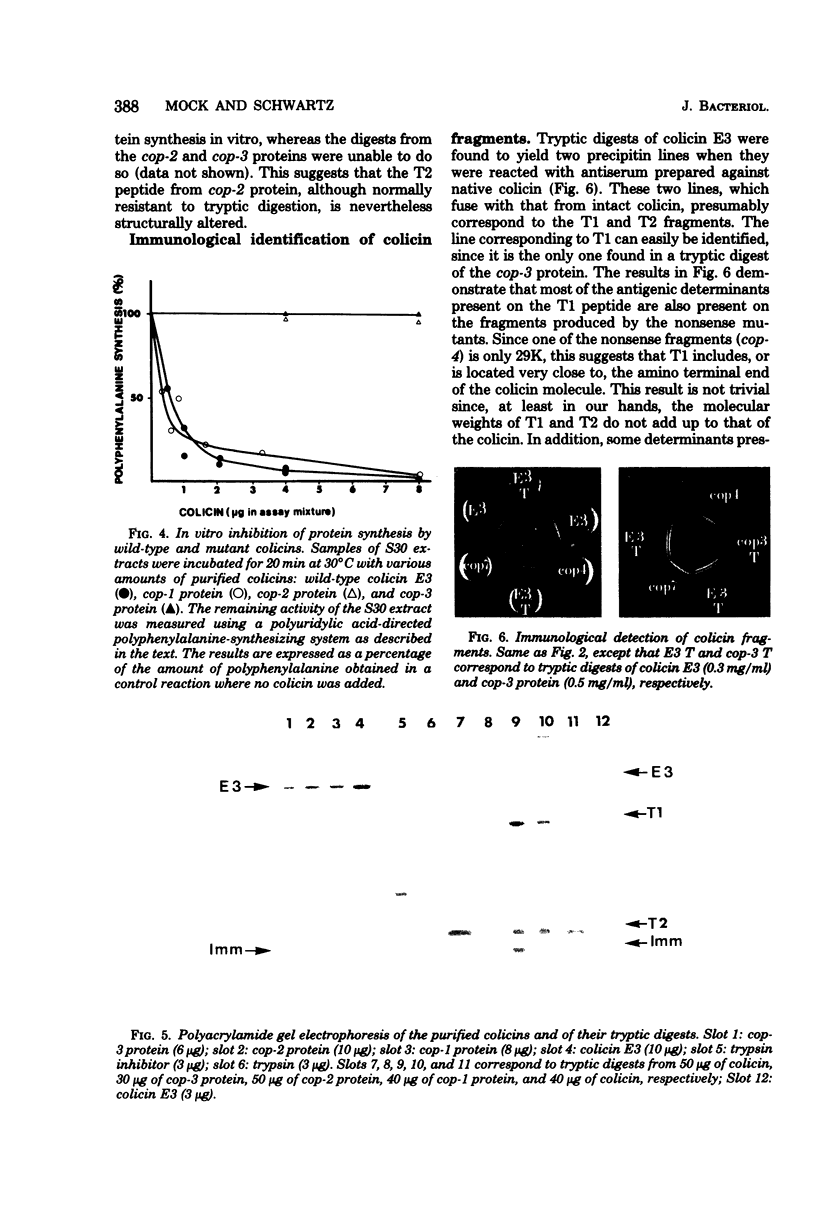
Among 69 ColE3 mutant plasmids selected on the basis of their inability to produce an active colicin, seven (cop-1 to cop-7) were found to bear a mutation affecting the structural gene for colicin. Three of these (cop-1, cop-2 and cop-3) lead to the production of an inactive colicin molecule which has the same molecular weight as wild-type colicin E3 (67,000). These three inactive colicins are still able to interact with the outer membrane receptor. The cop-1 protein retained the ability to inhibit protein synthesis in vitro and therefore seems specifically affected in it ability to penetrate the cell envelope. The cop-2 and cop-3 proteins lost the ability to inhibit protein synthesis in vitro, and activity which is normally associated with the C-terminal part of the colicin molecule. On the basis of this and further evidence, it is suggested that the cop-2 and cop-3 mutations affect the structure of the C-terminal part of the colicin molecule. The other four mutations (cop-4 to cop-7) lead to the production of colicin-related polypeptides of lower molecular weight (29,000 to 45,000) which display a reaction of partial immunological identity with wild-type colicin. These four polypeptides are unable to interact with the cell surface receptor. Three of these mutants are shown to carry a nonsense mutation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreoli P. M., Nijkamp H. J. Mutants of the Clo DF13 plasmid in Escherichia coli with a decreased bacteriocinogenic activity. Mol Gen Genet. 1976 Mar 22;144(2):159–170. doi: 10.1007/BF02428104. [DOI] [PubMed] [Google Scholar]
- Boon T. Inactivation of ribosomes in vitro by colicin E 3 . Proc Natl Acad Sci U S A. 1971 Oct;68(10):2421–2425. doi: 10.1073/pnas.68.10.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman C. M., Dahlberg J. E., Ikemura T., Konisky J., Nomura M. Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc Natl Acad Sci U S A. 1971 May;68(5):964–968. doi: 10.1073/pnas.68.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman C. M., Sidikaro J., Nomura M. Specific inactivation of ribosomes by colicin E3 in vitro and mechanism of immunity in colicinogenic cells. Nat New Biol. 1971 Dec 1;234(48):133–137. doi: 10.1038/newbio234133a0. [DOI] [PubMed] [Google Scholar]
- Gaastra W., Oudega B., de Graaf F. K. The use of mutants in the study of structure-function relationships in cloacin DF13. Biochim Biophys Acta. 1978 May 3;540(2):301–312. doi: 10.1016/0304-4165(78)90143-5. [DOI] [PubMed] [Google Scholar]
- Hardy K. G. Colicinogeny and related phenomena. Bacteriol Rev. 1975 Dec;39(4):464–515. doi: 10.1128/br.39.4.464-515.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman H. R., Helinski D. R. Purification and characterization of colicin E2 and colicin E3. J Biol Chem. 1967 Nov 25;242(22):5360–5368. [PubMed] [Google Scholar]
- Hirose A., Kumagai J., Imahori K. Dessociation and reconstitution of colicin E3 and immunity substance complex. J Biochem. 1976 Feb;79(2):305–311. doi: 10.1093/oxfordjournals.jbchem.a131073. [DOI] [PubMed] [Google Scholar]
- Jakes K. S., Zinder N. D. Highly purified colicin E3 contains immunity protein. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3380–3384. doi: 10.1073/pnas.71.9.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lau C., Richards F. M. Proteolytic and chemical modification of colicin E3 activity. Biochemistry. 1976 Aug 24;15(17):3856–3863. doi: 10.1021/bi00662a032. [DOI] [PubMed] [Google Scholar]
- Mock M., Schwartz M. Mechanism of colicin E3 production in strains harboring wild-type or mutant plasmids. J Bacteriol. 1978 Nov;136(2):700–707. doi: 10.1128/jb.136.2.700-707.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Ohno-Iwashita Y., Suzuki K., Imahori K. Purification and characterization of active component and active fragment of colicin E3. J Biochem. 1977 Oct;82(4):1045–1053. doi: 10.1093/oxfordjournals.jbchem.a131775. [DOI] [PubMed] [Google Scholar]
- Sabet S. F., Schnaitman C. A. Purification and properties of the colicin E3 receptor of Escherichia coli. J Biol Chem. 1973 Mar 10;248(5):1797–1806. [PubMed] [Google Scholar]
- Senior B. W., Holland I. B. Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc Natl Acad Sci U S A. 1971 May;68(5):959–963. doi: 10.1073/pnas.68.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilby M., Hindennach I., Henning U. Bypass of receptor-mediated resistance to colicin E3 in Escherichia coli K-12. J Bacteriol. 1978 Dec;136(3):1189–1191. doi: 10.1128/jb.136.3.1189-1191.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J., Sherratt D. J. Synthesis of E colicins in Escherichia coli. Mol Gen Genet. 1975 Oct 22;140(4):349–353. doi: 10.1007/BF00267325. [DOI] [PubMed] [Google Scholar]
- Watson D. H., Sherratt D. J. In vivo proteolytic cleavage of colicins requires specific receptor binding. Nature. 1979 Mar 22;278(5702):362–364. doi: 10.1038/278362a0. [DOI] [PubMed] [Google Scholar]
- Webster R. E., Engelhardt D. L., Zinder N. D., Konigsberg W. Amber mutants and chain termination in vitro. J Mol Biol. 1967 Oct 14;29(1):27–43. doi: 10.1016/0022-2836(67)90179-9. [DOI] [PubMed] [Google Scholar]
- de Graaf F. K., Stukart M. J., Boogerd F. C., Metselaar K. Limited proteolysis of cloacin DF13 and characterization of the cleavage products. Biochemistry. 1978 Mar 21;17(6):1137–1142. doi: 10.1021/bi00599a031. [DOI] [PubMed] [Google Scholar]






