Abstract
Human embryonic stem cell-derived cardiomyocytes (hESC-CMs) have demonstrated the ability to improve myocardial function following transplantation into an ischemic heart; however, the functional benefits are transient possibly due to poor cell retention. A diagnostic technique that could visualize transplanted hESC-CMs could help to optimize stem cell delivery techniques. Thus, the purpose of this study was to develop a labeling technique for hESCs and hESC-CMs with the FDA-approved contrast agent indocyanine green (ICG) for optical imaging (OI). hESCs were labeled with 0.5, 1.0, 2.0, and 2.5 mg/ml of ICG for 30, 45, and 60 min at 37°C. Longitudinal OI studies were performed with both hESCs and hESC-CMs. The expression of surface proteins was assessed with immunofluorescent staining. hESCs labeled with 2 mg ICG/ml for 60 min achieved maximum fluorescence. Longitudinal studies revealed that the fluorescent signal was equivalent to controls at 120 h postlabeling. The fluorescence signal of hESCs and hESC-CMs at 1, 24, and 48 h was significantly higher compared to precontrast data (p < 0.05). Immunocytochemistry revealed retention of cell-specific surface and nuclear markers postlabeling. These data demonstrate that hESCs and hESC-CMs labeled with ICG show a significant fluorescence up to 48 h and can be visualized with OI. The labeling procedure does not impair the viability or functional integrity of the cells. The technique may be useful for assessing different delivery routes in order to improve the engraftment of transplanted hESC-CMs or other stem cell progenitors.
Keywords: Optical imaging, Cell labeling, Stem cell, Cardiomyocytes, Indocyanine green, Contrast agent
INTRODUCTION
Stem cell therapies hold unprecedented potential to provide therapeutics for a wide variety of diseases. Due to the limited mitotic abilities of cells in many organs and thus a limited regeneration capacity, injury resulting from disease or trauma often leads to irreversible cell damage. Human embryonic stem cells (hESCs) have the unique ability to regenerate into cells of almost all human tissues (2, 5, 21). hESC-CMs are a particularly promising source of therapeutics for myocardial regeneration following cardiac ischemia. When compared to cardiac progenitors derived from human mesenchymal stem cells (hMSC), hESC-CMs have a distinct advantage because their in vitro differentiation model closely resembles in vivo cardiac development at a structural and electrophysiological level. Few studies, however, currently demonstrate this potential for hMSC (4, 11, 14, 19).
A major challenge for successful hESC-derived myocardial regeneration is the development of an efficient delivery technique. Previous studies have shown poor outcomes of hESC-CM delivery, such that the majority of the injected cells are either washed away to extra cardiac organs or die soon after injection (7, 17). More specifically, the present cell survival rate of hESC-CM transplants in cardiac infarcts is bleak, with only 10% of the cells surviving the injection. This poor survival rate is thought to be the primary reason behind the limited benefits observed in cardiac function following hESC-CM-based therapy (7). A diagnostic technique that could directly visualize the transplanted hESC-CM in vivo is needed in order to help direct and optimize stem cell delivery techniques and confirm successful stem cell deposition into the myocardium (20).
Among various available imaging techniques, optical imaging (OI) has the advantage of being quick, inexpensive, easy to perform, noninvasive, and does not involve ionizing radiation. The first clinical OI scanners have recently entered clinical applications and pilot studies show promising results for investigations of breast tissue in patients (13).
In order to track hESCs and/or their progeny with OI, cells must be labeled with an exogenous or endogenous marker. While many investigators have focused on bioluminescence OI techniques (e.g., firefly luciferase) for in vivo stem cell tracking, this technique has little clinical relevance because it involves the introduction of nonhuman virus substrates into the cells of interest (D-luciferin), which alters the host’s DNA permanently (7). Exogenous fluorescent markers represent a potentially clinically applicable alternative to firefly luciferase. Indocyanine green (ICG) is an exogenous fluorescent marker with a favorable near-infrared (NIR) absorption and emission spectrum (600–950 nm) and is FDA approved for investigations of cardiac perfusion in patients (23).
The purpose of this study was to establish a labeling technique of hESCs and hESC-CMs with ICG for subsequent OI. The major goal of this investigation was to develop an ICG-labeling protocol, which should provide maximal fluorescence of labeled hESCs and hESC-CMs on OI scans and, at the same time, preserve cell viability and expression of appropriate surface markers. If successful, this technique would be in principle readily applicable for monitoring stem cell-based therapies, particularly new injection techniques.
MATERIALS AND METHODS
Contrast Agent
ICG is an FDA-approved, hydrophilic, anionic, tricarbocyanine NIR dye with a molecular weight of 774.97 Da. The structure of ICG is composed of two polycyclic moieties connected by a carbon chain. ICG shows an absorbance peak at 780 nm and an emission peak at 830 nm (22). The ICG solution used in this study was prepared by dissolving ICG powder (laser grade I-25; Sigma-Aldrich Co., St. Louis, MO) in 20% dimethyl sulfoxide (DMSO, 99.9%, purchased from Sigma-Aldrich Co.) and 80% Dulbecco’s modified Eagles medium (D-MEM High Glucose; D5648) with resulting stock solutions at concentrations of 0.5, 1, 2, and 2.5 mg/ml.
Culture of Undifferentiated hESCs
All studies were in accordance with and approved by our institution’s Committee on Human Research. For conduction of the described experiments, HSF-6 hESCs (UC06, a federal line/NIH approved) were kept in an undifferentiated and pluripotent state, cultured on gelatin-coated (0.1% gel) 10-cm (Falcon) petri dishes, and plated on irradiated mouse embryonic fibroblasts (MEFs ~800,000 cells/plate). The feeder cells were mitotically inactivated using a cesium source gamma irradiator (3000 rad for 15 min). Culture medium [Knockout Serum Replacement (KSR) media] contained: Knockout DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 20% KSR (Invitrogen), 1% nonessential amino acids (Invitrogen), 1 mM L-glutamine (Invitrogen), 0.1 mM mercaptoethanol (Sigma), and 4 ng/ml human basic FGF (Invitrogen). Cells were maintained at 37°C in 5% CO2 and split by manual dissection at a ratio of 1:3 approximately every 4 days. hESCs were grown until 10-cm petri dishes were 80% confluent. Three days prior to labeling experiments the hESCs were dissected with the EZ tool (Invitrogen) to ensure that the colonies were divided into homogeneous subcolonies. Following dissection, an equal amount of cell solution was transferred to six-well plates (Corning Inc. VWR Int., West Chester, PA, USA) that were pregelatinized with 0.1% gel and plated with ~100,000 feeders. Once colonies formed, roughly 3–4 days later, the cells were ready to be labeled.
Optimization of hESC Labeling
hESCs were incubated with increasing concentrations of 0.5, 1.0, 1.5, 2.0, and 2.5 mg ICG/ml for either 30, 45, or 60 min in standard cell culture conditions. The stock cell labeling solution was added to cell culture dishes (60 μl/ml). Following incubation the cells were trypisinized (0.25% trypsin), washed three times in PBS, resuspended in 10 ml of prewarmed KSR, and transferred to gelatinized-coated (0.1% gel) 10-cm petri dishes. hESCs were separated from feeders after an incubation of 60 min at 37°C/5% CO2, at which time the feeder cells adhered tightly to the plate, while the labeled hESCs only adhered weakly. The hESCs were collected by pipetting and were transferred to a 15-ml centrifuge tube (26). hESCs were then counted and transferred into microcentrifuge tubes for OI (VWR Int.) where they contained 300,000 cells/1 ml KSR media.
Cardiomyocyte Differentiation
hESCs were allowed to spontaneously differentiate into beating cardiomyocytes using established methods (16). Briefly, embryoid bodies (EBs) were first produced in six-well low attachment plates. On day 7 EBs were transferred to a standard six-well plate precoated with 0.1% gelatin. Cardiomyocyte media was similar to the KSR used in the undifferentiated culture, but lacked bFGF and contained 20% defined FBS (Hyclone) instead of 20% KSR.
Beating cardiomyocytes (CMs) were labeled on day 25 with ICG under previously optimized conditions that were established using the hESCs. CMs were grown in six-well plates that were precoated with 0.10% gelatin. Following labeling beating CMs were dissected out with a P-100 micropipette and transferred to a 15-ml conical tube. The cell suspension was then enzymatically dispersed using TrypLE (GIBCO, 12604) for 5 min at 37°C and then neutralized with CM media containing 20% FBS. Cells were allowed to suspend by gravity and were washed with phosphate-buffered saline (PBS) three times and were then resuspended in fresh CM media for imaging.
Optical Imaging
In vitro fluorescence imaging was performed with an IVIS 50 Imaging System (Xenogen, Alameda, CA). The optical imager is an integrated fluorescence system (400–900 nm) that is composed of a light-tight specimen chamber (dark box) and a 0.5-inch charge-coupled device (CCD) camera. To minimize electronic background and maximize sensitivity, the CCD camera is thermoelectrically cooled to −70°C.
All in vitro experimental samples were imaged in 1 ml of KSR or CM media in microcentrifuge tubes (VWR Int.). All images were acquired using the filter setting preset for ICG with a background wavelength at 665–695 nm, an excitation wavelength at 710–760 nm, and an emission wavelength set at 810–875 nm. Consistent illumination parameters were used for all NIR fluorescent acquisitions. For all experiments, the field of view (FOV) was set at 7, exposure time was 2 s, f/stop was 2, lamp voltage was set to “high” and binning was kept on medium. Following acquisition, all images were normalized to units of average efficiency, displayed in the same scale of fluorescent intensity, and analyzed using the Living Image 2.5 software (Xenogen, Alameda, CA).
Optimization of the Cell Labeling Protocol
hESCs labeled with different ICG concentrations and incubation times underwent OI directly after the labeling process.
Longitudinal Follow-up Studies
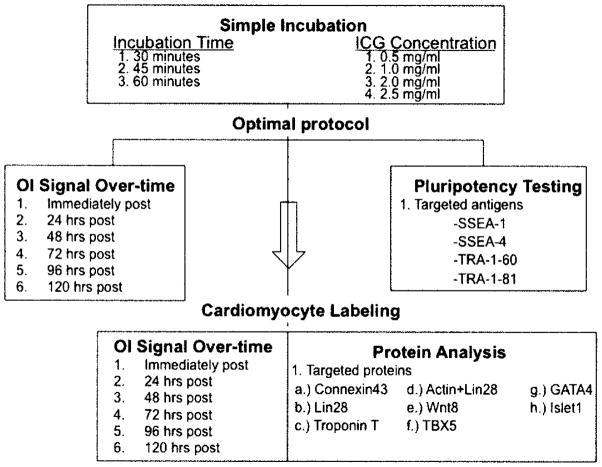
hESCs and hESC-CMs were labeled with the optical labeling protocol, determined based on experiments from the above protocol, and in separate tubes 100,000 hESCs or hESC-CMs were resuspended in 1 ml of fresh KSR or CM media and imaged until the fluorescent signal matched that of the nonlabeled control cells. All experiments were performed in triplicate (Fig. 1).
Figure 1.
Experimental scheme followed for the labeling with indocyanine green (ICG), imaging, and protein analysis of undifferentiated human embryonic stem cells (hESCs) and cardiomyocytes.
Optical Imaging Data Analysis
For quantitative analyses of OI data, regions of interest (ROI) were placed on each ICG test sample (micro-centrifuge tube). Quantitative measures of fluorescence intensity of labeled cells in test tubes were normalized to units of average efficiency, displayed in the same scale of fluorescent intensity, and analyzed using the Living Image 2.5 software (Xenogen, Alameda, CA). The unit of “efficiency” represents the fractional ratio of fluorescent-emitted photons per incident excitation photon (Xenogen).
Statistical Analysis
Means and SDs were used to describe the variation in fluorescent signal over time and viability for the following groups: 1) fluorescent signal for hESCs compared to nonlabeled controls, 2) fluorescent signal for CMs compared to nonlabeled controls, 3) fluorescent signal for hESCs compared to CMs, and 4) viability of CMs compared to nonlabeled controls. The multivariate analysis of variance (MANOVA) was used to describe the variation in fluorescent signal and viability for hESCs compared to nonlabeled controls for the various labeling protocols (various ICG concentrations and incubation times). Multiple comparison p-values were determined by the Tukey-Kramer test. All analyses were performed by SAS software (SAS V9.3; Cary, NC, USA).
Cell Viability Testing
Trypan blue tests were performed on all cells before and after the labeling procedure to verify viability. Cells were counted in a hemocytometer and then underwent subsequent optical imaging studies.
Immunohistochemistry
Analysis of hESC Pluripotency Postlabeling
To ensure that the hESCs retained marker expression patterns characteristic of undifferentiated hESCs postlabeling, the cells were analyzed using immunofluorescence techniques followed by fluorescent microscopy. hESC underwent labeling under the optimized conditions as described above. The ICG-labeled hESCs and unlabeled controls were manually dissected out, washed three times, and replated onto 24-well plates that were pregelatinized with 0.1% gel and contained 20,000 irradiated feeder cells per well. After colony formation occurred (approximately 4 days later) the cells were incubated with primary antibodies according to the manufacturer’s instructions (ES Cell Marker Sample Kit protocol; Chemicon, Billerica, MA) followed by incubation with the secondary antibody Alexa Fluor® 488 Goat anti-mouse IgG. Confocal images were acquired using a Leica microscope.
Assessment of Cardiomyocyte Protein Expression Postlabeling
Following labeling with ICG, the beating CM areas were dissected out with a P-100 micropipette and transferred to a 15-ml conical tube. The cell suspension was then enzymatically dispersed using TrypLE (GIBCO, 12604) for 5 min at 37°C and then neutralized with CM media containing 20% FBS. Cells were allowed to settle by gravity and were then resuspended in fresh CM media. ROC inhibitor was added at a concentration of 10 μM (EMD: y-27632) to prevent apoptosis. Cells were plated at a low density on pregelatinized-coated wells of Lab-Tek II eight-well glass chamber slide system and incubated for 24 h. Cells were fixed using 4% paraformaldyhyde for 15 min. Cells were washed two times for 1 min with PBS followed by washing three times for 1 min each with washing buffer 1 (0.10% Triton X-100 in PBS at pH 7.4). Cells were blocked with 10% PBS, 0.10% Triton X-100, 5% normal goat serum, and in PBS for 1 h at room temperature. Primary antibodies were added to chamber slides in a total of 200 μl/well of antibody buffer and incubated overnight at 4°C. Primary antibodies were Troponin T (Thermo Scientific, MS-295-PO), GATA4 (Abcam, ab61170), Connexin43 (Millipore, AB1728), Alexa Fluor® 488 Phalloidin (Invitrogen, A12379), Islet1 (Developmental Studies Hybridoma Bank), Lin28 (R&D Systems, AF3757), Wnt-8A (R&D Systems, AF2248), TBX5 (GenWay, 18-003-42369), SMA. All were used at 1:100. The next day cells were washed three times for 1 min each in washing buffer 2 (0.03% Triton X-100 in PBS at pH 7.4) followed by an additional wash in PBS for 1 min. Cells were incubated with proper secondary antibodies in 1× PBS. Secondary antibodies used were Alexa Fluor® anti-rabbit 555 (Molecular Probes, A21429), Alexa Fluor® anti-mouse IgG 488 (Molecular Probes, A11029), and Alexa Fluor® anti-goat 555 (Invitrogen Molecular Probes, A21431). All were used at 1:500. Slides were placed in humidifying trays/covered and left on benchtop for 1–2 h. Cells were washed in washing buffer 2 for 1 min and then washed again in PBS for 1 min, postfixed with 4% PFA for 1 min, followed by washing with PBS for 1 min. Samples were combined with VECTASHIELD® mounting medium containing DAPI (Vector Laboratories Inc., H-1500) and wells were sealed with coverslips. Samples were then imaged using a Zeiss AxioObserver Z1 microscope.
RESULTS
Optical Imaging
Optimization of hESC Labeling Protocol
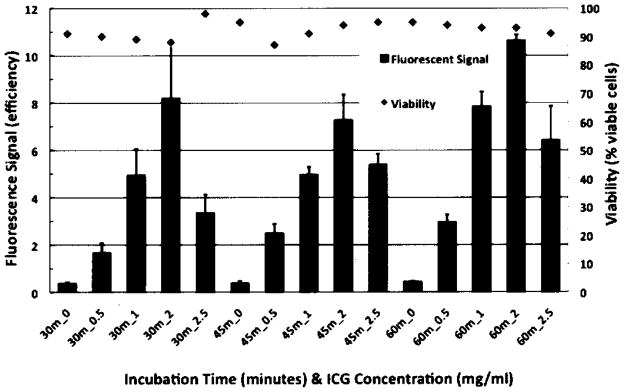
Labeling of hESCs with varying concentrations and incubation times showed a maximal fluorescence signal with the combination of the 60-min incubation time and an ICG concentration of 2.0 mg/ml (Fig. 2). The average fluorescence signal was significantly higher with the 60-min incubation time compared to lower incubation times (p < 0.0001), and also significantly higher with the ICG concentration of 2.0 mg/ml than lower or higher concentrations (p < 0.0001).
Figure 2.
Cell viability of human embryonic stem cells (hESCs) maintained postlabeling with indocyanine green (ICG). Fluorescent signal (units in efficiency) seen with optical imaging (OI) and viability (% viable cells) of hESCs, labeled with different ICG concentrations (0 = control as well as 0.5, 1.0, 1.5, 2.0, and 2.5 mg ICG/ml) for different incubation intervals (30, 45, and 60 min). Data are displayed as means and SD of triplicate samples containing 300,000 cells in a pellet per sample in 1 ml of KSR.
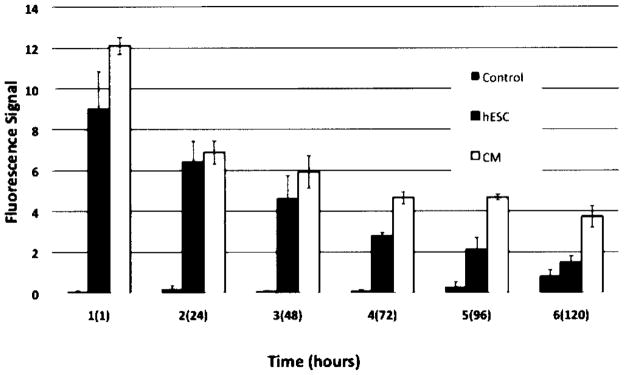
Longitudinal OI Studies
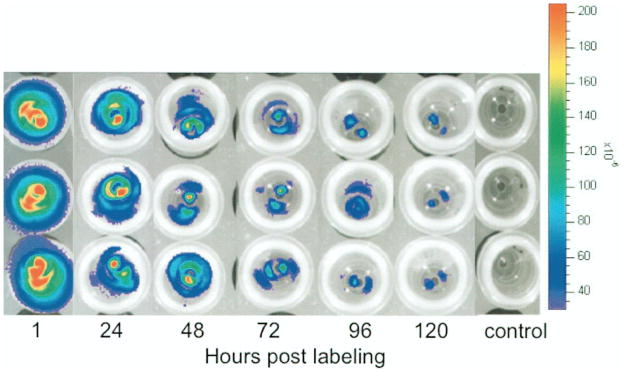
Labeling of hESCs with ICG at a concentration of 2.0 mg/ml after a 60-min incubation time revealed that the fluorescent signal slowly decreased over time, compatible with slow release of the contrast agent from the cells. The signal decreased by 50% within 48 h postlabeling and was equivalent to controls at 120 h postlabeling (Fig. 3). The fluorescence signal at 1, 24, and 48 h was significantly higher compared to precontrast data (p < 0.05). The signal at 120 h postlabeling was not significantly different from baseline (p > 0.05).
Figure 3.
Representative optical images of ICG-labeled hESCs in 1 ml of KSR using the optimized labeling protocol (2.0 mg ICG/ml, 60-min incubation) at different time points after labeling (1, 24, 48, 72, 96, and 120 h) showing decreasing fluorescent signal over time. The three rows represent triplicates.
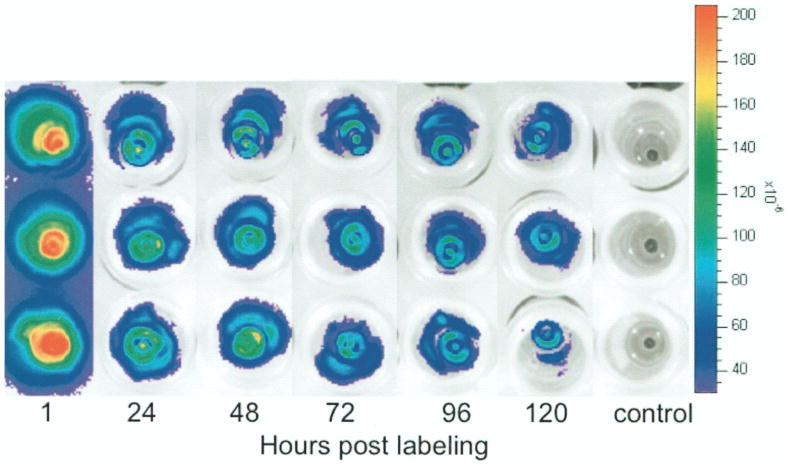
Twelve days after induction of hESC differentiation as described above (16), beating cardiomyocytes appeared. Longitudinal studies of CMs, labeled on day 25 with 2.0 mg ICG/ml for 60 min, revealed similar fluorescence signal kinetics compared to labeled hESCs (Fig. 4). The cells’ fluorescence signal was significantly elevated at 1, 24, and 48 h (p < 0.05), but showed no significant difference compared to baseline data at 120 h postlabeling (p > 0.05).
Figure 4.
Representative optical images of ICG-labeled hESC-CMs in 1 ml of CM media using the optimized labeling protocol (2.0 mg ICG/ml, 60-min incubation) at different time points after labeling (1, 24, 48, 72, 96, and 120 h) showing decreasing fluorescent signal over time. The three rows represent triplicates.
Fluorescence
The fluorescence signal of CMs was significantly higher compared to the fluorescence signal of hESCs at 1 h (p < 0.0001), 48 h (p < 0.05), 72 h (p < 0.01), and 96 h (p < 0.001) while there was no statistically significant difference in signal at 24 h postlabeling (Fig. 5).
Figure 5.
OI fluorescent signal (units in efficiency) of ICG-labeled human embryonic stem cells (hESC), ICG-labeled hESC-derived cardiomyocytes (CM), and nonlabeled controls at different time points after labeling (1, 24, 48, 72, 96, and 120 h). Data are displayed as means and SD of triplicate samples with 100,000 cells per sample.
Viability Testing
Trypan blue exclusion testing showed no significant difference in viability between labeled hESCs or labeled CMs compared to unlabeled controls. All groups exhibited viabilities greater than 88%, which were not significantly different between experimental groups (p > 0.05) (Fig. 2).
Immunocytochemistry
Analysis of hESC Pluripotency
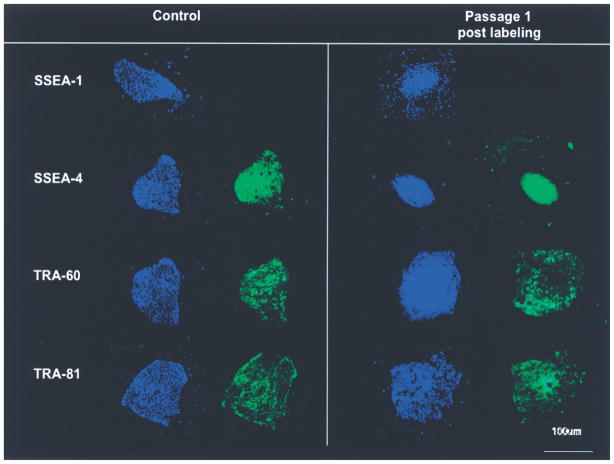
Fluorescent microscopy experiments following immunohistochemistry revealed that ICG-labeled and unlabeled HSF-6 cells expressed the surface antigens SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81 and did not express SSEA-1 (Fig. 6). These expression patterns are consistent with undifferentiated hESCs or “pluripotent” stem cells. Furthermore, there were no observed morphologic abnormalities of labeled hESCs (colonies had clearly defined borders and the cells within each colony were homogenously sized) compared to unlabeled controls.
Figure 6.
Fluorescent microscopy following immunohistochemistry revealed that the hESCs labeled under the optimized conditions expressed the hESC cell-specific surface antigens, including SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81, and did not express SSEA-1. These expression markers are consistent with undifferentiated hESCs. Scale bar: 100 μm.
Retention of Cardiomyocyte Protein Expression Post-labeling
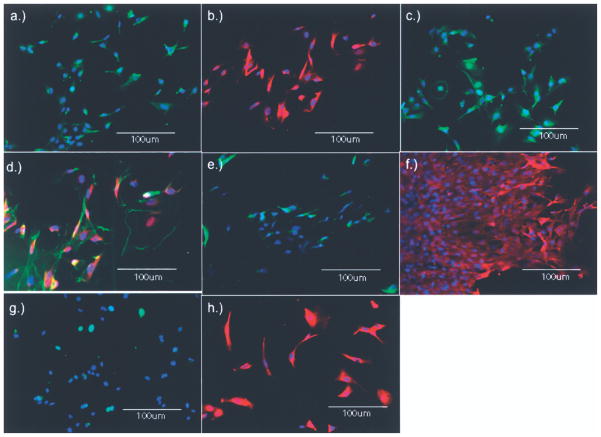
The presence of cardiac-specific proteins was studied using immunocytochemistry. Figure 7 shows positive staining for Connexin43, Lin28, Troponin T, Actin + Lin28, Wnt8a, TBX5, GATA4, and Islet1. The staining patterns recapitulate previous publications on the location of the targeted proteins (Fig. 7). There was no difference in antigen expression patterns between la- beled and unlabeled CMs.
Figure 7.
Cardiomyocyte protein expression maintained post-ICG labeling. Positive staining was observed for (a) Connexin43, (b) Wnt8a, (c) Troponin T, (d) GATA4, (e) Lin28, (f) TBX5, (g) Actin, (h) Islet1. Scale bar: 100 μm.
DISCUSSION
The results of this study indicate that hESCs and hESC-CMs can be labeled by simple incubation with the FDA-approved fluorescent marker ICG without impairing the cells viability, hESCs pluripotency, or cardiomyocyte function. The ICG-labeled hESCs and cardiomyocytes can be detected up to 48 h postlabeling with optical imaging.
Our study is novel with regard to two aspects. 1) To the best of our knowledge, this is the first study that utilized the FDA-approved fluorochrome ICG for the labeling of hESCs and hESC-derived CMs. Murine ESC-derived hepatocytes have been previously labeled with ICG and adult mesenchymal stem cells have been previously labeled with other organic fluorescent markers, such as PKH67, PKH26 (1), GFP (6), Gadophrin-2 and luc (27), but this is the first study that has utilized ICG for the purpose of tracking hESCs and hESC-derived CMs. 2) Previous studies used a stereomicroscope for the visualization of ICG-labeled cells in vitro (28), while the current study is the first that utilized ICG labeling for stem cell depiction with optical imaging.
While the ICG-labeling technique is in principle ready to be translated to the in vivo arena, there are specific obstacles that need to be addressed for successful in vivo applications. One example of this is the current poor survival rate of transplanted stem cells, which could be improved by optimizing stem cell delivery techniques, such as delivery mode (direct myocardial injection vs. coronary vessel delivery), catheter types, needle size, timing of cell delivery with regard to an ischemic event, and optimization of scaffolds that can “anchor” the cells to the area of interest.
The described data confirm previous studies about the feasibility of cell labeling with ICG (3, 12, 28). ICG labeling of murine ESC-derived hepatocytes was originally optimized by Yamada et al. and subsequent studies have followed the original labeling protocol, which included a 15-min incubation with ICG at a concentration of 1.0 mg/ml (3, 12, 28). Our optimized labeling protocol for human hESCs and hESC-CMs included longer incubation periods and higher ICG concentrations compared to Yamada et al.’s (28) protocol. These discrepancies are most likely due to the difference in goals of the two studies. Yamada et al. was optimizing a protocol to distinguish hepatocytes from the rest of the EBs based on the fact that hepatocytes have a higher affinity for the dye and internalize ICG more rapidly compared to the other cell types. Conversely, the present study aimed to load high quantities of ICG into hESCs and hESC-CMs in order to achieve the highest possible fluorescent signal without affecting the cells viability or function.
Our studies are in accordance with previous reports by other investigators who found that ICG does not alter the viability or function of the labeled cells. We found an unaltered protein expression of ICG-labeled hESCs and hESC-CMs. Other investigators reported an unaltered morphology, tissue integration, function, and immunoreactivity of ICG-labeled hepatocytes compared to unlabeled controls in vivo (28).
Our ICG-labeled hESC-CMs demonstrated a stronger fluorescence signal compared to ICG-labeled hESCs at all time points of observation. This phenomenon can be explained by the increased network of gap junctions in the CMs, which permit the transfer of molecules under 1,000 Da from cell to cell (18). Additionally, the increased signal seen in the CMs is due to their relatively larger diameter of 20–30 μm compared to undifferentiated hESCs with a diameter of about 10 μm (22) (Fig. 6).
We observed an excretion of ICG from the hESCs and hESC-CMs within 120 h. While this limits the potential for extensive long-term-follow-up studies, the rapid excretion of ICG suggests a safe elimination. Of note, the cellular excretion of ICG varies with cell type. Hepatocyte-like cells (HLCs) excreted ICG even faster, within 20 h postlabeling (12).
The mechanism by which ICG is taken up into cells and excreted has been studied extensively and has been shown to follow an active saturable carrier-mediated transport process, according to the Michaelis-Menten equation (8, 9). The release of ICG from cells is due to cytoplasmic and biliary excretion mechanisms. Furthermore, excretion relies on the membrane potential-dependent transport system established by an ATP-utilizing pump (24).
Major efforts have recently focused on improving cell engraftment following a myocardial infarct (MI) by assessing different modes of delivery (25). One possible route of cell delivery to myocardium in a clinical setting is cell administration through an intracoronary injection. Limited benefits from this approach have prompted investigators to assess different delivery methods, such as direct intramyocardial injections. A recent study investigated the distribution patterns of transplanted bone marrow stem cells transfected with firefly luciferase following three delivery methods: intra-aortic, intravenous, and intramyocardial. Within 48 h of delivery, the large majority of the implanted cells localized to extracardiac organs with all of these delivery methods. Interestingly, this study also revealed the importance of the timing of delivery with regard to the ischemic event. When cells were injected into an acute infarct there was a 50% survival rate at 2 h, but improved retention was observed when cells were injected into a more mature infarct (15). These results confirm the critical need of a cell tracking technique in the early phase after delivery of therapeutic cells, which would be possible with our ICG-labeling technique.
The ICG-labeling technique is advantageous over other techniques being developed because it is cost effective, technically straightforward, and rapid (<2 h). The most notable advantage of the ICG-labeling technique compared to others in the field, namely labeling with iron oxide-based nanoparticles for MR imaging, is the magnitudes increased sensitivity that one achieves with OI. While MRI has the advantage of high anatomical resolution, it provides limited sensitivity. Single cell sensitivity is especially important in the field of stem cell research due to the unpredictable migrational dynamics of the cells, even after local injections. While labeling cells for PET imaging does offer sensitive cell detection, PET techniques involve radiation exposure with unknown side effects on the stem cells. More specifically, causing damage to the DNA of the cells from repeated or even acute radiation exposure is extremely risky in a field where the behavioral dynamics of the cells are not currently well understood. Other advantages of the ICG-labeling technique are its compatibility with conventional fluorescent microscopy, which enables direct correlations of imaging findings with pathological specimens (10).
We recognize that there are several limitations to our study. Firstly, our data showed that the fluorescent marker is slowly released from the labeled cells. The cells could be detected with OI for at least 48 h; however, the fluorescent signal rapidly decayed after this time interval. Thus, potential applications of our proposed technique would be limited to this time interval. Our ICG-based cell tracking technique could contribute to short-term posttransplant investigations by assessing factors that may increase stem cell engraftment, such as different delivery modes, the timing of delivery, various scaffolds and growth factors, and different stem cell types. In addition, several investigators noted that the transplanted stem cells do not engraft into the desired area but migrate to other areas in the early time period after transplantation (7). The ICG-labeling method could be used to monitor such migrational dynamics in a fast, inexpensive, and relatively easy manner.
Our applied marker ICG is FDA approved for cardiac imaging and thus would be in theory readily accessible to patients under “off label use” conditions. Clinical scanners for breast imaging have just entered clinical practice. However, clinical OI scanners for evaluations of patients’ hearts have yet to be developed; therefore, potential future clinical applications of the ICG-labeling technique await further development of OI scanners for cardiac applications (13). Our studies were obtained in vitro and future studies are being conducted to investigate in vivo applications of the proposed cell tracking technique, such as tracking of hESC-CMs after implantation into ischemic myocardium of experimental animals.
CONCLUSIONS
In summary, the derived data establish an optimized technique for depiction of the labeled cells with optical imaging and illustrates the feasibility of hESC-based labeling with the FDA-approved fluorescent dye ICG. Other potential applications of the ICG-labeling technique include, but are not limited to, investigations of other stem cell lineages and progenitors, such as neural precursors, chondrocytes, osteoblasts, and neutrophils.
Acknowledgments
This study was supported by a research grant from the California Institute for Regenerative Medicine (CIRM), grant #RS1-00381-1. The Islet-1 monoclonal antibody 40.2D6 was developed by Thomas Jessel and was obtained from the Developmental Studies Hybridoma Bank developed with support of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA.
References
- 1.Askenasy N, Farkas DL. Optical imaging of PKH-labeled hematopoietic cells in recipient bone marrow in vivo. Stem Cells. 2002;20(6):501–513. doi: 10.1634/stemcells.20-6-501. [DOI] [PubMed] [Google Scholar]
- 2.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc Natl Acad Sci USA. 2007;104(30):12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baharvand H, Hashemi SM, Kazemi Ashtiani S, Farrokhi A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol. 2006;50(7):645–652. doi: 10.1387/ijdb.052072hb. [DOI] [PubMed] [Google Scholar]
- 4.Bettiol E, Clement S, Krause KH, Jaconi ME. Embryonic and adult stem cell-derived cardiomyocytes: Lessons from in vitro models. Rev Physiol Biochem Pharmacol. 2006;157:1–30. doi: 10.1007/112_0508. [DOI] [PubMed] [Google Scholar]
- 5.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16(5):455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Brazelton TR, Blau HM. Optimizing techniques for tracking transplanted stem cells in vivo. Stem Cells. 2005;23(9):1251–1265. doi: 10.1634/stemcells.2005-0149. [DOI] [PubMed] [Google Scholar]
- 7.Cao F, Wagner RA, Wilson KD, Xie X, Fu JD, Drukker M, Lee A, Li RA, Gambhir SS, Weissman IL, Robbins RC, Wu JC. Transcriptional and functional profiling of human embryonic stem cell-derived cardiomyocytes. PLoS ONE. 2008;3(10):e3474. doi: 10.1371/journal.pone.0003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmettre T, Devoisselle JM, Mordon S. Fluorescence properties and metabolic features of indocyanine green (ICG) as related to angiography. Surv Ophthalmol. 2000;45(1):15–27. doi: 10.1016/s0039-6257(00)00123-5. [DOI] [PubMed] [Google Scholar]
- 9.Fickweiler S, Szeimies RM, Baumler W, Steinbach P, Karrer S, Goetz AE, Abels C, Hofstadter F, Landthaler M. Indocyanine green: Intracellular uptake and phototherapeutic effects in vitro. J Photochem Photobiol B. 1997;38(2–3):178–183. doi: 10.1016/s1011-1344(96)07453-2. [DOI] [PubMed] [Google Scholar]
- 10.Frangioni JV, Hajjar RJ. In vivo tracking of stem cells for clinical trials in cardiovascular disease. Circulation. 2004;110(21):3378–3383. doi: 10.1161/01.CIR.0000149840.46523.FC. [DOI] [PubMed] [Google Scholar]
- 11.Furlani D, Li W, Pittermann E, Klopsch C, Wang L, Knopp A, Jungebluth P, Thedinga E, Havenstein C, Westien I, Ugurlucan M, Li RK, Ma N, Steinhoff G. A transformed cell population derived from cultured mesenchymal stem cells has no functional effect after transplantation into the injured heart. Cell Transplant. 2009;18(3):319–331. doi: 10.3727/096368909788534906. [DOI] [PubMed] [Google Scholar]
- 12.Hay DC, Zhao D, Ross A, Mandalam R, Lebkowski J, Cui W. Direct differentiation of human embryonic stem cells to hepatocyte-like cells exhibiting functional activities. Cloning Stem Cells. 2007;9(1):51–62. doi: 10.1089/clo.2006.0045. [DOI] [PubMed] [Google Scholar]
- 13.Intes X. Time-domain optical mammography SoftScan: Initial results. Acad Radiol. 2005;12(8):934–947. doi: 10.1016/j.acra.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 14.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25(9):1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 15.Li SH, Lai TY, Sun Z, Han M, Moriyama E, Wilson B, Fazel S, Weisel RD, Yau T, Wu JC, Li RK. Tracking cardiac engraftment and distribution of implanted bone marrow cells: Comparing intra-aortic, intravenous, and intramyocardial delivery. J Thorac Cardiovasc Surg. 2009;137(5):1225–1233. doi: 10.1016/j.jtcvs.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Maltsev VA, Wobus AM, Rohwedel J, Bader M, Hescheler J. Cardiomyocytes differentiated in vitro from embryonic stem cells developmentally express cardiac-specific genes and ionic currents. Circ Res. 1994;75(2):233–244. doi: 10.1161/01.res.75.2.233. [DOI] [PubMed] [Google Scholar]
- 17.Martens TP, Godier AF, Parks JJ, Wan LQ, Koeckert MS, Eng GM, Hudson BI, Sherman W, Vunjak-Novakovic G. Percutaneous cell delivery into the heart using hydrogels polymerizing in situ. Cell Transplant. 2009;18(3):297–304. doi: 10.3727/096368909788534915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson BJ. Gap junctions—from cell to molecule. J Cell Sci. 2003;116(Pt. 22):4479–4481. doi: 10.1242/jcs.00821. [DOI] [PubMed] [Google Scholar]
- 19.Nistor GI, Totoiu MO, Haque N, Carpenter MK, Keirstead HS. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia. 2005;49(3):385–396. doi: 10.1002/glia.20127. [DOI] [PubMed] [Google Scholar]
- 20.Patel AN, Sherman W. Cardiac stem cell therapy: Advances from 2008. Cell Transplant. 2009;18(3):243–244. doi: 10.3727/096368909788534960. [DOI] [PubMed] [Google Scholar]
- 21.Pellettieri J, Sanchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annu Rev Genet. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- 22.Satin J, Itzhaki I, Rapoport S, Schroder EA, Izu L, Arbel G, Beyar R, Balke CW, Schiller J, Gepstein L. Calcium handling in human embryonic stem cell-derived cardiomyocytes. Stem Cells. 2008;26(8):1961–1972. doi: 10.1634/stemcells.2007-0591. [DOI] [PubMed] [Google Scholar]
- 23.Saxena V, Sadoqi M, Shao J. Degradation kinetics of indocyanine green in aqueous solution. J Pharm Sci. 2003;92(10):2090–2097. doi: 10.1002/jps.10470. [DOI] [PubMed] [Google Scholar]
- 24.Shinohara H, Tanaka A, Kitai T, Yanabu N, Inomoto T, Satoh S, Hatano E, Yamaoka Y, Hirao K. Direct measurement of hepatic indocyanine green clearance with near-infrared spectroscopy: Separate evaluation of uptake and removal. Hepatology. 1996;23(1):137–144. doi: 10.1053/jhep.1996.v23.pm0008550033. [DOI] [PubMed] [Google Scholar]
- 25.Silva SA, Sousa AL, Haddad AF, Azevedo JC, Soares VE, Peixoto CM, Soares AJ, Issa AF, Felipe LR, Branco RV, Addad JA, Moreira RC, Tuche FA, Mesquita CT, Drumond CC, Junior AO, Rochitte CE, Luz JH, Rabischoffisky A, Nogueira FB, Vieira RB, Junior HS, Borojevic R, Dohmann HF. Autologous bone-marrow mononuclear cell transplantation after acute myocardial infarction: Comparison of two delivery techniques. Cell Transplant. 2009;18(3):343–352. doi: 10.3727/096368909788534951. [DOI] [PubMed] [Google Scholar]
- 26.Tompers DM, Labosky PA. Electroporation of murine embryonic stem cells: A step-by-step guide. Stem Cells. 2004;22(3):243–249. doi: 10.1634/stemcells.22-3-243. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Rosol M, Ge S, Peterson D, McNamara G, Pollack H, Kohn DB, Nelson MD, Crooks GM. Dynamic tracking of human hematopoietic stem cell engraftment using in vivo bioluminescence imaging. Blood. 2003;102(10):3478–3482. doi: 10.1182/blood-2003-05-1432. [DOI] [PubMed] [Google Scholar]
- 28.Yamada T, Yoshikawa M, Kanda S, Kato Y, Nakajima Y, Ishizaka S, Tsunoda Y. In vitro differentiation of embryonic stem cells into hepatocyte-like cells identified by cellular uptake of indocyanine green. Stem Cells. 2002;20(2):146–154. doi: 10.1634/stemcells.20-2-146. [DOI] [PubMed] [Google Scholar]