Abstract
Histone deacetylase inhibitors (HDACi) induce growth arrest and apoptosis in colon cancer cells, and have anti-tumor efficacy in vivo. The precise mechanism by which HDACi induce apoptosis in tumor cells is unknown, with both transcription-dependent and independent mechanisms implicated. To determine whether HDACi-induced apoptosis is linked to a defined transcriptional response a panel of 30 colon cancer cell lines was screened for sensitivity to HDACi-induced apoptosis and sensitive and resistant lines identified. Differences in gene expression induced by HDACi in the 5 most sensitive and resistant lines were determined by microarray profiling. A robust transcriptional response involving coordinate induction of multiple immediate-early (fos, jun, egr1, egr3, atf3, arc, nr4a1) and stress response genes (Ndrg4, Mt1B, Mt1E, Mt1F, Mt1H), were selectively induced in HDACi sensitive cells. Bioinformatic analysis of the promoters of these genes demonstrated a higher GC content and frequency of KLF4/Sp1/Sp3 binding sites compared to a control set. While over and underexpression of KLF4 failed to modulate the HDACi-induced transcriptional or apoptotic response, Sp1/Sp3 reporter activity was preferentially induced by HDACi in sensitive cell lines. Furthermore, pharmacological and molecular inhibition of Sp1/Sp3 attenuated both the HDACi-induced transcriptional response and apoptosis. Remarkably, a significant percentage of the HDACi induced genes were basally repressed in colon tumors. HDACi induced apoptosis is therefore linked to initiation of a defined Sp1/Sp3-mediated transcriptional response, which involves de-repression of a transcriptional network basally repressed in colon cancer.
Keywords: HDACi, microarray, colon cancer cell lines
Introduction
Histone deacetylase inhibitors (HDACi) are a class of targeted therapeutics with promising anti-tumor efficacy in multiple malignancies. HDACi’s inhibit the HDAC family of transcriptional co-repressors, which catalyze the deacetylation of lysine residues within target proteins (1). HDACi treatment likely regulates gene expression by at least two mechanisms. First, inhibition of HDAC activity results in hyperacetylation of DNA bound histones, generating a chromatin environment more permissive to transcriptional activation (2, 3). Second, HDACi induce hyper-acetylation of sequence-specific transcription factors (4–8), which can either increase or decrease transcriptional activity (8, 9).
Multiple structurally distinct HDACi have been described, including short-chain fatty acids (butyrate, valproic acid), hydroxamic acids (trichostatin A, SAHA), cyclic tetrapeptides, tetrapeptides and benzamidines (2). Of these, SAHA is approved for the treatment of cutaneous T-cell lymphoma, while several others are in phase I and II clinical trials. These HDACi primarily inhibit the class I (HDACs 1, 2, 3 and 8) and class II HDACs (HDACs 4, 5, 6, 7, 9 and 10) (2).
HDACi induce growth arrest, differentiation and apoptosis in a variety of tumor cell lines, including colon cancer cells (2). Depending on the cell type, HDACi-induced apoptosis can be mediated by both the intrinsic/mitochondrial (10) or extrinsic/death receptor pathways (11, 12). In colon cancer cells, most studies implicate the intrinsic pathway, with a sequential series of events including loss of mitochondrial membrane potential, mitochondrial Bax localization, cytochrome c release and caspase-9 activation observed upon HDACi treatment (12–14). However, HDACi have also been shown to sensitize colon cancer cells to TRAIL and Fas-ligand induced apoptosis (15, 16), suggesting activation of the two pathways may occur in parallel.
While the downstream effectors of HDACi-induced apoptosis have been studied in detail (17, 18), the molecular events which initiate the process are less well defined. One postulated transcription-dependent mechanism is alteration in the balance in expression of pro and anti-apoptotic genes in favor of apoptosis (2). For example, in colon cancer cells HDACi upregulate expression of the pro-apoptotic genes, Bax (19) and Bak (20), and downregulate expression of the anti-apoptotic Bcl-XL gene (13).
Alternatively, HDACi can initiate apoptosis via transcription independent mechanisms (21). For example, HDACi can promote apoptosis via hyperacetylation of HSP90 and subsequent depletion of pro-survival HSP90 client proteins (22, 23). HDACi-induced apoptosis has also been linked to aberrant mitosis (22, 24, 25) (26, 27).
Therefore, whether HDACi induced apoptosis is initiated primarily in a transcription-dependent or independent manner, is currently unclear. In the present study we identify a robust and reproducible transcriptional response involving selective induction of 48 genes and repression of 44 genes in HDACi-sensitive cell lines was identified. The induced genes were highly enriched for immediate early (IE) and stress response (SR) genes. We demonstrate that this transcriptional response and subsequent apoptosis are coordinately induced by HDACi in a Sp1/Sp3-dependent manner. Notably, a significant percentage of the HDACi-induced genes are basally repressed in colon tumors, suggesting HDACi induced apoptosis involves deregulation of a complex transcriptional network involved in the survival of colon cancer cells.
Materials and Methods
Cell lines and cell culture
The source and the methods of maintenance of the 30 colon cancer cell lines utilized have been previously described (28). RKO-EcR and RKO-EcR-KLF4 (also referred to as RKO-RG24) were kindly provided to us by Dr. Vincent Yang and used as previously described (29).
Drug sensitivity assays
Methods used to determine apoptosis, clonogenic survival and response of xenografts to HDACi treatment in vivo are described in the supplementary text.
Microarray experiments, bioinformatics analysis
Microarray and data analysis methods used are described in supplementary text.
qRT-PCR, western blotting, transient transfection, siRNA knockdown, ChIP analysis
Detailed methods are described in supplementary text.
Immunohistochemistry
We utilized the Protein Atlas online immunohistochemistry resource (www.proteinatlas.org) for images of metallotheonin and ATF3 staining in normal colon and colon tumor sections (30, 31).
Results
Identification of colon cancer cell lines sensitive and refractory to HDACi-induced apoptosis
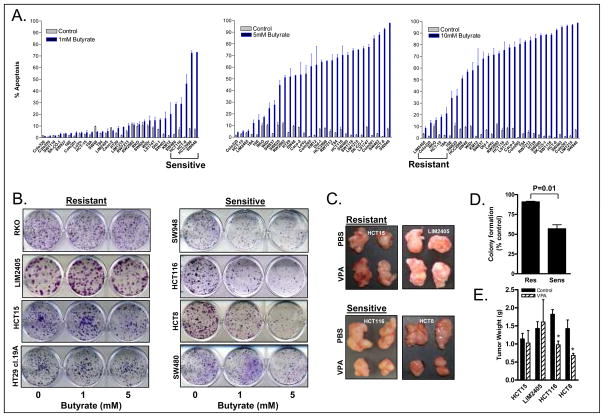
To identify colon cancer cell lines sensitive and refractory to HDACi, a panel of 30 colon cancer cell lines was screened for HDACi induced apoptosis by treatment with low (1 mM), intermediate (5 mM) or high dose (10 mM) sodium butyrate for 72 hours. As shown in Figure 1, a continuum of response to butyrate-induced apoptosis was observed. The 5 cell lines with the highest apoptotic response to low dose butyrate treatment were defined as butyrate “sensitive” (SW480, HCT116, HCT8, HCC2998 and SW948), and the 5 cell lines with the lowest apoptotic response to high dose butyrate treatment defined as butyrate “resistant” (LIM2405, Colo320, RKO, HCT15 and HT29 cl.19A), for subsequent studies.
Figure 1.
(A) Relative response of 30 colon carcinoma cell lines to HDACi-induced apoptosis. Cells lines were treated with 1, 5 or 10 mM butyrate for 72 hours (Mean±SEM, n=3). (B). Clonogenic survival of sensitive and resistant lines following 24h HDACi treatment (n=3–4). (C). Response of 2 sensitive and 2 resistant cell lines to VPA treatment in vivo. (D) Quantitation of colony formation shown in panel (C). Values shown are mean±SEM percentage colony formation following 5 mM butyrate treatment relative to control for the sensitive and resistant cell lines. (E). Quantitation of tumor size following HDACi treatment in vivo (n=4–5, *P<0.05).
To validate these findings using an independent method, clonogenic survival assays were performed following 24h butyrate treatment on the adherent sensitive and resistant cell lines. Consistent with the apoptosis data, colony formation was reduced to a significantly greater extent in sensitive compared with resistant lines (Figure 1B&C, P=0.01, unpaired t test). To confirm the differential response of colon cancer cells to HDACi in vivo, two sensitive (HCT116 and HCT8) and two resistant (LIM2405, HCT15) cell lines, were grown as xenografts in SCID mice and treated with the HDACi, VPA, for 2 weeks. VPA was used in these studies as it is structurally similar to butyrate but not metabolised as rapidly in vivo. Consistent with the in vitro findings, VPA treatment significantly inhibited growth of HCT116 and HCT8 xenografts, but had minimal effects on growth of LIM2405 and HCT15 cells (Figure 1D & E).
To confirm that the apoptotic effects of butyrate were due to the inhibition of HDAC activity, the cell lines were re-screened for apoptotic response to VPA, and to the structurally distinct hydroxamic acid-based HDAC inhibitors, Trichostatin A (TSA), and SAHA. As shown in Supplementary Figure 1, cell lines sensitive and resistant to butyrate-induced apoptosis were likewise differentially sensitive to each of the other HDACi. Notably, the differential sensitivity of these cell lines to HDACi was specific to this class of agent and was not due to an inherent differential sensitivity of these cell lines to apoptosis, as a number of cell lines refractory to HDACi-induced apoptosis were sensitive to apoptosis induced by the mechanistically distinct chemotherapeutic agents, 5FU, CPT and oxaliplatin (Supplementary Figure 1B).
Differential sensitivity of cell lines to HDACi-induced apoptosis is not linked to the degree of HDAC inhibition or HDACi-induced growth arrest
Interestingly, HDACi treatment resulted in robust induction of histone H3 and histone H4 acetylation to a similar extent in both sensitive and resistant cell lines (P=0.44 and P=0.37 for Ac.H3 and Ac.H4 respectively, Supplementary Figure 2), indicating differential sensitivity to HDACi was not due to differences in the degree of HDAC inhibition.
Consistent with this finding, HDACi treatment resulted in significant growth inhibition in all 10 cell lines (Supplementary Figure 3). The percentage of cells in S phase was decreased by 73±8.2% and 62±1.8% in resistant and sensitive cell lines, respectively (P=0.59), and the percentage of cells in G2/M was induced 359±95 and 388±112% in resistant and sensitive cell lines respectively (P=0.85) 24h post HDACi treatment. Consistent with HDACi-induced growth inhibition, expression of p21WAF1/CIP1, a regulator of cell cycle progression and known HDACi target gene, was consistently induced by butyrate treatment in each of the 10 cell lines (Supplementary Figure 4A&B).
HDACi-induced apoptosis is inhibited by actinomycin D
To determine the importance of transcription in HDACi-induced apoptosis, HCT116 cells were co-treated with HDACi and the RNA polymerase inhibitor, actinomycin D. Actinomycin D significantly attenuated HDACi-induced apoptosis demonstrating HDACi-induced apoptosis in colon cancer cells is dependent upon de novo transcription (Supplementary Figure 5A&B).
Immediate early (IE) and stress response (SR) genes are preferentially induced by HDACi in sensitive colon cancer cell lines
Having identified sensitive and resistant cell lines and established the dependency of HDACi-induced apoptosis upon de novo transcription, the transcriptional basis for this differential sensitivity was determined. The 5 sensitive and 5 resistant cell lines were treated with 5 mM butyrate for 24 hours and changes in gene expression profiled utilizing 27,000-feature cDNA microarrays (Entire database provided as Supplementary Table 5). Notably, the overall number of genes changed in response to HDACi treatment (p>0.05, t test) and the range of transcriptional changes in terms of fold-change was similar for sensitive and resistant cell lines.
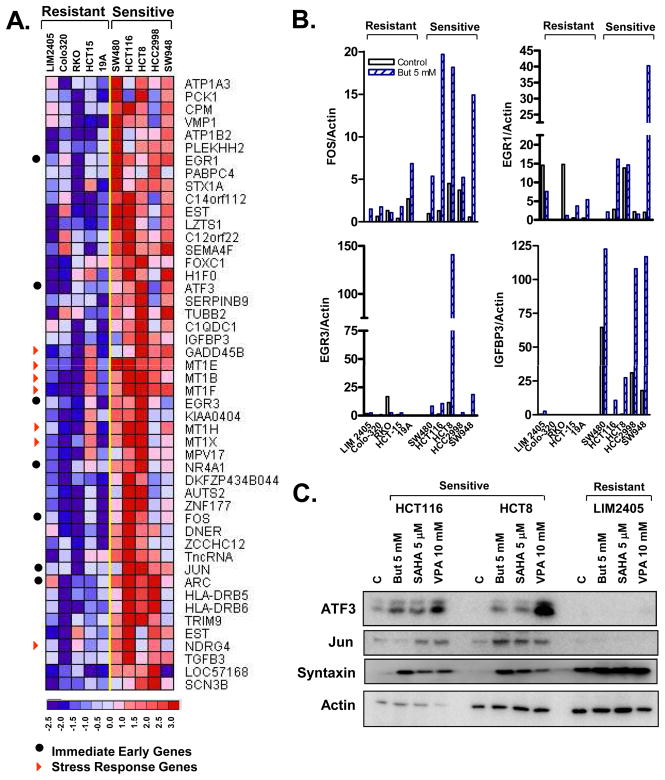
Genes differentially induced by HDACi in sensitive versus resistant cell lines were identified using a stringent supervised analysis as described in the methods. Satisfying these criteria, 48 sequences were identified as significantly and preferentially induced by butyrate in sensitive cell lines (Figure 2, Supplementary Table 4). Notably, 7 of these 48 genes, Fos, Jun, Atf3, Arc, Nr4a1 (Nur77), Egr1 and Egr3 are “immediate early” (IE) genes while an additional 7 genes have previously been classified as stress response (SR) genes: Gadd45b, Ndrg4, Mt1B, Mt1E, Mt1F, Mt1H and MtIX (Figure 2A).
Figure 2.
(A). Heat map of the 48 genes selectively induced by butyrate in sensitive cell lines following 24h treatment. (B) QPCR validation of select gene expression changes following 24 h treatment with 5 mM butyrate (But 5). (C) Western blot validation of differential induction of select proteins in sensitive (HCT8 and HCT116) and resistant cell lines (LIM2405).
A similar number of genes (44) preferentially repressed by HDACi in sensitive lines were also identified (Supplementary Table 4). These included several genes involved in organization of microtubules and the actin cytoskeleton (TRIP6, SRHML, PLXNB1, MAP7, LASP1 and LAD1), cell adhesion (OCLN, DSC2), transcriptional repression (NCoR2, SET) and apoptosis (FLIP, DAXX).
Validation of transcriptional changes
To validate the differential induction of HDACi induced genes in sensitive and resistant lines we performed qRT-PCR and western blot analyses. As shown in Figure 2B, Fos, EGR1, EGR3 and IGFBP3 mRNA induction by butyrate demonstrated strong concordance with the microarray data, with robust and preferential induction in the sensitive cell lines. Preferential induction in sensitive cell lines of 3 additional genes, ATF3, Jun and syntaxin was confirmed at the protein level (Figure 2C). To confirm that the transcriptional changes induced by butyrate were largely due to HDAC inhibition, gene expression changes induced in response to butyrate were compared to changes induced by VPA and SAHA in HCT116 cells by microarray analysis. Examination of the gene expression changes across the entire microarray revealed a marked similarity in the transcriptional changes induced by each of these agents (Supplementary Figure 6A&B), while specific examination of the genes comprising the apoptosis signature revealed an even stronger correlation (r=0.83 or greater), indicating the butyrate-induced changes are highly likely to be the consequence of HDAC inhibition.
Induction of IE genes is specific to HDACi-induced apoptosis
To confirm that the transcriptional response observed was specific to HDACi-induced apoptosis, and not a non-specific reflection of cells undergoing apoptosis, HCT116 cells were treated with HDACi or the mechanistically distinct chemotherapeutic agents 5FU and oxaliplatin for 24h. As shown in Supplementary Figure 7, expression of Fos, EGR1, EGR3 and IGFBP3 mRNA, and ATF3, Jun and syntaxin protein expression were induced primarily in response to HDACi treatment. In contrast, p21 which is induced both by HDACi and in a p53-dependent manner in response to DNA damage was induced by all agents tested (Supplementary Figure 7).
Promoters of genes selectively induced by HDACi in sensitive cell lines are GC rich and are enriched for KLF4/Sp1/Sp3 and binding sites
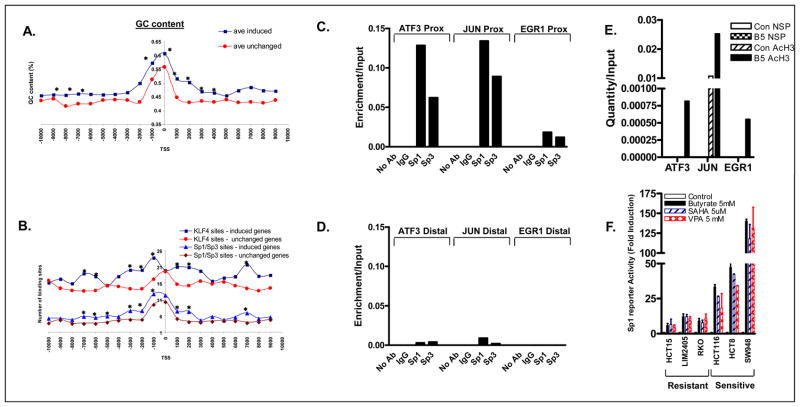
IE and SR gene expression has previously been shown to be coordinately induced in response to serum, TPA and growth factor treatment, suggesting a common transcription factor or set of transcription factors likely regulate their expression. To determine the identity of such a factor(s) a systematic bioinformatic search of the promoter regions of each of these genes was performed. As a control, the promoters of an equal number of genes not induced by HDACi were also analyzed. First, a nucleotide composition analysis of the region 10 kb upstream through 10 kb downstream of the transcription start site (TSS), measured in 1 kb windows, demonstrated that the GC content of HDACi-induced gene promoters was significantly higher than that of the control set. As shown in Figure 3A, promoter GC content increased significantly in both groups with increasing proximity to the TSS, however the magnitude of this increase was significantly higher in HDACi-induced genes. To determine whether the promoters of HDACi induced genes were enriched for specific transcription factor binding sites, the frequency of occurrence of all established transcription factor binding sites in the 1.5 kb promoter region upstream and 200 bp downstream of the TSS were computed using the MATCH database. As shown in Supplementary Table 5, the frequency of occurrence of multiple transcription factor binding sites was significantly different between HDACi-induced versus uninduced genes, with the vast majority of sites underrepresented in the promoters of genes induced by HDACi. The transcription factor binding sites most significantly enriched in HDACi induced genes were the related GC-rich KLF4, Sp1 and Sp3 binding sites. To further confirm this finding and to examine the distribution of KLF4/Sp1/Sp3 binding sites within the promoters of these genes in more detail, we computed the frequency of occurrence of these sites in 1 kb increments beginning 10 kb upstream through 10 kb downstream of the TSS. As shown in Figure 3B, the frequency of KLF4 and Sp1/Sp3 sites was significantly higher in HDACi-induced genes.
Figure 3.
(A) Mean GC content of 48 genes preferentially induced by HDACi in sensitive cell lines and an equal number of control genes. (B). Frequency of KLF4/Sp1/Sp3 binding sites in the promoters of HDACi-induced genes. *indicates P<0.05. (C-D) Quantitative ChIP analysis in untreated HCT116 cells demonstrating Sp1 and Sp3 localization to proximal promoters of HDACi-induced genes (C) but not distal promoter regions (D). (E) ChIP analysis demonstrating induction of histone H3 hyperacetylation in the proximal promoter regions of 3 genes following 24 hour treatment of HCT116 cells with butyrate 5 mM (B5). (F) Preferential induction of Sp1/Sp3 reporter activity in HDACi-sensitive cell lines treated with butyrate, SAHA or VPA for 24h (n=2).
KLF4 is not a key regulator of HDACi-induction of the IE/SR gene signature and apoptosis
To address the role of KLF4 in mediating the HDACi-induced transcriptional and apoptotic response we first determined the effect of KLF4 overexpression on HDACi induced apoptosis and IE gene expression in the HDACi-resistant RKO cell line. RKO cells have low levels of KLF4 due to hemizygous deletion of a portion of the KLF4 locus as well as promoter methylation (32). RKO-EcR-KLF4 cells are an isogenic derivative of RKO cells containing a stably integrated ponasterone-inducible KLF4 gene (29). Ponasterone treatment induced robust induction of KLF4 expression in RKO-EcR-KLF4 cells after 24h, however no change in the magnitude of HDACi-induced apoptosis or HDACi induction of IE gene expression was observed (Supplementary Figure 8). Second, KLF4 mRNA was selectively downregulated in HDACi-sensitive HCT116 cells. Despite greater than 70% knockdown of KLF4 mRNA expression, no change in HDACi-induced apoptosis was observed (Supplementary Figure 8B), collectively indicating KLF4 does not play a central role in HDACi induction of IE/SR gene expression or in HDACi-induced apoptosis in colon cancer.
Activity of Sp1 and Sp3 is functionally implicated in HDACi-induction of the IE/SR gene signature and apoptosis
Next, we determined the role played by Sp1 and Sp3 transcription factors in HDACi-induced gene expression and apoptosis utilizing multiple independent approaches. First, to determine whether Sp1 and Sp3 were physically localized to the promoters of HDACi-responsive genes, we performed ChIP analyses in HCT116 cells interrogating the Sp1/Sp3 sites within the proximal promoter regions of the ATF3, EGR1 and JUN promoters. Pulldowns performed using anti-Sp1 or anti-Sp3 antibodies resulted in significant enrichment of the proximal promoter fragments of these genes, relative to IP’s performed using No Ab or IgG controls (Figure 3C). In comparison, for each of these genes no enrichment of distal promoter regions, devoid of Sp1/Sp3 sites was observed (Figure 3D). To determine the functionality of these sites in terms of facilitating transcription in response to HDACi treatment, we also examined AcH3 levels at these sites pre and post HDACi treatment by ChIP analysis. As shown in Figure 3E, AcH3 levels were increased approximately 2-fold 24h post HDACi treatment consistent with the transcriptional induction of these genes. Notably, enrichment of Sp1 or Sp3, or of the class I HDAC, HDAC3, at the Jun promoter was not significantly altered in response to HDACi treatment (data not shown), indicating these factors are retained at the promoter during transcription of the gene.
Second, HDACi sensitive and resistant colon cancer cell lines were transfected with WT or mutant Sp1/Sp3-luciferase reporter constructs, and treated with HDACi for 24h. As shown in Figure 3F, the magnitude of induction of Sp1/Sp3 reporter activity was 2–10-fold higher in HDACi-sensitive compared to resistant lines indicating preferential induction of Sp1/Sp3 driven transcription in sensitive cells. Minimal effects were observed on the mutant Sp1/Sp3-luciferase reporter construct in both sensitive and resistant lines (data not shown).
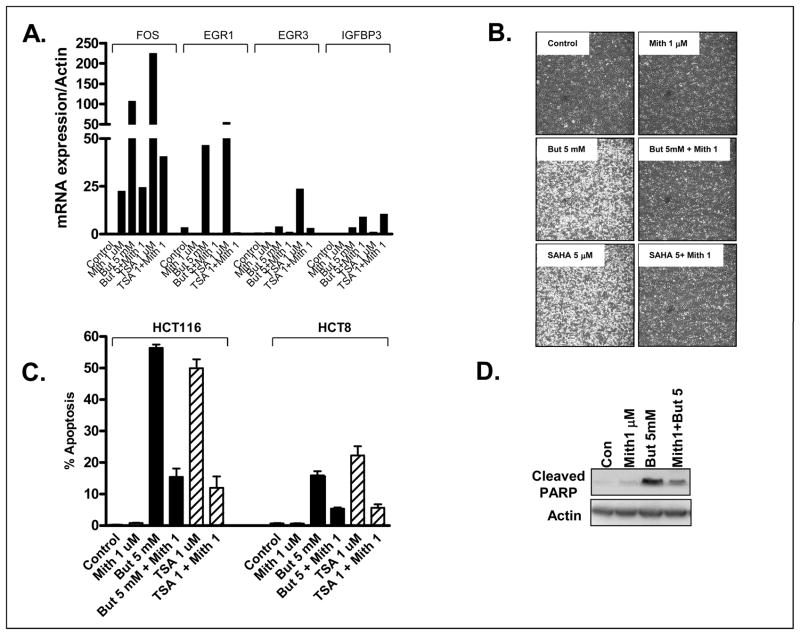
Third, whether pharmacological inhibition of Sp1/Sp3 binding abrogated HDACi-induction of IE genes and apoptosis was examined using the anitibiotic mithramycin, which inhibits Sp1/Sp3 transcription by competitively binding to GC-rich elements (33). Co-treatment of HCT116 cells with mithramycin markedly attenuated butyrate and TSA-mediated induction of 3 of the 4 genes tested: FOS, EGR1 and, EGR3 (Figure 4A). In contrast, mithramycin treatment failed to inhibit HDACi-induction of IGFBP3, instead potentiating the HDACi effect (Figure 4A). Likewise, mithramycin markedly attenuated HDACi-induced apoptosis in both HCT116 and HCT8 cells at the 24 hour time point as assessed by cell monolayer morphology, the percentage of cells with a sub-diploid DNA content or PARP cleavage (Figure 4B-D). However, this effect was lost at later time points (48 and 72h) when mithramycin alone induced significant apoptosis (data not shown).
Figure 4.
The Sp1/Sp3 inhibitor, mithramycin (Mith, M, 1 μM), attenuates HDACi-induction of (A) target gene expression (HCT116), (B) cell viability (HCT116), (C) sub-diploid DNA content (HCT116 and HCT8) or (D) and PARP cleavage (HCT116). Cells were treated with HDACi (Butyrate 5 mM, TSA 1 μM) for 24 hours.
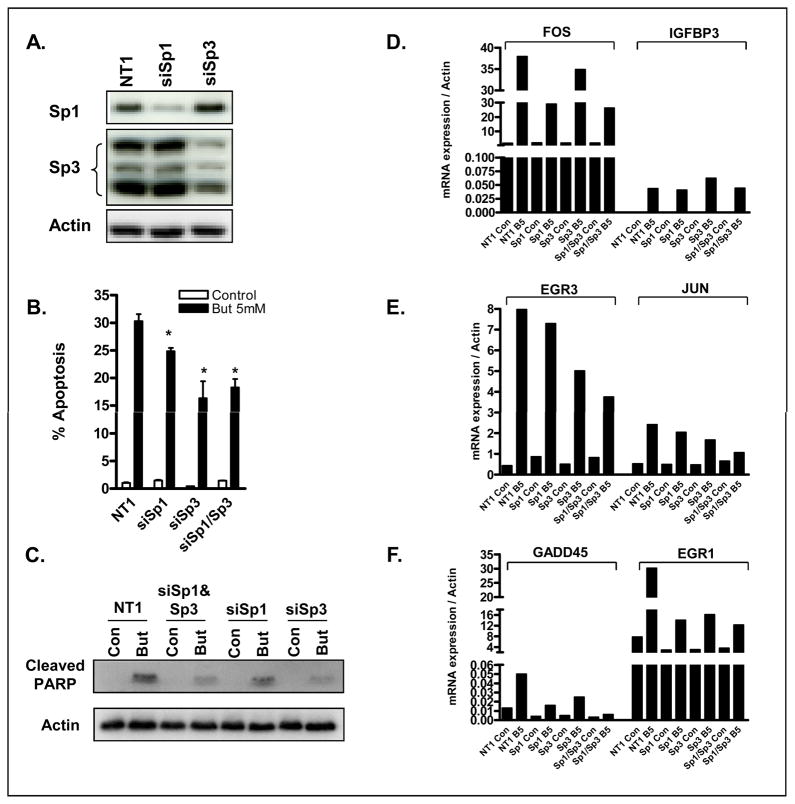
Finally, to directly determine the role of Sp1 and Sp3 in the HDACi-induced transcriptional and apoptotic response, Sp1 and Sp3 were downregulated in HCT116 cells using Sp1 and Sp3 targeting siRNAs. Effective downregulation of Sp1 and all three isoforms of Sp3 were confirmed by western blot (Figure 5A). As shown in Figure 5B, downregulation of Sp1 and Sp3 resulted in a 17% and 46% inhibition of HDACi-induced apoptosis, respectively, and in the extent of HDACi-induced PARP cleavage (Figure 5C).
Figure 5.
(A) Silencing efficiency of Sp1 and Sp3 protein expression. (B-C) Silencing of Sp1 and Sp3 in HCT116 cell partially attenuates HDACi-induced apoptosis at 24h as assessed by (B) sub-diploid DNA content and (C) PARP cleavage, and (D-F) HDACi-induced transcriptional response.
Downregulation of Sp1 and Sp3, alone, and in combination, resulted in a heterogeneous pattern of effects on HDACi-induced gene expression, which fell into 3 categories. First, for a subset of the genes (FOS, IGFBP3), Sp1 and Sp3 silencing had no effect on HDACi-induced gene expression (Figure 5D). For a second subset (EGR3, JUN), silencing of Sp3 but not Sp1 partially attenuated HDACi-mediated transcriptional induction (Figure 5E). For the final subset (GADD45, EGR1), individual silencing of both Sp1 and Sp3 inhibited HDACi-mediated transcriptional induction (Figure 5F). Collectively, these results indicate that Sp1 and Sp3 regulate a significant, although not the entire component of the HDACi-induced transcriptional response associated with promotion of apoptosis in colon cancer cells.
Genes selectively induced by HDACi in sensitive cell lines are basally repressed in colon tumors
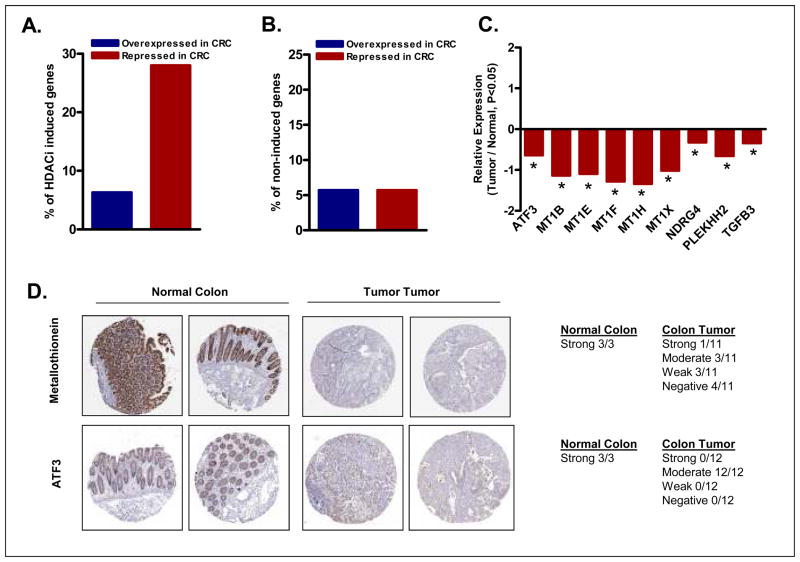
To independently assess the biological significance of the subset of genes preferentially induced by HDACi in sensitive cell lines, we compared the relative expression of this gene subset in colon tumors and normal colonic mucosa. For this, we generated a microarray database comparing gene expression differences between 12 matched normal colon and colon tumor pairs. Of the 48 genes preferentially induced by HDACi in sensitive cells, corresponding gene expression data was available for 32 genes in the matched primary colon tumor/normal database. Notably, expression of 9 of these 32 genes (28%) was significantly repressed (>1.5-fold and P<0.05) in colon tumors compared to normal colonic mucosa. In contrast, only 2/32 (6.3%) were upregulated in colon tumors by these same criteria (Figure 6A). As a control, the expression pattern in colon tumor and normal tissue of an equal number of genes not induced by HDACi treatment was also examined. As shown in Figure 6B, 5.7% of the genes on this control list were upregulated and 5.7% downregulated (>1.5-fold and P<0.05) in colon tumors relative to normal colonic mucosa (P=0.004, Chi Square test).
Figure 6.
Expression of multiple HDACi-induced genes is reduced in primary colon tumors relative to adjacent normal mucosa. (A) The percentage of HDACi-induced and (B) un-induced (control) sequences significantly over or underexpressed in colon tumors. (C) Identity of the 9 genes induced by HDACi and significantly repressed in colon tumors relative to normal colonic epithelium (*P<0.05). (D) Immunohistochemical validation of reduced expression of ATF3 and metallothionein in normal colon and colon tumors. Images downloaded from the publicly available protein atlas database (www.proteinatlas.org) as per the image use policy of protein atlas.
The HDACi induced genes downregulated in colon tumors were, ATF3, MT1B, MT1E, MT1F, MT1H, MT1X, NDRG4, PLEKHH2 and TGFB3 (Figure 6C). As shown in Figure 6D, the downregulation of MT1F and ATF3 in colon tumors was independently validated using the protein atlas IHC database (31, 34). These data demonstrate that the subset of genes identified as selectively induced during HDACi-induced apoptosis were enriched for genes that are basally downregulated in colon tumors. These findings are also consistent with a previous report indicating downregulation of a number of immediate-early genes in colon tumors (35).
Discussion
HDAC-inhibitors induce growth arrest, differentiation and apoptosis in tumor cells of diverse origins, and their efficacy for cancer treatment was recently underscored by the approval of SAHA for the treatment of cutaneous T-Cell lymphoma. While the downstream effectors of HDACi induced apoptosis in tumor cells have been studied in detail (3, 17), the molecular events which initiate HDACi induced apoptosis are unknown. Possibilities include transcription-dependent effects such as altered expression of pro and anti-apoptotic molecules, or transcription-independent effects such as induction of aberrant mitosis, ROS production, or altered acetylation of pro-survival molecules such as HSP90.
In the present study, the importance of de novo transcription in HDACi-induced apoptosis in colon cancer cell lines was established. Through gene expression profiling of multiple colon cancer cell lines sensitive and resistant to HDACi, we identified a reproducible transcriptional response preferentially induced in response to HDACi treatment in sensitive cell lines. Importantly, this transcriptional response was observed in response to multiple, structurally distinct, HDACi, and across a range of colon cancer cell lines with extensive molecular and mutational heterogeneity including differences in microsatellite instability status, ploidy, and the mutation and methylation status of multiple tumor suppressor genes and oncogenes (28).
A striking feature of the HDACi-induced transcriptional response is that a number of the induced genes, particularly the IE and SR genes, are coordinately induced in response to several other stimuli. This includes the coordinate induction of IE gene expression in response to serum stimulation, phorbol ester and growth factor treatment (36, 37), and the coordinate induction of the metallothionein gene family, clustered within the q13 region of chromosome 16, in response to metal induction and oxidative stress (38). The coordinate induction of these genes in response to HDACi treatment suggested they likely reflected the modulation of a specific transcription factor or factors which co-ordinately regulate their expression.
Several findings collectively indicated a key role for Sp1/Sp3 transcription factors in driving this transcriptional response. First, the promoters of HDACi induced genes had a high GC content and were significantly enriched for Sp1/Sp3 binding sites. Second, localization of Sp1 and Sp3 at the promoters of several of these genes was detectable by ChIP analysis. Third, induction of Sp1/Sp3 reporter activity was preferentially induced by HDACi in sensitive cell lines. Finally, disruption of Sp1/Sp3 DNA binding with mithramycin or siRNA-mediated downregulation of Sp1 or Sp3 attenuated induction of a number of HDACi targets. Importantly, both mithramycin and Sp1/Sp3 siRNA attenuated HDACi induced apoptosis, establishing a direct link between Sp1/Sp3 mediated transcriptional response and initiation of HDACi-induced apoptosis. It is important to note however, that expression of a subset of the genes induced by HDACi, including FOS and IGFBP3 were not modulated by Sp1 and or Sp3 silencing, indicating additional transcription factors also likely participate in HDACi-induced transcriptional changes.
While the precise mechanism by which initiation of this transcriptional response triggers apoptosis remains to be determined, it is noteworthy that several genes comprising the HDACi transcriptional response have previously been linked with HDACi-induced apoptosis through over and underexpression studies. These include IGFBP3 (39), EGR1 (40), NR4A1 (TR3/Nur77) (14) and GADD45B (41). However, in each case, induction of these genes only partially accounts for the overall apoptotic response, suggesting that their collective induction, which may sufficiently alter the cellular transcriptional equilibrium in favor of apoptosis, may be the critical determinant of HDACi-induced apoptosis. Lending support to this concept is the finding that a number of these transcripts are basally repressed in colon tumors compared to normal colon tissue. In the context of tumorigenesis therefore, selective repression of these transcripts may alter the transcriptional balance in favor of cell survival.
While HDACi treatment uniformly induces expression of anti-proliferative genes such as p21 in both sensitive and resistant cell lines, it is notable that several of the genes preferentially induced by HDACi in sensitive cells, including the IE genes FOS, JUN, EGR1 and EGR3 are typically linked to the promotion of cell proliferation such as in response to serum or growth factor treatment (42). HDACi treatment therefore clearly induces a conflicting transcriptional response comprising both pro and anti-proliferative signals, which is magnified in HDACi sensitive cells. HDACi induction of IE gene expression is also sustained for over 24h, whereas growth factor induction is typically maximal 30 minutes to 1 hour post stimulation and rapidly returns to baseline (35). Whether these conflicting transcriptional signals or the sustained induction of IE gene expression are perceived by the cells as a stress response, and whether this is ultimately the stimulus for apoptosis induction is worthy or further investigation. In support of this hypothesis, it is notable that co-treatment of colon cancer cells with agents such as PMA which independently induce IE gene expression, potentiate HDACi-induced apoptosis (43).
In conclusion, this study demonstrates that HDACi-induced apoptosis in colon cancer cells is linked to a consistent transcriptional response involving induction of a transcriptional network enriched for IE and SR genes, which are regulated, to a large extent, by the Sp1/Sp3 transcription factors. Importantly, several components of this transcriptional response are basally repressed in colon tumors suggesting HDACi-induced apoptosis may be initiated through induction of a transcriptional network whose basal repression is linked to tumor cell survival.
Supplementary Material
Acknowledgments
We thank Dr. Geoff Childs and Aldo Massimi at the AECOM cDNA microarray facility for generation of cDNA microarrays and assistance with array scanning, and David Reynolds at the AECOM DNA sequencing facility.
Grant support: These studies were supported by NIH grants CA123316, CA100823 and CA88104.
References
- 1.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;2637:5541–52. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 2.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;59:769–84. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 3.Mariadason JM. HDACs and HDAC inhibitors in colon cancer. Epigenetics. 2008;31:28–37. doi: 10.4161/epi.3.1.5736. [DOI] [PubMed] [Google Scholar]
- 4.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;904:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 5.Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;27515:10887–92. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 6.Ammanamanchi S, Freeman JW, Brattain MG. Acetylated sp3 is a transcriptional activator. J Biol Chem. 2003;27837:35775–80. doi: 10.1074/jbc.M305961200. [DOI] [PubMed] [Google Scholar]
- 7.Ferrante RJ, Kubilus JK, Lee J, et al. Histone deacetylase inhibition by sodium butyrate chemotherapy ameliorates the neurodegenerative phenotype in Huntington’s disease mice. J Neurosci. 2003;2328:9418–27. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun H, Koop R, Ertmer A, Nacht S, Suske G. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 2001;2924:4994–5000. doi: 10.1093/nar/29.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imhof A, Yang XJ, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;79:689–92. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 10.Heerdt BG, Houston MA, Augenlicht LH. Short-chain fatty acid-initiated cell cycle arrest and apoptosis of colonic epithelial cells is linked to mitochondrial function. Cell Growth Differ. 1997;85:523–32. [PubMed] [Google Scholar]
- 11.Insinga A, Monestiroli S, Ronzoni S, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005;111:71–6. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- 12.Nebbioso A, Clarke N, Voltz E, et al. Tumor-selective action of HDAC inhibitors involves TRAIL induction in acute myeloid leukemia cells. Nat Med. 2005;111:77–84. doi: 10.1038/nm1161. [DOI] [PubMed] [Google Scholar]
- 13.Ruemmele FM, Schwartz S, Seidman EG, Dionne S, Levy E, Lentze MJ. Butyrate induced Caco-2 cell apoptosis is mediated via the mitochondrial pathway. Gut. 2003;521:94–100. doi: 10.1136/gut.52.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson AJ, Arango D, Mariadason JM, Heerdt BG, Augenlicht LH. TR3/Nur77 in colon cancer cell apoptosis. Cancer Res. 2003;6317:5401–7. [PubMed] [Google Scholar]
- 15.Hernandez A, Thomas R, Smith F, et al. Butyrate sensitizes human colon cancer cells to TRAIL-mediated apoptosis. Surgery. 2001;1302:265–72. doi: 10.1067/msy.2001.115897. [DOI] [PubMed] [Google Scholar]
- 16.Bonnotte B, Favre N, Reveneau S, et al. Cancer cell sensitization to fas-mediated apoptosis by sodium butyrate. Cell Death Differ. 1998;56:480–7. doi: 10.1038/sj.cdd.4400371. [DOI] [PubMed] [Google Scholar]
- 17.Lindemann RK, Newbold A, Whitecross KF, et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci U S A. 2007;10419:8071–6. doi: 10.1073/pnas.0702294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruefli AA, Ausserlechner MJ, Bernhard D, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci U S A. 2001;9819:10833–8. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bandyopadhyay D, Mishra A, Medrano EE. Overexpression of histone deacetylase 1 confers resistance to sodium butyrate-mediated apoptosis in melanoma cells through a p53-mediated pathway. Cancer Res. 2004;6421:7706–10. doi: 10.1158/0008-5472.CAN-03-3897. [DOI] [PubMed] [Google Scholar]
- 20.Chirakkal H, Leech SH, Brookes KE, Prais AL, Waby JS, Corfe BM. Upregulation of BAK by butyrate in the colon is associated with increased Sp3 binding. Oncogene. 2006;2554:7192–200. doi: 10.1038/sj.onc.1209702. [DOI] [PubMed] [Google Scholar]
- 21.Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer cell. 2003;41:13–8. doi: 10.1016/s1535-6108(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 22.Park JH, Jong HS, Kim SG, et al. Inhibitors of histone deacetylases induce tumor-selective cytotoxicity through modulating Aurora-A kinase. Journal of molecular medicine (Berlin, Germany) 2008;861:117–28. doi: 10.1007/s00109-007-0260-8. [DOI] [PubMed] [Google Scholar]
- 23.Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;28029:26729–34. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 24.Xu WS, Perez G, Ngo L, Gui CY, Marks PA. Induction of polyploidy by histone deacetylase inhibitor: a pathway for antitumor effects. Cancer Res. 2005;6517:7832–9. doi: 10.1158/0008-5472.CAN-04-4608. [DOI] [PubMed] [Google Scholar]
- 25.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nature cell biology. 2001;32:114–20. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 26.Dowling M, Voong KR, Kim M, Keutmann MK, Harris E, Kao GD. Mitotic spindle checkpoint inactivation by trichostatin a defines a mechanism for increasing cancer cell killing by microtubule-disrupting agents. Cancer Biol Ther. 2005;42:197–206. [PubMed] [Google Scholar]
- 27.Blagosklonny MV, Robey R, Sackett DL, et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Molecular cancer therapeutics. 2002;111:937–41. [PubMed] [Google Scholar]
- 28.Mariadason JM, Arango D, Shi Q, et al. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;6324:8791–812. [PubMed] [Google Scholar]
- 29.Dang DT, Chen X, Feng J, Torbenson M, Dang LH, Yang VW. Overexpression of Kruppel-like factor 4 in the human colon cancer cell line RKO leads to reduced tumorigenecity. Oncogene. 2003;2222:3424–30. doi: 10.1038/sj.onc.1206413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berglund L, Bjorling E, Oksvold P, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;710:2019–27. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Uhlen M, Bjorling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;412:1920–32. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;232:395–402. doi: 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray R, Snyder RC, Thomas S, Koller CA, Miller DM. Mithramycin blocks protein binding and function of the SV40 early promoter. J Clin Invest. 1989;836:2003–7. doi: 10.1172/JCI114110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjorling E, Lindskog C, Oksvold P, et al. A web-based tool for in silico biomarker discovery based on tissue-specific protein profiles in normal and cancer tissues. Mol Cell Proteomics. 2008;75:825–44. doi: 10.1074/mcp.M700411-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Tice DA, Soloviev I, Polakis P. Activation of the Wnt pathway interferes with serum response element-driven transcription of immediate early genes. J Biol Chem. 2002;2778:6118–23. doi: 10.1074/jbc.M111255200. [DOI] [PubMed] [Google Scholar]
- 36.Sassone-Corsi P. Goals for signal transduction pathways: linking up with transcriptional regulation. Embo J. 1994;1320:4717–28. doi: 10.1002/j.1460-2075.1994.tb06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. Embo J. 1999;1817:4779–93. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haq F, Mahoney M, Koropatnick J. Signaling events for metallothionein induction. Mutation research. 2003;5331–2:211–26. doi: 10.1016/j.mrfmmm.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 39.Collard TJ, Guy M, Butt AJ, et al. Transcriptional upregulation of the insulin-like growth factor binding protein IGFBP-3 by sodium butyrate increases IGF-independent apoptosis in human colonic adenoma-derived epithelial cells. Carcinogenesis. 2003;243:393–401. doi: 10.1093/carcin/24.3.393. [DOI] [PubMed] [Google Scholar]
- 40.Pan L, Lu J, Wang X, et al. Histone deacetylase inhibitor trichostatin a potentiates doxorubicin-induced apoptosis by up-regulating PTEN expression. Cancer. 2007;1098:1676–88. doi: 10.1002/cncr.22585. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Clark S, Birkeland M, et al. Induction and superinduction of growth arrest and DNA damage gene 45 (GADD45) alpha and beta messenger RNAs by histone deacetylase inhibitors trichostatin A (TSA) and butyrate in SW620 human colon carcinoma cells. Cancer Lett. 2002;1881–2:127–40. doi: 10.1016/s0304-3835(02)00322-1. [DOI] [PubMed] [Google Scholar]
- 42.Kovary K, Bravo R. The jun and fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;119:4466–72. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahmani M, Dai Y, Grant S. The histone deacetylase inhibitor sodium butyrate interacts synergistically with phorbol myristate acetate (PMA) to induce mitochondrial damage and apoptosis in human myeloid leukemia cells through a tumor necrosis factor-alpha-mediated process. Exp Cell Res. 2002;2771:31–47. doi: 10.1006/excr.2002.5548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.