Abstract
Class 1 integrons play a role in the emergence of multi-resistant bacteria by facilitating the recruitment of gene cassettes encoding antibiotic resistance genes. 512 E. coli strains sourced from humans (n = 202), animals (n = 304) and the environment (n = 6) were screened for the presence of the intI1 gene. In 31/79 integron positive E. coli strains, the gene cassette regions could not be PCR amplified using standard primers. DNA sequence analysis of 6 serologically diverse strains revealed atypical integrons harboured the dfrA5 cassette gene and only 24 bp of the integron 3′-conserved segment (CS) remained, due to the insertion of IS26. PCR targeting intI1 and IS26 followed by restriction fragment length polymorphism (RFLP) analysis identified the integron-dfrA5-IS26 element in 27 E. coli strains of bovine origin and 4 strains of human origin. Southern hybridization and transformation studies revealed the integron-dfrA5-IS26 gene arrangement was either chromosomally located or plasmid borne. Plasmid location in 4/9 E. coli strains and PCR linkage of Tn21 transposition genes with the intI1 gene in 20/31 strains, suggests this element is readily disseminated by horizontal transfer.
Introduction
The emergence and spread of antibiotic resistance is an escalating global health concern. Multi-drug resistant bacteria are the principal cause of failure in the treatment of infectious diseases, resulting in increases in the term and magnitude of morbidity, higher rates of mortality, and a greater health cost burden [1]. Monitoring and surveillance of antibiotic resistance genes in the community is essential for developing strategies to minimize the spread of antimicrobial resistance and evaluating their impact. Class 1 integrons play an important role in the emergence of multi-resistant bacteria via the stockpiling of resistance determinants [2], [3], [4]. Integrons, while not mobile themselves are often carried by mobilizing elements such as conjugative plasmids, transposons or phages, which ensure their horizontal transfer [5].
Class 1 integrons are usually comprised of two conserved segments, which flank the variable gene cassette region. Essential features of the 5′-CS are the integrase gene (intI1), the product of which mediates orientation specific integration and excision of gene cassettes [6], a promoter region, containing the potential promoter, Pc to allow expression of the inserted cassette genes [7], and an integration site (attI1) for the site-specific insertion of gene cassettes [8]. The 3′-CS of class 1 integrons typically contains the qacEΔ1 gene, mediating low-level resistance to quaternary ammonium compounds [9], the sulfonamide resistance gene, sul1 [10] and ORF5 of unknown function [3].
An accurate assessment of the importance of integrons in the dissemination of antibiotic resistance genes is compounded by the detection of class 1 integrons in which gene cassette regions containing antibiotic resistance genes are unable to be amplified using standard PCR methods [11]–[15]. This shortfall in standard PCR screening methods prevents the true breadth of resistance gene carriage in the community from being revealed. In this study, serologically diverse E. coli strains from a range of sources were screened for the presence of class 1 integrons. Cassette arrays were characterized in all of the integrons, including those that could not be amplified using standard PCR methods.
Materials and Methods
E. coli strains used in this study
512 E. coli strains were isolated from a variety of sources (Table 1). None of the E. coli strains sourced from either animals or humans were from a known outbreak. E. coli isolates from clinical cases of human infection were selected for screening on the basis of having resistance to at least one antibiotic. There was no selection criteria applied to the other E. coli strains and all strains received were screened for class 1 integrons. E. coli isolates from cattle and swine with clinical cases of infection had been submitted to the Elizabeth Macarthur Agricultural Institute, NSW, Australia. Canine-derived E. coli strains were sourced from the Faculty of Veterinary Science, University of Melbourne, Victoria, Australia. E. coli strains from Australian native animals were obtained from the School of Botany and Zoology, The Australian National University, ACT, Australia.
Table 1. E. coli strains examined for class 1 integron carriage.
| Source of strains | Number of strains | ||
| Animal | Diagnostic specimens | Bovine | 177 |
| Canine | 46 | ||
| Porcine | 20 | ||
| Healthy | Native animals | 54 | |
| Porcine | 4 | ||
| Healthy sheep | 1 | ||
| Unknown symptoms | Porcine | 1 | |
| Parrot | 1 | ||
| Total no. of animal-derived E. coli strains | 304 | ||
| Human | Diagnostic specimens | UTI | 95 |
| SIDS | 21 | ||
| Diarrhoea | 22 | ||
| Bloody diarrhoea | 7 | ||
| Suspected diarrhoea | 5 | ||
| Gastroenteritis | 4 | ||
| Infantile gastroenteritis | 3 | ||
| Enteritis | 1 | ||
| Suspected traveler's diarrhoea/infantile enteritis | 1 | ||
| Septicemia | 5 | ||
| HUS | 2 | ||
| Suspected HUS | 1 | ||
| Neonatal meningitis | 1 | ||
| Appendicitis | 1 | ||
| Healthy | Infants | 24 | |
| Human | 2 | ||
| Unknown symptoms | Human | 7 | |
| Total no. of human-derived E. coli strains | 202 | ||
| Environmental-derived E. coli strains | 6 | ||
| Total number of E. coli strains | 512 | ||
Abbreviations: UTI, Urinary tract infection; SIDS, sudden infant death syndrome; HUS, hemolytic uremic syndrome.
Of the human-derived E. coli strains, 190 were collected as part of the work of the Microbiological Diagnostic Unit (MDU, Public Health Laboratory, Department of Microbiology and Immunology, University of Melbourne, Victoria, Australia) from 1994 to 2003, and 12 were archived strains from patients in the UK, USA, Denmark, Austria, Germany, Indonesia, Thailand and Japan. The 24 strains isolated from healthy infants were part of a previous study of antibiotic resistance in verocytotoxigenic E. coli (VTEC) and non-VTEC isolated from animals and humans [16]. These strains were collected between 1989 and 1992 from infants less than 1 year of age who had not suffered from gastrointestinal symptoms or been treated with antibiotics in the 2 weeks prior to collection.
Characterization of E. coli strains
Each of the strains was identified as E. coli based on conformity to species description when cultured in various specialized media. E. coli strains were O-serotyped and H-serotyped, as previously described [16]. The plate/replicator method was used to test the sensitivity of strains to the following antibiotics: ampicillin (32 µg/ml), streptomycin (25 µg/ml), tetracycline (20 µg/ml), chloramphenicol (10 µg/ml), sulfathiazole (550 µg/ml) trimethoprim (50 µg/ml), kanamycin (10 µg/ml), nalidixic acid (50 µg/ml), spectinomycin (50 µg/ml), gentamicin (2.5 µg/ml) and ciprofloxacin (2 µg/ml) [16]. Resistance profiling, strain identification and serotyping were performed at MDU.
PCR amplification and RFLP analysis
Bacterial DNA used as template for PCR was extracted by suspending one or two fresh colonies in 1 ml of sterile distilled water, which was subsequently washed, resuspended in 500 µl of water and heated at 100°C for 7 min. A volume of 3–5 µl of this DNA preparation was used per 50 µl PCR reaction mix. PCR targeting the intI1 gene was performed to identify by inference, E. coli strains that contained class 1 integrons using the primers L2 and L3 [12]. PCR amplification of the E. coli-specific universal stress protein A (uspA) gene using the primers EC2 [17] and FD-uspA (5′ AAA GTT TCT CTG ATC CAC GTA G) served to confirm the identification of E. coli isolates and DNA integrity. The E. coli strain UB1637 (pR388, In3) was used as a positive control for PCR. The intI1 and uspA PCR was performed simultaneously using the following cycling conditions: 94°C for 4 min, followed by 30 cycles of 94°C for 30 sec, 57°C for 1 min and 72°C for 1 min, and finally 72°C for 10 min. Gene cassette regions of class 1 integrons were amplified using the primers L1 and R1 [12] using conditions described previously [18]. The primer pair L1 and JL-D2 (5′ CGC ATC ACC TCA ATA CCT T) (X. Liu, unpublished results) was used to amplify integron-IS26 elements.
PCR amplification of the intI1 gene linked to the Tn21 transposase genes (tnpM, tnpR and tnpA) was achieved using the primer pair FD-tnpA (5′ GGT CGG TAT CGT TGA ATG TGT) and FD-IntI1 (5′ GTT ACG ACA TTC GAA CCG TG). The following PCR conditions were used to amplify the integron-IS26 gene arrangement and intI1 linked to Tn21 transposase genes: 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min and 72°C for 4 min, and finally 72°C for 10 min.
Gene cassette regions amplified using the standard primer pair L1 and R1 were classified according to their restriction profile, following restriction digestion with RsaI and AluI. RFLP analysis of the integron-IS26 gene arrangement was performed by digestion with RsaI. RFLP analysis of the intI1-Tn21 amplicon was undertaken with EcoRI.
Transformation studies and Southern Hybridization
In order to identify an E. coli isolate with the integron situated on a plasmid to facilitate DNA sequencing of the intI1 gene and flanking sequence, transformation studies were performed. The presence of the intI1 gene on a plasmid was determined in seven of the E. coli isolates (D18, D22, D79, D87, D111, D398 and D318) where cassette arrays could not be amplified using the standard primer pair L1 and R1. These E. coli isolates, with differing serotypes and resistance phenotypes, were randomly selected from the collection. Plasmid-associated intI1 gene was detected by the electroporation of plasmid DNA isolated from these strains into E. coli JM109, followed by selection with trimethoprim (50 µg/ml), sulfathiazole (550 µg/ml), or streptomycin (25 µg/ml) and intI1 PCR of DNA from the resultant colonies.
Southern hybridizations were also were undertaken on four E. coli isolates (D21, D22, D23 and D87) to identify an E. coli isolate located on a plasmid and facilitate DNA sequencing of the integron and flanking sequence. E. coli isolates D21 and D23 were randomly selected from the collection while D22 and D87 were selected from the subset of E. coli isolates previously tested by transformation. Southern hybridizations involved observing the hybridization of a DIG-labelled integrase probe to plasmid DNA extractions compared with that of total genomic DNA. The primers L2 and L3 [12] were used to synthesize the class 1 integrase (intI1) probe.
DNA sequence analysis
Plasmid DNA isolated from the E. coli strain D22 was transformed [19] into E. coli JM109 and the DNA sequence of the integron and flanking regions of the plasmid was determined. DNA sequence analysis was performed by the dideoxy chain termination method [20] using an ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The nucleotide sequence of one representative amplicon from each of the other RFLP cassette array groups described in this study was also determined.
Results
PCR detection of the intI1 gene and characterization of cassette arrays
PCR screening for intI1 identified 79/512 (15.4%) E. coli strains contained class 1 integrons. The intI1 gene was detected in 32/304 (10.5%) of E. coli strains sourced from animals and 47/202 (23.3%) strains sourced from healthy and ill humans. The intI1 gene was not detected in E. coli strains from Australian native animals or the environment.
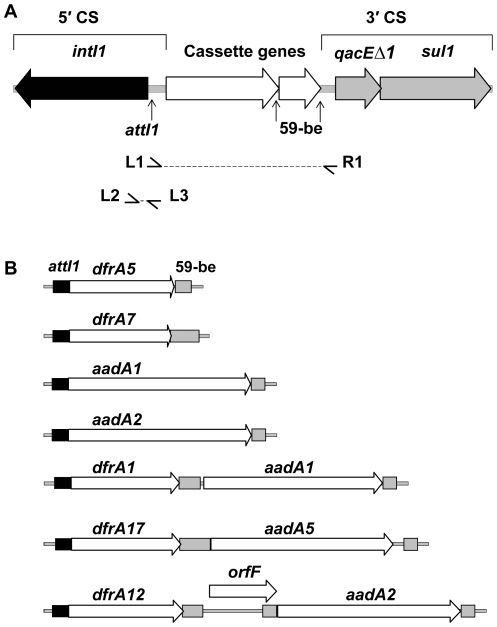
Gene cassette arrays within class 1 integrons were PCR amplified using standard L1 and R1 primers. RFLP identified 7 distinct RFLP groups. DNA sequence analysis of a selected representative of each RFLP group showed 100% identity to sequences previously deposited in GenBank. The following cassette arrays were identified: dfrA5, (AJ419169), n = 9; dfrA7, (EU250577); n = 2, aadA1, (AB188267), n = 30; aadA2, (DQ238100), n = 1; dfrA1/aadA1, (AJ884723), n = 2; dfrA17/aadA5, (AY748452), n = 4 and dfrA12/orfF/aadA2, (AB297450), n = 1 (Figure 1). In 31/79 integron positive E. coli strains, the cassette arrays could not be PCR amplified using standard L1 and R1 primers. These E. coli strains were sourced from cattle with clinical cases of infection (n = 27) and human patients suffering from UTI (n = 2) and bloody diarrhoea (n = 2).
Figure 1. Features and characterization of typical class 1 integrons.
(A) Structure of a typical class 1 integron showing the 5′- and 3′-CS. The location and the direction of transcription of genes are indicated. The class 1 integrase gene intI1 (black arrow) and attI1 integron-integration site (vertical arrow) are located in the integron 5′-CS. The qacEΔ1 gene and the sul1 gene (grey arrows), and the open reading frame orf5 (not shown) are located in the 3′-CS. Inserted gene cassettes are represented by unfilled arrows and their associated 59-be are indicated by vertical arrows. L1, R1 and L2, L3 primer annealing sites are indicated by small horizontal arrows. (B) A diagram showing the genetic structure of the gene cassette arrays amplified using standard PCR primers that target the 5′- and 3′-CS of typical class 1 integrons (cassette arrays detected: dfrA5, dfrA7, aadA1, aadA2, dfrA1/aadA1, dfrA17/aadA5 and dfrA12/orfF/aadA2). The following features are indicated: attI1 recombination sites (black filled boxes), gene cassette arrays (unfilled arrows) and 59-bes (grey filled boxes). All diagrams are drawn to scale.
Characterization of a representative class 1 integron gene cassette array that could not be PCR amplified using standard L1 and R1 primers
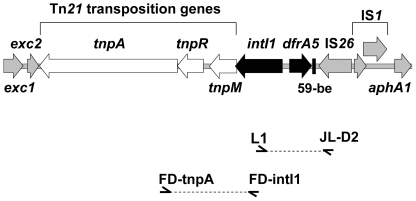
Transformation studies revealed the integron was located on a plasmid in E. coli strains D18, D22 and D79 and located on chromosomal DNA in strains D87, D111, D298 and D318. Southern hybridization using the intI1 gene as a probe demonstrated that the intI1 gene was plasmid-borne in E. coli strains D21 and D22 and chromosomally located in strains D23 and D87 (results not shown). E. coli strain D22, containing a plasmid-associated intI1 gene identified by Southern hybridization and transformation studies, was selected for sequencing of the integron and flanking regions. The bovine-derived E. coli strain, D22 contained an integron located on a plasmid of approximately 22 kb. Approximately 8.7 kb of the plasmid, encompassing the integron and flanking regions was subjected to DNA sequence analysis (GenBank accession number EU914098). This analysis revealed a class 1 integron with the typical 5′-CS and a complete dfrA5 gene cassette, but lacked the usual class 1 integron-associated 3′-CS due to the insertion of IS26 (Figure 2). The insertion of IS26 presumably caused the deletion of all but 24 bp of the 3′-CS. The Tn21 transposition genes tnpM, tnpR and tnpA were located downstream of the intI1 gene.
Figure 2. An atypical class 1 integron with an IS26-mediated deletion in the integron 3′-CS.
Genetic structure of an atypical class 1 integron located on a plasmid isolated from the bovine-derived E. coli strain, D22. The genetic map represents 8756 bp of nucleotide sequence. All genes are indicated by arrows and shaded in grey, except the class 1 integron intI1 gene and dfrA5 cassette gene (black) and the Tn21 transposition genes tnpM, tnpR and tnpA (unfilled). Other genes identified include the entry exclusion proteins exc1 and exc2 (truncated) and the kanamycin resistance gene, aphA1 (partial sequence). Features involved in recombination include the 59-be (black filled box), IS26 transposase gene, tnpA, and IS1 genes insA and insB. The position of PCR primers used to screen E. coli strains for this atypical class 1 integron is shown.
Distribution of dfrA5-IS26 configuration and linkage of intI1 with Tn21 transposition genes
Identification of the unique class 1 integron-IS26 gene arrangement in the bovine-derived strain D22 prompted the PCR interrogation of all strains with “non-amplifiable” gene cassette arrays in class 1 integrons using primers that target intI1 and IS26. The presence of an integron-IS26 gene arrangement was detected using primers L1 and JL-D2. A single 0.8 kb amplification product was detected in 27/27 E. coli isolates sourced from cattle and 4/4 isolates from humans. Subsequent RFLP analysis using the restriction enzyme RsaI revealed the presence of the dfrA5-IS26 configuration in each instance (Table 2). Nucleotide sequencing and clustalW alignment of the 848 bp amplicons in 2 human-derived and 4 bovine-derived E. coli strains confirmed that in each instance IS26 was inserted in an identical position as that described for E. coli strain D22. Linkage of Tn21 transposition genes and intI1 was detected using the PCR primers FD-tnpA and FD-intI1. A single 1.5 kb PCR amplification product was detected in 18/27 E. coli isolates sourced from cattle and 2/4 isolates from humans (Table 2). RFLP analysis indicated these PCR amplification products were identical and nucleotide sequencing of the 1490 bp amplicon identified the integrase gene, intI1 and the Tn21 transposition genes, tnpM, tnpR and tnpA.
Table 2. Features of E. coli strains harbouring integron-IS26 elements.
| Strain no. | Year | Serotype | Source | A | S | T | C | Su | Tm | K | Na | G | Cp | Sp | Tn21 tnpM, tnpR, tnpA |
| D18 | 2002 | O123:H11 | B - P7 | + | + | + | − | + | + | + | − | − | − | − | + |
| D21 | 2002 | O123:H11 | B - P7 | + | + | + | − | + | + | + | − | − | − | − | + |
| AD22 | 2002 | O26:H11 | B - P3 | + | + | + | − | + | + | + | − | − | − | − | + |
| D23 | 2002 | O111:H11 | B - P4 | + | + | + | − | + | − | − | − | − | − | − | − |
| D48A | 2002 | O111:H11 | B - P2 | + | + | + | − | + | + | + | − | − | − | − | − |
| D49 | 2002 | O111:H11 | B - P2 | + | B+ | + | − | + | + | + | − | − | − | − | − |
| D52 | 2002 | O111:H- | B - P2 | + | B+ | − | − | + | + | + | − | − | − | − | − |
| D53 | 2002 | O111:H- | B - P2 | + | B+ | + | − | + | + | + | − | − | − | − | − |
| D55 | 2002 | O123:H11 | B - P8 | + | + | + | − | + | + | + | − | − | − | − | + |
| D79 | 2002 | O111:H- | B - P6 | + | + | + | − | + | + | + | − | − | − | − | + |
| D81 | 2002 | O111:H- | B - P6 | + | + | + | − | + | + | + | − | − | − | − | + |
| D87 | 2002 | O111:H11 | B - P5 | + | + | − | − | + | + | + | − | − | − | − | − |
| D101 | 2002 | O111:H- | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D103 | 2002 | O111:H- | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D105 | 2002 | Ont:H9 | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D111 | 2002 | O177:H11 | B - P1 | − | + | + | − | + | + | + | − | − | − | − | + |
| D120 | 2002 | O111:H- | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D252 | 2002 | O177:H11 | B - P1 | − | + | + | − | + | + | + | − | − | − | − | − |
| D257 | 2002 | O111:H- | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D260 | 2002 | O111:H- | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D263 | 2002 | O111:H- | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D272 | 2002 | O111:H11 | B - P5 | + | + | − | − | + | + | + | − | − | − | − | − |
| D275 | 2002 | O111:H11 | B - P5 | + | + | − | − | + | + | + | − | − | − | − | + |
| D298 | 2002 | O180:H- | B - P9 | + | + | + | − | + | + | + | − | − | − | − | + |
| D305 | 2002 | O111:H- | B - P1 | + | + | + | − | + | + | + | − | − | − | − | + |
| D318 | 2002 | Ont:H32 | B - P2 | + | + | + | − | + | + | + | − | − | − | − | + |
| D319 | 2002 | O162:H9 | B - P1 | + | + | + | − | + | + | + | − | − | − | − | − |
| 20 | 1999 | Ont:H32 | H - UTI | + | − | − | − | − | + | − | − | − | − | − | − |
| 11604 | 1999 | O11:H- | H - UTI UTIUUUTI | − | − | − | − | − | + | − | − | − | − | − | − |
| 6877 | 1998 | O26:H- | H - BD | + | + | + | − | + | + | + | − | − | − | − | + |
| 6878 | 1998 | O26:H- | H - BD | + | + | + | − | + | + | + | − | − | − | − | + |
The genetic structure of the integron-dfrA5-IS26 element and flanking regions of E. coli strain D22 is illustrated in Figure 2.
Moderate level of resistance. Resistance to antibiotics is indicated by (+) and susceptibility is indicated by (−). Abbreviations of antibiotics: A, ampicillin (32 µg/ml); S, streptomycin (25 µg/ml); T, tetracycline (20 µg/ml); C, chloramphenicol (10 µg/ml); Su, sulfathiazole (550 µg/ml); Tm, trimethoprim (50 µg/ml); K, kanamycin (10 µg/ml); Na, nalidixic acid (50 µg/ml); Sp, spectinomycin (50 µg/ml); G, gentamicin (2.5 µg/ml); and Cp, ciprofloxacin (2 µg/ml). The source of E. coli strains is indicated by B, Bovine or H, human. The location of cattle properties is indicated by P1, Kameruka; P2, Cowra; P3, Eden; P4, Dungog; P5, Finley; P6, Gerringong; P7, Bega; P8, Canowindra and P9, Richmond. Human-derived E. coli strains were sourced from the Melbourne Diagnostic Unit (MDU) from patients with bloody diarrhoea (BD) or urinary tract infections (UTI).
Serotypes
Features of E. coli strains containing the dfrA5-IS26 configuration are summarized in Table 2. Variation in serotypes was observed in E. coli isolates from animal and human sources that contained the novel dfrA5-IS26 configuration, suggesting that the horizontal transfer of this integron had occurred. Nine E. coli strains, with the serotype O111:H- that were isolated between 2002 and 2003 and sourced from cattle geographically separated by up to 319 km, harbored the dfrA5-IS26 configuration. Multiple strains with identical cassette arrays and serotypes, suggests the clonal spread of integron-containing E. coli had occurred between hosts.
Discussion
In this study we have characterized antibiotic resistance gene cassette arrays in class 1 integrons that could not be amplified using standard PCR primers, by using primer pairs that target the intI1 gene and IS26. Characterization of previously “non-amplifiable” cassette arrays resulted in the identification a unique dfrA5-IS26 configuration in E. coli strains isolated from both animal and human sources. In each instance, IS26 was inserted beyond the 59-base element of the dfrA5 gene cassette and after the first 24 bp of the 3′-CS.
The distribution of the dfrA5-IS26 configuration was widespread with regard to geographical location. The dfrA5-IS26 configuration was detected in E. coli strains sourced from cattle located at nine NSW properties separated by up to 900 km. The distribution of the dfrA5-IS26 configuration described in this study is not likely to be due to IS26 insertion into a “hot spot” in the class 1 integron 3′-CS, as no marked target site specificity has been previously observed for IS26 [21]. This study also established a linkage between the intI1 and Tn21 transposition genes, in 18/27 E. coli isolates from bovine sources and 2/4 isolates from human sources, which implicates Tn21 in the distribution of this novel integron.
Variation in E. coli serotypes containing the dfrA5-IS26 configuration supports the contention that horizontal transfer of this gene arrangement has occurred. Identification of this novel dfrA5-IS26 configuration in E. coli strains of different serotypes that were sourced from animals and humans, suggests that the horizontal transfer of integron-related antibiotic resistance genes has occurred between these hosts. DNA sequence analysis of the 111 kb IncI1 EHEC virulence plasmid from the human-derived enterohemorrhagic E. coli strain, 6877, by Venturini et al. found the dfrA5-IS26 configuration in this isolate was within a Tn21-derivative [22], which confirms PCR results indicating the integron-IS26 configuration is located adjacent to Tn21 transposition genes in this strain (Table 2). Identification of the dfrA5-IS26 configuration in a virulence plasmid, highlights the possibility of co-selection of antibiotic resistance and virulence determinants under antibiotic selective pressures [22]. This dfrA5-IS26 configuration, widespread in E. coli isolates of bovine origin and also found in E. coli of human origin, may act as a conduit for the transfer of integron-related resistance genes to human pathogens.
Integron-related resistance gene transfer among E. coli with different serotypes sourced from both humans and animals underlies the importance of developing surveillance systems and research to further define food-chain, veterinary and environmental factors involved in the spread of antibiotic-resistant bacteria to humans. Epidemiological evidence will be needed to confirm the horizontal transfer of the dfrA5-IS26 configuration in E. coli between animal and human hosts. Information derived from antibiotic resistance surveillance studies is crucial when assessing the risks to public health and determining the impact of regulatory guidelines concerning the responsible use of antibiotics in human and veterinary medicine.
Acknowledgments
We thank Dr. J. Iredell (Centre for Infectious Diseases and Microbiology, Westmead Hospital) and Prof. R. Hall (University of Sydney) for the positive control E. coli strain. We also thank Assoc. Prof. G. Hogg, director of MDU Public Health Laboratory, Prof. G. F. Browning for E. coli strains isolated from dogs (Faculty of Veterinary Science, University of Melbourne) and Dr. D. M. Gordon for the strains from native animals (School of Botany and Zoology, The Australian National University) and X. Liu (Elizabeth McArthur Agricultural Institute) for the IS26 primer.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by an Australian Research Council Linkage grant. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Goldmann DA. The epidemiology of antimicrobial resistance. Ecosys Health. 1999;5:158–163. [Google Scholar]
- 2.Leverstein-van Hall MA, Blok HEM, Donders RT, Paauw A, Fluit AC, et al. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J Infect Dis. 2003;187:251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- 3.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 4.Recchia GD, Hall RM. Gene cassettes: a new type of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 5.Toussaint A, Merlin C. Mobile elements as a combination of functional modules. Plasmid. 2002;47:26–35. doi: 10.1006/plas.2001.1552. [DOI] [PubMed] [Google Scholar]
- 6.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 7.Lévesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 8.Partridge SR, Recchia GD, Scaramuzzi C, Collis CM, Stokes HW, et al. Definition of the attI1 site of class 1 integrons. Microbiology. 2000;146:2855–2864. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- 9.Paulsen IT, Littlejohn TG, Rådström P, Sundström L, Sköld O, et al. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sul1 and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 11.White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob Agents Chemother. 2001;45:2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maguire AJ, Brown DFJ, Gray JJ, Desselberger U. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob Agents Chemother. 2001;45:1022–1029. doi: 10.1128/AAC.45.4.1022-1029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thungapathra M, Amita Sinha KK, Chaudhuri SR, Garg P, et al. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class 1 integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob Agents Chemother. 2002;46:2948–2955. doi: 10.1128/AAC.46.9.2948-2955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz F-J, Hafner D, Geisel R, Follmann P, Kirschke C, et al. Increased prevalence of class I integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in a German university hospital. J Clin Microbiol. 2001;39:3724–3726. doi: 10.1128/JCM.39.10.3724-3726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallego L, Towner KJ. Carriage of class 1 integrons and antibiotic resistance in clinical isolates of Acinetobacter baumannii from Northern Spain. J Med Microbiol. 2001;50:71–77. doi: 10.1099/0022-1317-50-1-71. [DOI] [PubMed] [Google Scholar]
- 16.Bettelheim KA, Hornitzky MA, Djordjevic SP, Kuzevski A. Antibiotic resistance among verocytotoxigenic Escherichia coli (VTEC) and non-VTEC isolated from domestic animals and humans. J Med Microbiol. 2003;52:155–162. doi: 10.1099/jmm.0.04903-0. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Griffiths MW. PCR differentiation of Escherichia coli from other gram-negative bacteria using primers derived from the nucleotide sequences flanking the gene encoding the universal stress protein. Lett Appl Microbiol. 1998;27:369–371. doi: 10.1046/j.1472-765x.1998.00445.x. [DOI] [PubMed] [Google Scholar]
- 18.Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. Molecular cloning: A laboratory manual. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mollet B, Iida S, Arber W. Gene organization and target specificity of the prokaryotic mobile genetic element IS26. Mol Gen Genet. 1985;201:198–203. doi: 10.1007/BF00425660. [DOI] [PubMed] [Google Scholar]
- 22.Venturini C, Beatson SA, Djordjevic SP, Walker MJ. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 2009:fj.09–144972. doi: 10.1096/fj.09-144972. [DOI] [PubMed] [Google Scholar]