Abstract
The essential eukaryotic molecular chaperone Hsp90 operates with the help of different co-chaperones, which regulate its ATPase activity and serve as adaptors to recruit client proteins and other molecular chaperones, such as Hsp70, to the Hsp90 complex. Several Hsp90 and Hsp70 co-chaperones contain the tetratricopeptide repeat (TPR) domain, which interacts with the highly conserved EEVD motif at the C-terminal ends of Hsp90 and Hsp70. The acidic side chains in EEVD interact with a subset of basic residues in the TPR binding pocket called a ‘carboxylate clamp’. Since the carboxylate clamp residues are conserved in the TPR domains of known Hsp90/Hsp70 co-chaperones, we carried out an in silico search for TPR proteins in Arabidopsis and rice comprising of at least one three-motif TPR domain with conserved amino acid residues required for Hsp90/Hsp70 binding. This approach identified in Arabidopsis a total of 36 carboxylate clamp (CC)-TPR proteins, including 24 novel proteins, with potential to interact with Hsp90/Hsp70. The newly identified CC-TPR proteins in Arabidopsis and rice contain additional protein domains such as ankyrin, SET, octicosapeptide/Phox/Bem1p (Phox/PB1), DnaJ-like, thioredoxin, FBD and F-box, and protein kinase and U-box, indicating varied functions for these proteins. To provide proof-of-concept of the newly identified CC-TPR proteins for interaction with Hsp90, we demonstrated interaction of AtTPR1 and AtTPR2 with AtHsp90 in yeast two-hybrid and in vitro pull down assays. These findings indicate that the in silico approach used here successfully identified in a genome-wide context CC-TPR proteins with potential to interact with Hsp90/Hsp70, and further suggest that the Hsp90/Hsp70 system relies on TPR co-chaperones more than it was previously realized.
Introduction
The eukaryotic Hsp90 performs key roles in signal transduction by regulating maturation, localization, stability and protein interactions of a large number of signaling proteins [1], [2]. Due to its role in chaperoning oncogenic protein kinases, Hsp90 has recently been surfaced as a drug target for inhibiting cancer progression in humans [3]. Additional roles of Hsp90 lie in chromatin remodeling, epigenetic regulation, and morphological evolution [2]. Eukaryotic Hsp90 functions as a homodimer with each monomer comprising of an N-terminal ATP-binding domain that is responsible for the ATPase activity of Hsp90, a middle domain that harbors the client protein binding site and can also regulate the ATPase activity of the N-terminal domain, and a C-terminal dimerization domain [4]. Hsp90-specific inhibitors such as geldanamycin (GA) bind to the N-terminal domain of Hsp90 and inhibit its in vivo functions [3]. The C-terminus of cytosolic Hsp90 contains a conserved pentapeptide MEEVD, which is responsible for binding with the tetratricopeptide repeat (TPR) domain present in several co-chaperones of Hsp90 [1], [5].
Hsp90 has an ATP-driven chaperone cycle [2]. The open V-shaped conformation of dimeric Hsp90 allows for loading of the client protein. Following binding of ATP, a conformational change in the middle domain leads to a closed conformation in which the monomers are twisted around each other and the two N-terminal domains are dimerized. With the coordinated assistance of Hsp70 and a range of co-chaperones in dynamic protein heterocomplexes, the final maturation of the client protein takes place. Upon ATP hydrolysis, the client protein and the heterocomplex dissociate and Hsp90 enters a new chaperone cycle [4].
The various co-chaperones assist Hsp90 by influencing its ATPase activity and by linking it to other proteins and providing some measure of specificity to the client protein-Hsp90 interaction [6]. On a structural basis the Hsp90 co-chaperones can be broadly divided into two categories: TPR domain proteins and non-TPR proteins. The TPR domain is a protein-protein interaction domain, comprised of a loosely conserved 34 amino acid sequence that is repeated several times [7]. Several Hsp90 co-chaperones, such as Hsp70-Hsp90 organizing protein (Hop), high molecular weight immunophilins [cyclophilin 40 (Cyp40) and FK506-binding proteins 51 and 52 (FKBP51 and FKBP52)], protein phosphatase 5 (PP5) and the carboxyl terminus of Hsc70 interacting protein (CHIP) contain a three-motif TPR domain with conserved carboxylate clamp residues [5]. Within a TPR motif, eight amino acids at positions 4 (W/L/F), 7 (L/I/M), 8 (G/A/S), 11 (Y/L/F), 20 (A/S/E), 24 (F/Y/L), 27 (A/S/L), and 32 (P/K/E) have a higher frequency of conservation and these are important in maintaining the α-helical structures within a motif. Clustering of several α-helices within a tandem array of TPR motifs generates an amphipathic channel with a large amount of surface area, which allows the TPR domain to recognize its target protein [7].
A good understanding of the interaction of the TPR domain with molecular chaperones Hsp90 and Hsp70 came from cocrystalization study of TPR domains in Hop with pentapeptide MEEVD and octapeptide GPTIEEVD, which correspond to the C-termini of Hsp90 and Hsp70, respectively [8]. The side chains of several basic amino acids in the TPR groove were seen to establish interactions with the acidic side chains of the EEVD motif. These basic amino acids are referred to as ‘carboxylate clamp’ residues. With reference to the positions of the clamp residues in a three-motif TPR domain, the consensus is Lys5 and Asn9; Asn6; Lys2 and Arg6. These amino acids are conserved and functionally important in most three-motif TPR containing co-chaperones of Hsp90 and Hsp70. Additional contacts between EEVD and the TPR groove involve hydrophobic interactions [9].
Hsp90 has been studied widely in animal and yeast model systems, but little is known by comparison about plant Hsp90. The Hsp90 family in the model plant Arabidopsis thaliana comprises of seven members: four closely related isoforms are cytosolic (AtHsp90-1 to AtHsp90-4), one is chloroplastic (AtHsp90-5), one mitochondrial (AtHsp90-6), and one is localized to the endoplasmic reticulum (ER) (AtHsp90-7) [10]. The occurrence of multiple Hsp90 isoforms that display both developmental and stress-responsive gene expression, suggests a range of specific functions for these proteins. The recent demonstration of the involvement of Hsp90 and co-chaperones RAR1 (required for M1a12 resistance) and SGT1 (suppressor of G2 allele of skp1), in plant disease resistance [11], [12] has provided impetus for further investigation of the roles of Hsp90 in plant signaling pathways. Apart from RAR1 and SGT1, Hsp70 [13], high molecular weight immunophilins [14], Hop-like protein [15], PP5 [16], and p23-like proteins [17] have been found to associate with plant Hsp90. Plant orthologs of the previously characterized Hsp90/Hsp70 co-chaperones with TPR domains include Hop [15], PP5 [16], immunophilins like Cyp40 (SQUINT) [18], Rotamase AtFKBP62 (AtROF1) and AtFKBP65 (AtROF2) [19], PASTICCINO1 (AtPAS1) [20], AtFKBP42 (AtTWD1) [21], [22], Translocon of the outer envelope of chloroplast (TOC64) [23] and AtCHIP [24]. Given the conservation and preponderance of the TPR domain in established and potential co-chaperones of the plant Hsp90/Hsp70 system, we set out to uncover all carboxylate clamp (CC) type TPR proteins (CC-TPR proteins) in Arabidopsis using an in silico approach and following the residues conserved for interaction with the EEVD motif of cytosolic Hsp90/Hsp70. Here we report the identification of 24 new CC-TPR proteins in Arabidopsis, and similar as well as distinct CC-TPR proteins in rice (Oryza sativa), as putative Hsp90/Hsp70 interactors. Some of the newly uncovered proteins contain additional protein domains such as ankyrin, SET, octicosapeptide/Phox/Bem1p (Phox/PB1), DnaJ, thioredoxin, FBD and F-box, and protein kinase and U-box, indicating novel functions that may add new dimensions to the Hsp90/Hsp70 chaperone complex in plants.
Methods
Identification of CC-TPR proteins in Arabidopsis
The term ‘TPR’ submitted as query at the InterPro home page: http://www.ebi.ac.uk/interpro/ [25] retrieved 31 entries of which IPR013026 was found to best conform to TPR structure and function. The ‘Taxonomic coverage’ of IPR013026 revealed 235 TPR proteins in the Arabidopsis proteome (taxon ID 3702). The sequences of all proteins and the Arabidopsis Genome Initiative Identifier (AGI ID) were recorded and confirmed against the database of National Centre for Biotechnology Information (NCBI) and each sequence was analyzed for the presence of TPR domain using InterProScan (http://www.ebi.ac.uk/InterProScan/) [26]. Since the known co-chaperones that interact with MEEVD of Hsp90 consist of three-motif TPR domain, proteins with one or more TPR domains comprising of three motifs were short-listed. Subsequently, these motifs were analyzed for the presence of the conserved residues (K5N9-N6-K2R6). A second round of search was carried out where protein sequences with one or two TPR motifs were manually mapped for the presence of the second and/or third motif and the conserved residues (K5N9-N6-K2R6). Following selection of the CC-TPR proteins, these proteins were searched for additional known domains and localization signal sequences. Multiple sequence alignment for each of the three motifs of the CC-TPR proteins identified in Arabidopsis (above-described search) and human Hop TPR2a as the reference sequence was used to build a statistical model of the corresponding motif using the software package HMMER 3.0 (http://hmmer.org/). Each of these three models was queried on the basis of HMM profile against TAIR9_pep_20090619 (Arabidopsis annotation from TAIR) and Rice Genome Annotation Project (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/O_sativa/annotation_dbs/pseudomolecules/version_6.0). The HMMER-selected proteins (E-value <10) were scanned for conserved residues (K5N9-N6-K2R6), and BLAST and manual editing were used to remove redundancy.
Phylogenetic tree construction
Full-length sequences of CC-TPR proteins of Arabidopsis and rice were aligned using Clustal X 2.0.10. The phylogenetic tree was derived from the sequence comparisons using the neighbor-joining method in Clustal X.
Protein localization prediction
The electronic Fluorescent Pictograph (eFP) Browser [27] from the Bio-Array Resource (BAR) (http://bbc.botany.utoronto.ca/) was used to predict the subcellular localization (SL) of the CC-TPR proteins. The information retrieved was based on computational prediction and/or experimental documentation compiled in SUBA; the Arabidopsis SUBcellular localization database [28]. If experimental documentation for SL was available for a protein, it was considered as significant. However, for proteins lacking experimental documentation we inferred data available from several prediction algorithms such as iPSORT (http://hc.ims.u-tokyo.ac.jp/iPSORT/), LOCtree (http://cubic.bioc.columbia.edu/cgi-bin/var/nair/loctree/query), MitoPred (http://bioapps.rit.albany.edu/MITOPRED/), MitoProt II (http://ihg2.helmholtz-muenchen.de/ihg/mitoprot.html), MultiLoc (http://www-bs.informatik.uni-tuebingen.de/Services/MultiLoc/), PeroxP, Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html), SubLoc (http://www.bioinfo.tsinghua.edu.cn/SubLoc/), Target P (http://www.cbs.dtu.dk/services/TargetP/) and WOLFPSORT (http://wolfpsort.org/).This data was cross-checked with the actual eFP Browser data that was generated using heuristic prediction algorithms [27].
Expression analyses using AtGenExpress Visualization Tool
Affymetrix microarray data provided by Weigel World (http://www.weigelworld.org/) was accessed using AtGenExpress Visualization Tool (AVT) (http://www.weigelworld.org/resources/microarray/AtGenExpress/) [29], [30] to analyze the developmental expression of the newly identified CC-TPR genes by inputting their AGI IDs on the homepage. Absolute expression values were retrieved and used to develop the figure.
Expression analysis using MPSS database
The MPSS database (http://mpss.udel.edu/rice/) was searched (opting 17-nt signature sequences) using the locus ID given in the Rice Genome Annotation Project database to generate the expression profiles of rice CC-TPR proteins in different tissues and stress.
Electronic Northern Analysis
The expression profiles of Hsp90 and CC-TPR genes were examined against the AtGenExpress extended tissue and abiotic stress data sets using the Expression Browser tool available at the Botany Array Resource (BAR) (http://bbc.botany.utoronto.ca/affydb/cgi-bin/affy_db_exprss_browser_in.cgi) [31]. The output option was selected as average of replicate treatments relative to the average of appropriate control. The output files were formatted into heatmaps using the Data MetaFormatter tool hosted at the BAR website by selecting ‘View Graphical Representation of Log Transformed Cluster Data’. The colour scale indicates the log2-level of expression above or below the median. Strong red indicates more than four-fold above the median, while dark blue indicates four-fold below the median.
Expression Angler
The Expression Angler tool at BAR identifies genes that are co-expressed with a gene of interest [31]. Using the cut-off r-value between 0.75–1.00, 156 genes were found against the AtGenExpress stress series dataset to be co-expressed with AtHsp90-1 (AT5G52640), a bona fide heat stress (HS)-induced gene. This list of genes was checked against the Arabidopsis list of 36 CC-TPR proteins to identify those that are co-expressed with AtHsp90-1 under HS.
Plant growth and treatments
Arabidopsis seeds were surface sterilized and plated on 1X Murashige and Skoog (MS) medium (Sigma) supplemented with B5 vitamins, 1% (w/v) agar and 1% sucrose. The plates were kept for 3 days in the dark at 4°C to encourage synchronized germination and then transferred to a growth chamber maintained at 22°C with a 16/8 h photoperiod (80 µmol m-2 s-1). For HS treatment, 10 day-old seedlings were exposed to 38°C for 1 h and 3 h in an incubator, following which the plant tissue above the medium was collected and quick-frozen for RNA isolation. For treatment with brassinosteroid (BR), 21 day-old seedlings grown on MS medium were submerged in water containing either 1 µM 24-epibrassinolide (EBR) or 0.01% ethanol (solvent for EBR) for 3 h and 12 h. After the treatment, the plant material was quick-frozen.
RNA extraction and Quantitative real-time RT-PCR
Total RNA was prepared using SV Total RNA Isolation System (Promega). First strand cDNA was synthesized from 1 µg of total RNA using QuantiTect Reverse Transcription Kit (Qiagen) and used as a template for quantitative RT-PCR (qRT-PCR). qRT-PCR was performed in 200 µl tubes with a Rotor-Gene RG-3000 real-time thermal cycling system (Corbett Research) using SYBR green to monitor double-strand DNA synthesis. Three independent biological samples were used with gene-specific primers (Table S1). Data were analyzed using Rotor-Gene 6.0.16 software (Corbett Research). Values were normalized using ubiquitin as the internal reference, and fold change in the expression level was calculated [32].
Yeast two-hybrid assay
To determine interaction of the two newly identified proteins, AtTPR1 and AtTPR2, with AtHsp90-2 by the yeast two-hybrid approach, AtHsp90-2 and AtTPR1/AtTPR2 coding sequences were cloned into bait and prey vector, respectively. AtHsp90-2 was amplified by PCR using the Arabidopsis Biological Resource Center (ABRC) cDNA clone C105057 and primers attB1-Hsp90-2F 5′- GGGG ACA AGT TTG TAC AAA AAA GCA GGC TCC ATG GCG GAC GCT GAA ACC TTT GCT TTC-3′ and attB2-Hsp90-2R 5′- GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC GTC GAC TTC CTC CAT CTT GCT ACC TTC -3′. The PCR product was cloned into pDONR221 by in vitro BP recombination (Invitrogen) to generate pDONRHsp90-2, which was used in LR reaction with pDestDB (bait vector) to generate pDBHsp90-2. AtTPR1 and AtTPR2 cDNAs were synthesized from total RNA from Arabidopsis leaf using gene-specific gateway primers (attB1-TPR1F 5′- GGGG ACA AGT TTG TAC AAA AAA GCA GGC TCC ATG GTA CTG ATC GAA TCA AGT GAG AG-3′ and attB2-TPR1R 5′- GGGG GAC CAC TTT GTA CAA GAA AGC TGG GTC TA TGG CTC TTC CAC TAA ACC CG-3′; attB1-TPR2F 5′-GGGG ACA AGT TTG TAC AAA AAA GCA GGC TCC ATG GCG CTA TGG ATG GAC GCT GG-3′ and attB2-TPR2R 5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTC GTT TGG TGG AGT CCA TTT TCC AGC G-3′). AtTPR1 and AtTPR2 PCR products were cloned, as described for AtHsp90-2, into pDestAD (prey vector) to generate pADTPR1 and pADTPR2. Following sequence verification, pDBHsp90-2 and pADTPR1/pADTPR2 were transformed into Y8930 and Y8800 yeast strains, respectively. Transformants were selected on synthetic complete (SC) media lacking either leucine (bait vector) or tryptophan (prey vector). Bait and prey transformants were then mated and selected on SC media lacking leucine and tryptophan. The interaction of pDBHsp90-2 with pADTPR1/pADTPR2 was assayed based on the ability of cells to grow on SC-Leu-Trp-His plus 3 mM 3-aminotriazole (3AT).
In vitro protein-binding assay
Plasmid expressing AtHsp90-2 with a cleavable N-terminal glutathione-S-transferase (GST) tag in pGEX-6p-1 (Hsp90-GST) was kindly provided by Dr. David Hubert, University of North Carolina. Plasmid expressing Hsp90ΔMEEVD-GST was generated by PCR amplification and cloning of the PCR product into pGEX-6p-1 (GE Healthcare). The chitin-binding domain (CBD) affinity tag was fused to the C-terminus of TPR1 by cloning TPR1 cDNA in frame into pTYB2 (New England Biolabs) to prepare TPR1-CBD. Similarly, the GST tag was fused to the N-terminus of TPR2 by cloning TPR2 cDNA in frame into pGEX-6p-1 to prepare TPR2-GST fusion protein.
Hsp90-GST and Hsp90ΔMEEVD-GST were expressed in BL21 cells. Expressed proteins were purified on Glutathione-Sepharose 4B beads as per the protocol provided by GE Healthcare. The beads were washed once with PBS buffer containing 500 mM NaCl, five times with PBS buffer, and once with cleavage buffer (50 mM Tris, pH 7.0, 150 mM NaCl, 1 mM EDTA and 1 mM DTT). The beads were incubated at 4°C overnight with cleavage buffer containing PreScission Protease to cleave the GST tag. The cleaved protein was eluted with the cleavage buffer. Fresh Glutathione-Sepharose 4B beads were added to the eluate (cleaved Hsp90 and Hsp90ΔMEEVD) to remove the PreScission Protease. Purified Hsp90 and Hsp90ΔMEEVD were concentrated, dialyzed against 10 mM HEPES, pH 7.5, and 50 mM NaCl, and stored at −80°C until further use.
TPR1-CBD and TPR2-GST proteins were produced in E. coli strains ER2566 and BL21 cells and immobilized on chitin beads and Glutathione-Sepharose 4B beads, respectively. For in vitro binding assays, 50 µl aliquot of beads with immobilized proteins were incubated for 60 min on ice with 1 µg of purified Hsp90 or Hsp90ΔMEEVD in reaction buffer (20 mM HEPES, pH 7.5, 20 mM KCl, 1 mM MgCl2, 0.01% NP-40) containing either no nucleotide, 5 mM ADP or 5 mM ATP. After incubation the supernatant was removed and the beads were washed thrice with the reaction buffer containing either no nucleotide, 5 mM ADP or 5 mM ATP. Proteins retained on the beads were extracted into SDS-sample buffer, separated by SDS-PAGE and immunoblotted with the R2 anti-Hsp90 antibody [33].
Results
Identification of CC-TPR proteins in Arabidopsis
Orthologs of most of the known CC-TPR co-chaperones of animal and yeast Hsp90 have also been identified in plants. To understand the extensiveness of CC-TPR proteins in the model plant Arabidopsis, the InterPro database was first searched for TPR proteins. The initial big list of 235 TPR proteins was narrowed down to 52 proteins on the criterion of proteins possessing a Hop TPR2a-like three-motif domain. The start and end sites of each of the three motifs in the 52 proteins were identified and the motifs were then searched for the conserved residues (K5N9-N6-K2R6) responsible for interacting with the MEEVD motif of cytosolic Hsp90. This led to the identification of 24 proteins, including SQUINT, which appeared in the database as a two-motif TPR domain. Following from this example, proteins showing one or two motifs of TPR were manually searched for the conserved residues, leading to another 12 proteins being identified as CC-TPR proteins with potential to interact with Hsp90/Hsp70. Thus, a total of 36 CC-TPR proteins were identified in Arabidopsis (Table 1). Analyses of these genes in TAIR, which provides structural and functional annotation as well as links to different databases containing information of specific gene, transcript, and/or protein, led us to infer that 24 of the 36 genes in the list were novel, while 12 genes had been characterized previously in Arabidopsis or another plant species (Table 1).
Table 1. Properties of the Arabidopsis CC-TPR proteins.
| AGI ID | Name | Number of amino acids | Additional functional domains | Subcellular localization | mRNA species |
| One TPR domain | |||||
| AT4G30480 | AtTPR1 | 161/208/277 | C/N | 3 | |
| AT1G04130 | AtTPR2 | 360 | C/N | 1 | |
| AT1G04190 | AtTPR3 | 328 | C/N/M | 1 | |
| AT1G04530 | AtTPR4 | 310 | C | 1 | |
| AT1G56440 | AtTPR5 | 476 | N/P | 1 | |
| AT1G58450 | AtTPR6 | 164 | M/C | 1 | |
| AT5G21990 | AtTPR7 | 554 | M/N | 1 | |
| AT4G08320 | AtTPR8 | 426/427 | C/M/N/P | 2 | |
| AT1G33400 | AtTPR9 | 798 | SET | N | 1 |
| AT3G04710 | AtTPR10 | 455/456 | Ankyrin | P/C | 2 |
| AT2G25290 | AtPhox1* | 697 | Phox/PB1 | C/N | 1 |
| AT1G62390 | AtPhox2* | 751 | Phox/PB1 | C/M/N/P | 1 |
| AT5G20360 | AtPhox3* | 809 | Phox/PB1 | N | 1 |
| AT4G32070 | AtPhox4* | 811 | Phox/PB1 | C/N | 1 |
| AT4G22670 | AtTPR11 | 441 | Heat shock chaperonin-binding | V | 1 |
| AT2G15790 | AtSquint | 361 | Cyclophilin | C | 1 |
| AT3G54010 | AtPAS1 | 635/545 | Peptidyl-prolyl-cis-trans isomerase | C/N | 2 |
| AT3G25230 | AtROF1 | 551/562 | Peptidyl-prolyl-cis-trans isomerase | C | 2 |
| AT5G48570 | AtROF2 | 578 | Peptidyl-prolyl-cis-trans isomerase | C/Pe | 1 |
| AT3G21640 | AtTWD1 | 365 | Peptidyl-prolyl-cis-trans isomerase | PM | 1 |
| AT3G07370 | AtCHIP | 278 | U-box | C | 1 |
| AT2G42810 | AtPP5 | 484/538 | PP5 | C/N/ER | 2 |
| AT3G17970 | AtToc64-III | 589 | Amidase | P/M | 1 |
| AT5G09420 | AtToc64-V | 603 | Amidase | M | 1 |
| More than one TPR domain | |||||
| AT1G78120 | AtTPR12 | 530 | M | 1 | |
| AT5G10090 | AtTPR13 | 594 | M/N/P | 1 | |
| AT5G65160 | AtTPR14 | 593 | M/N | 1 | |
| AT2G41520 | AtTPR15 | 1077/1108 | DnaJ | P/N | 2 |
| AT5G12430 | AtTPR16 | 1165 | DnaJ | P/N | 1 |
| AT1G53300 | AtTTL1 | 699 | Thioredoxin | N/M/C | 1 |
| AT3G14950 | AtTTL2 | 721 | Thioredoxin | M/N/P | 1 |
| AT2G42580 | AtTTL3 | 691 | Thioredoxin | M/N/P | 1 |
| AT3G58620 | AtTTL4 | 682 | Thioredoxin | P/M/N | 1 |
| AT1G12270 | AtHop1 | 572 | Heat shock chaperonin-binding | C/N | 1 |
| AT1G62740 | AtHop2 | 571 | Heat shock chaperonin-binding | PM | 1 |
| AT4G12400 | AtHop3 | 530/558 | Heat shock chaperonin-binding | M/P/N | 2 |
The 24 new CC-TPR proteins are referred to as AtTPR1-16, AtPhox1-4 and AtTTL1-4, while the known CC-TPR proteins are referred to by their names.
*represents new names of the proteins. For subcellular localization the bolded represent experimental documentation, while the italicized represent the most significant according to computational predictions. C, cytoplasm; ER, endoplasmic reticulum; M, mitochondria; N, nucleus; P, plastid; Pe, peroxisome, PM, plasma membrane; V, vacuole.
As an additional search of TPR proteins in Arabidopsis, we used the HMMER program, which aims to detect remote homologs on the basis of mathematical models, to generate a list of proteins against each motif of the CC-TPR proteins. The number of proteins generated against each motif varied and each list of proteins (data not shown) could be divided into four categories: 1) All 36 proteins described in Table 1 were among the top hits identified for all three motifs. An additional 2-6 proteins were detected in this list of proteins, which on analysis fell in the second category of proteins; 2) True TPR domain proteins (entry no. IPR013026), but lacking a three-motif TPR domain and/or the conserved carboxylate clamp residues. These proteins had been analysed earlier and rejected; 3) TPR-like domain containing proteins (InterPro entry no. IPR011990). The motif structure of this domain varied in the number of amino acids in different proteins and there was no conservation of the carboxylate clamp residues; 4) Proteins with no TPR domain. In conclusion, the HMMER search identified the same 36 CC-TPR proteins, lending further support to the results shown in Table 1.
Previously known CC-TPR proteins, such as Hop (AtHop1, AtHop2 and AtHop3), immunophilins (AtROF1, AtROF2, PAS1, AtTWD1 and SQUINT), AtCHIP, TOC64 (AtTOC64-III, and AtTOC64-V) and AtPP5 were all present in the list represented in Table 1. Hop, immunophilins, CHIP and PP5 are known co-chaperones [5], while Translocase of the mitochondrial outer membrane (Tom70), the functional homolog of TOC64, is a known interactor of Hsp90 in yeast and animal systems [34]. AtROF1and AtROF2 [19], AtTWD1 [21], and pea TOC64 [23] have been demonstrated to interact with Hsp90, while soybean Hop [15] and tomato PP5 [16] have been shown to act as co-chaperones of Hsp90. Of the newly identified proteins, TTL1 has been associated with salt sensitivity and altered abscisic acid (ABA) responses [35], but no links have been made of this protein to the Hsp90/Hsp70 chaperone machinery. The closest protein to TTLs in other organisms is Tpr2, a relatively new co-chaperone of mammalian Hsp90 that contains a DnaJ-like domain instead of the thioredoxin domain [35]–[37].
In addition to identifying members of a gene family, transcript data from NCBI also provides information on the number of splice variants for each gene family member. It is noted in Table 1 that there are two splice variants for PAS1, AtROF1, AtPP5, AtHop3, AtTPR8, AtTPR10 and AtTPR15, and three splice variants for AtTPR1. The DnaJ-domain containing AtTPR15 has two CC-TPR domains both of which are present in the first variant, but the second variant appears to be missing the first motif of the second TPR domain. With the exception of AtPP5 that has been shown to contain two splice variants localized in the cytoplasm/nucleus and ER, respectively [16], the splice variants of all other genes remain to be verified experimentally.
Multiple sequence alignments of excised TPR motifs of proteins listed in Table 1 against human Hop TPR2a as reference sequence showed high degree of conservation of the consensus residues K5N9-N6-K2R6 (Table 2). In the case of proteins with more than one TPR domain (AtTTLs, AtTPR12-16), the domain with highest conservation was used for alignment. It can be seen in Table 2 that with one exception, the substitutions for K5 and K2 in the first and third motif, respectively, are mostly conservative, whereby Lys (K) is replaced with Arg (R). Since this substitution occurs in confirmed Hsp90 interactors such as Hop, it is concluded that this substitution does not interfere with the binding of the TPR domain to Hsp90/Hsp70. The substitution of Asn (N9) with Gln (Q) in the first motif is also conservative. All others replacements are radical according to classification of amino acids by volume and polarity, with the K5 to Glu (E) replacement in AtTPR4 being most noteworthy. With the exception of AtPhox1 and AtPhox4, the consensus residue N6 of the second motif is highly conserved in Arabidopsis proteins. It should be noted that Ala (A) and Met (M) substitute R6 in the third motif of known co-chaperones AtHop1-3 and AtCHIP, respectively, indicating that some radical changes are accommodated in the TPR:Hsp90/Hsp70 interaction. How such substitutions affect interaction with Hsp90/Hsp70 remains to be seen.
Table 2. Multiple sequence alignment of excised TPR motifs of Arabidopsis proteins.
| Name | AGI ID | Motif I | Motif II | Motif III | |||
| Helix 1A Helix 1B | Helix 2A Helix 2B | Helix 3A Helix3B | |||||
| Human Hop | 225 | ALKEKELGNDAYKKKDFDTALKHYDKAKELDPTN | 259 | MTYITNQAAVYFEKGDYNKCRELCEKAIEVGREN | 300 | AKAYARIGNSYFKEEKYKDAIHFYNKSLAEHRTP | |
| AtTPR1 | AT4G30480 | 105 | ANEAKAEGNKLFVNGLYEEALSKYAFALELVQEL | 146 | SICYLNRGVCFLKLGKCEETIKECTKALELNPTY | 180 | NKALVRRAEAHEKLEHFEDAVTDLKKILELDPSN |
| AtTPR2 | AT1G04130 | 31 | AIEFKEEGNECVRKGKKHYSEAIDCYTKAISQGV | 71 | SILFSNRSHVNLLLGNYRRALTDAEESMRLSPHN | 105 | VKAVYRAAKASMSLDLLNEAKSYCEKGIENDPSN |
| AtTPR3 | AT1G04190 | 15 | EKSLKEKGNEFFKAGNFLKAAALYTQAIKLDPSN | 49 | ATLYSNRAAAFLSLVKLSKALADAETTIKLNPQW | 83 | EKGYFRKGCVLEAMEKYEDALAAFEMALQYNPQS |
| AtTPR4 | AT1G04530 | 136 | PLLLKNYAKFLEYKGDLSGAEEYYHKCTVVEPSD | 170 | GVALANYGRLVMKLHQDEAKAMSYFERAVQASPD | 260 | GETLCRYAKAFWSINNDHEKALFYFEKAVEASPN |
| AtTPR5 | AT1G56440 | 84 | SSSEKEQGNEFFKQKKFNEAIDCYSRSIALSPNA | 118 | VTYANRAMAYLKIKRYREAEVDCTEALNLDDRY | 151 | IKAYSRRATARKELGMIKEAKEDAEFALRLEPES |
| AtTPR6 | AT1G58450 | 10 | ANRKKEEGNLLYKTQKYERAAKKYNKAAECIENG | 58 | VSCFLNGAACSLKLKNFLETIVLCSEVLDIEFQN | 92 | VKALYRRAQSYIEVGDLISAEMDINRALEADPEN |
| AtTPR7 | AT5G21990 | 103 | AQMLKKQGNELHSRGNFSDAAEKYLRAKNNLKEI | 146 | LACSLNLMSCYLKTNQHEECIKEGSEVLGYDARN | 180 | VKALYRRGQAYRDLGLFEDAVSDLSKAHEVSPED |
| AtTPR8 | AT4G08320 | 175 | AETLKCQGNKAMQSNLYLEAVELYSFAIALTDKN | 209 | AVFYCNRAAAYTQINMCSEAIKDCLKSIEIDPNY | 243 | SKAYSRLGLAYYAQGKYAEAIEKGFKKALLLDPH |
| AtTPR9 | AT1G33400 | 63 | SLDLKRRGNHCFRSRDFDEALRLYSKALRVAPLD | 106 | ASLFLNRANVLHNLGLLKESLRDCHRALRIDPYY | 140 | AKAWYRRGKLNTLLGNYKDAFRDITVSMSLESSL |
| AtTPR10 | AT3G04710 | 327 | AAEAKARGQDAFHRKDFQMAIDAYTQAIDFDPTD | 361 | HTLFSNRSLCWLRLGQAEHALSDAKACRELNPDW | 395 | PKGCFREGAALRLLQRFDEAANAFYEGVLLSPES |
| AtPhox1 | AT2G25290 | 52 | ALELKEEGNKLFQKRDYEGAMFRYDKAVKLLPRD | 90 | AYLRTSMASCYMQMGLGEYPNAINECNLALEASP | 126 | SKALLKRARCYEALNKLDFAFRDSRVVLNMEPEN |
| AtPhox2 | AT1G62390 | 51 | AHELKEEGNKKFQARDYVGALEQYENGIKLIPKS | 89 | AVFHSNRAACLMQMKPIDYESVISECSMALKSQP | 125 | TRALLRRARAFEAVGKFDLAVQDVNVLLGSDPNH |
| AtPhox3 | AT5G20360 | 126 | AQGLKEEGNKLFQKRDYDGAMFKYGEAIKILPKD | 164 | SHVRANVASCYMQLEPGEFAKAIHECDLALSVTP | 200 | NKALLKRARCYEALNKLDLALRDVCMVSKLDPKN |
| AtPhox4 | AT4G32070 | 51 | ALELKEEGNKLFQKRDHEGAMLSFDKALKLLPKD | 89 | AYLRTSMASCYMQMGLGEYPNAISECNLALEASP | 125 | SKALVRRSRCYEALNKLDYAFRDARIVLNMEPGN |
| AtTPR11 | AT4G22670 | 123 | AQEAKGKAMEALSEGNFDEAIEHLTRAITLNPTS | 157 | AIMYGNRASVYIKLKKPNAAIRDANAALEINPDS | 191 | AKGYKSRGMARAMLGEWAEAAKDLHLASTIDYDE |
| AtSquint | AT2G15790 | 212 | VDFVKAHGNEHFKKQDYKMALRKYRKALRYLDIC | 264 | SQIFTNSAACKLKFGDAKGALLDTEFAMRDEDNN | 298 | VKALFRQGQAYMALNNVDAAAESLEKALQFEPND |
| AtPAS1 | AT3G54010 | 310 | ADKIRSTGNRLFKEGKFELAKAKYEKVLREFNHV | 359 | NMLHLNVAACLLKMGEWRKSIETCNKVLEAKPGH | 393 | VKGLYRRGMAYIAGGEYDDARNDFNMMIKVDKSS |
| AtROF1 | AT3G25230 | 400 | ASKKKEEGNSKFKGGKYSLASKRYEKAVKFIEYD | 449 | VACNLNDAACKLKLKDYKQAEKLCTKVLELESTN | 483 | VKALYRRAQAYMELSDLDLAEFDVKKALEIDPNN |
| AtROF2 | AT5G48570 | 410 | AGKKKEEGNVLFKAGKYARASKRYERGVKYIEYD | 459 | IACNLNDAACKLKLKDYKEAAKLSTKVLEMDSRN | 493 | VKAMYRRAHAYLETADLDLAELDIKKALEIDPDN |
| AtTWD1 | AT3G21640 | 179 | ADRRKMDGNSLFKEEKLEEAMQQYEMAIAYMGDD | 230 | NPCHLNIAACLIKLKRYDEAIGHCNIVLTEEEKN | 264 | PKALFRRGKAKAELGQMDSARDDFRKAQKYAPDD |
| AtCHIP | AT3G07370 | 10 | AERLKEDGNNCFKKERFGAAIDAYTEAIALSPNV | 44 | PAYWTNRALCHMKRKDWTKVEEDCRKAIQLVHNS | 78 | VKAHYMLGLALLQKKEFTNGVKELQRALDLGRCS |
| AtPP5 | AT2G42810 | 13 | AEEFKSQANEAFKGHKYSSAIDLYTKAIELNSNN | 47 | AVYWANRAFAHTKLEEYGSAIQDASKAIEVDSRY | 80 | SKGYYRRGAAYLAMGKFKDALKDFQQVKRLSPND |
| AtToc64III | AT3G17970 | 474 | AEIAKEKGNQAFKEKLWQKAIGLYSEAIKLSDNN | 508 | HVLFSNRSAAHASLNHYDEALSDAKKTVELKPDW | 542 | VKAYLRRGTAREMLGDCKGAIEDFRYALVLEPNN |
| AtToc64V | AT5G09420 | 488 | SEVMKEKGNAAYKGKQWNKAVNFYTEAIKLNGAN | 522 | ATYYCNRAAAFLELCCFQQAEQDCTKAMLIDKKN | 556 | VKAYLRRGTARESLVRYKEAAADFRHALVLEPQN |
| AtTPR12 | AT1G78120 | 159 | PETLKKMGNEEYCRGRFGQALVFYERAISADPKT | 193 | PTYWPNKSAALISLGRLLEASDACEEALRLNPTY | 227 | ERAHQRLASLQLRLGEVEKALCHYNEAGKYTETK |
| AtTPR13 | AT5G10090 | 237 | PETLKIMGNEDYKNGNFAEALALYEAAISIDPKK | 271 | ASYRSNKSAALTALGRILEAVFECREAIRIDPHY | 305 | HRAHHRLANLYLRLGEVENSIYHFKHAGPEADQE |
| AtTPR14 | AT5G65160 | 470 | VTEARFKGNELFKSGRFQEACAAYGEGLDHDPRN | 504 | SVLLCNRAACRSKLGQFDKSIEDCTAALSVRPGY | 538 | GKARLRRADCNAKIEKWELAVGDYEILKKESPED |
| AtTPR15 | AT2G41520 | 533 | CEVWRLRGNQAYKNGYMSKAEECYTHGINSSPSK | 599 | ALCYGNRAAARISLGRLREAISDCEMAASLDPSY | 631 | IKAYMRAANCHLVLGELGSAVQYFNKCMKSTSSV |
| AtTPR16 | AT5G12430 | 608 | CEKWRLRGNNAYKIGDLSRAEESYTQGIDSVPRI | 652 | MLCYSNRAATRMALGRMREAIADCTMASSIDSNF | 686 | LKVQVRAANCYLSLGEIEDASRYFKKCLQSGSDI |
| AtTTL1 | AT1G53300 | 465 | VARARARGNDLYKSERYTEASSAYAEGLRLDPCN | 499 | AILYCNRAACWFKLGMWERSIEDCNQALRYQPSY | 533 | TKPLLRRAASNSKMERWGAAVSDYEALIRELPHD |
| AtTTL2 | AT3G14950 | 258 | PEEVKRFGNEMFRKGCFAEALKLYDRAIELSPSN | 292 | ATYHSNRAAALSSLGQIGEAVNECEIAIKLDPNF | 326 | ARAHHRLASLLLRLGYVDNAGIHLYSVEEPLDPT |
| AtTTL3 | AT2G42580 | 458 | VVRARTRGNELFSSGRFSEACVAYGDGLKQDDSN | 492 | SVLYCNRAACWYKLGLWEKSVEDCNHALKSQPSY | 526 | IKALLRRAASYGKLGRWEDAVKDYEFLRRELPGD |
| AtTTL4 | AT3G58620 | 449 | VAKARTRGNELFSSGRYSEASVAYGDGLKLDAFN | 483 | SVLYCNRAACWFKLGMWEKSVDDCNQALRIQPSY | 517 | TKALLRRAASYGKLGRWEDAVRDYEVLRKELPGD |
| AtHop1 | AT1G12270 | 244 | AKKEKELGNAAYKKKDFETAIQHYSTAIEIDDED | 278 | ISYLTNRAAVYLEMGKYNECIEDCNKAVERGREL | 323 | TRKGTALTKMAKCSKDYEPAIEAFQKALTEHRNP |
| AtHop2 | AT1G62740 | 243 | AQKEKELGNAAYKKKDFETAIQHYSTAMEIDDED | 277 | ISYITNRAAVHLEMGKYDECIKDCDKAVERGREL | 322 | TRKGTALGKMAKVSKDYEPVIQTYQKALTEHRNP |
| AtHop3 | AT4G12400 | 230 | ALKEKGEGNVAYKKKDFGRAVEHYTKAMELDDED | 264 | ISYLTNRAAVYLEMGKYEECIEDCDKAVERGREL | 309 | TRKGSALVKMARCSKDFEPAIETFQKALTEHRNP |
The conserved residues are indicated in bold, and the substitutions in bold-italics. The numbers on the left of each motif refer to the amino acid positions in the sequences of proteins.
A notable feature of AtTPR5 is that the conserved N is detected in the fifth position of the second motif as opposed to the sixth position. If, however, the second motif is initiated at residue 117 instead of 118 in the protein sequence, then the N residue falls in the right place. The perfect match of residues in the first and third TPR motifs of AtTPR5 to the consensus K5N9 and K2R6, respectively, as well as the presence of a relatively conserved R residue after N in the second motif, suggest that AtTPR5 qualifies as a CC-TPR protein.
In silico characterization of the newly identified CC-TPR proteins in Arabidopsis
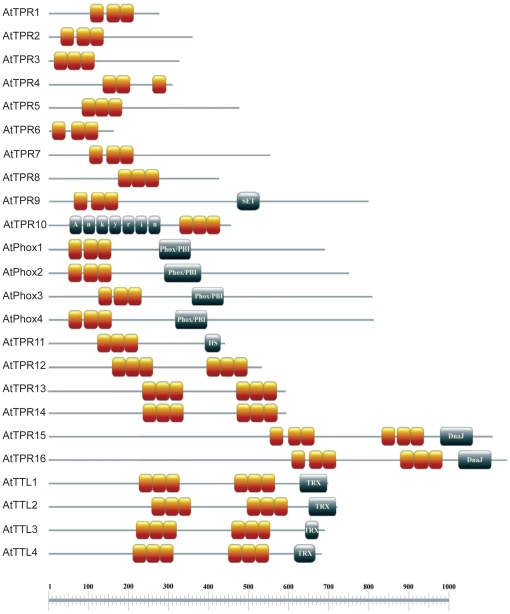
To obtain clues about the functions of the newly identified CC-TPR proteins, InterProScan against InterPro database was used to identify additional functional domains as well as subcellular localization signal sequences within these proteins. The known functional domains were highlighted on the output display page for each input FASTA sequence. Using this information the schematics of domain architecture of the new proteins were prepared (Figure 1). Eight of the proteins contained a single TPR domain, seven contained a single TPR domain plus another functional domain, three contained two TPR domains, and six contained two TPR domains plus another functional domain. The second functional domains in these proteins include SET, ankyrin, octicosapeptide/Phox/Bem1p (PB1), DnaJ, and thioredoxin (Figure 1). With the exception of DnaJ, these domains have not previously been linked with CC-TPR proteins.
Figure 1. Domain schematics of the newly identified CC-TPR proteins in Arabidopsis.
Each orange box represents one motif of the TPR domain, and the grey box represents another domain. The scale below indicates the size of the protein in number of amino acids. The schemes were generated using MyDomains from PROSITE (http://www.expasy.ch/tools/mydomains/).
The ankyrin repeat is a protein-protein interaction motif comprised of a tandemly repeated 33 amino acid sequence. Each repeat folds into a helix-loop-helix structure [38]. Ankyrin repeats are present in functionally diverse proteins involved in transcription initiation, cell-cycle regulation, ion transportation and signal transduction. This domain was identified in AtTPR10, which is predicted to localize to the plastid (Table 1).
As a conserved sequence motif made up of 130–140 amino acids, the SET domain occurs as a part of multidomain proteins involved in histone methylation, which regulates chromatin structure and gene transcription [39]. These enzymes use S-adenosylmethionine (AdoMet) as a donor substrate and add methyl groups to lysine residues of histone H3. SET domain proteins can also methylate non-histone proteins [40]. Recently it was shown that a human protein called SMYD2 interacts with Hsp90α, and that this interaction enhances the histone methyltransferase activity of SMYD2 [41]. The presence of the SET domain in AtTPR9 and its predicted localization to the nucleus (Table 1) suggest that this protein may be involved in methylating histone or non-histone proteins.
AtPhox1 to AtPhox4 constitute a gene family (Figure S1). In addition to the TPR domain, the AtPhox proteins contain the Phox/PB1 domain (Figure 1), which is known to mediate protein-protein interactions in cell processes such as cell polarity, pheromone signaling, cytoskeletal organization, osteoclastogenesis, angiogenesis and early cardiovascular development [42]. Heterodimerization of two PB1 domains is important in the formation of macromolecular signaling complexes. PB1 domains can also interact with other protein domains. The presence of Ser (S) in place of the consensus N6 in the second TPR motif of AtPhox1 and AtPhox4 (Table 2) warrants investigation of this substitution in the context of Hsp90/Hsp70 binding.
The TTLs (AtTTL1-4) are a novel protein family (Figure S2) unique to plants that in addition to the TPR domain contain a motif in the C-terminus with homology to thioredoxins. The thioredoxin fold is involved in the regulation of protein activity by changes in the redox state of thiol groups (S2 to SH2) [43]. The localization of these proteins to plastid/mitochondria where redox reactions are common is consistent with their possessing a thioredoxin domain.
Co-presence of TPR and DnaJ domains in a CC-TPR protein has been noted before in the human Tpr2 co-chaperone [36]. Two of the newly identified CC-TPR proteins AtTPR15 and AtTPR16 contain the DnaJ domain, which mediates protein-protein interactions and various chaperone functions [44].
The heat shock chaperonin-binding motif found in Hop is for binding heat shock proteins. This domain is found singly or duplicated in proteins [45]. The newly identified protein AtTPR11 contains this domain. As would be expected, transcripts of proteins with this domain are induced by high temperature.
Identification of CC-TPR proteins in rice
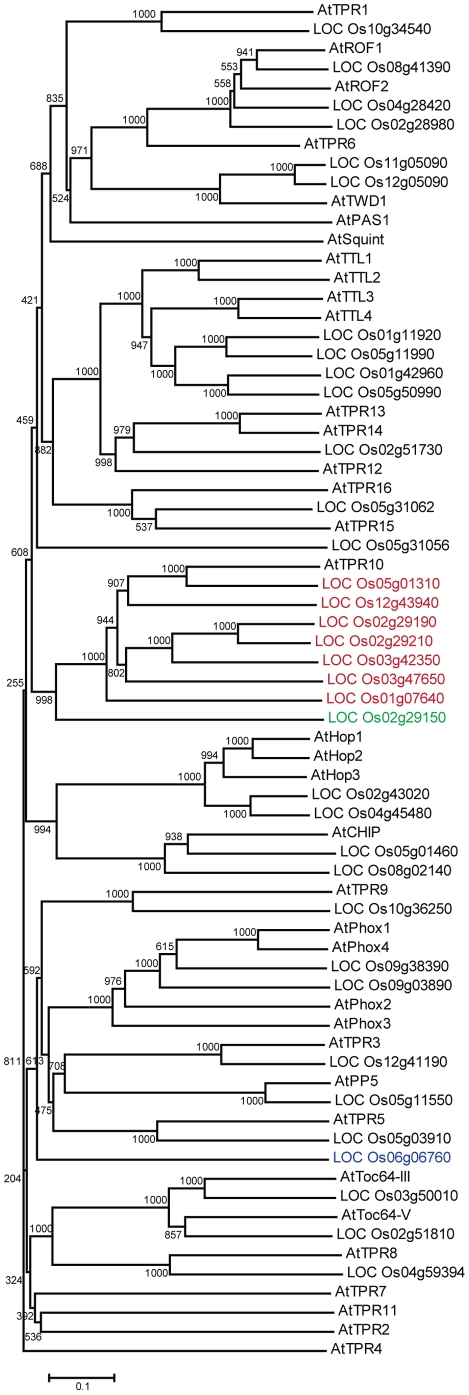
The HMM profile created with Arabidopsis sequences was used to search for CC-TPR proteins in rice. The top hits for each of the motifs were searched for the conserved carboxylate clamp residues. Sequence alignments of each of the three motifs of the top scoring CC-TPR proteins in rice are shown in Table 3. In case of proteins with more than one three-motif TPR domain, the most conserved domain was used for alignment. Proteins with both conservative and non-conservative amino acid substitutions in the carboxylate clamp residues were identified. The protein sequences of CC-TPR proteins identified in Arabidopsis (Table 1) and rice (Table 3) were used to generate a phylogenetic tree (Figure 2). The various additional domains detected in Arabidopsis CC-TPR proteins were also found in rice CC-TPR proteins. In addition, we found that unlike in Arabidopsis, rice contains numerous ankyrin containing CC-TPR proteins (Figure 2, highlighted in red), one CC-TPR protein with FBD and F-box (highlighted in green), and one CC-TPR protein with protein kinase (STYKc) and U-box (highlighted in blue). The latter two proteins in rice likely expand the ability of the Hsp90/Hsp70 system to link cellular proteins to the ubiquitin proteasome system.
Table 3. Multiple sequence alignment of excised TPR motifs of rice proteins.
| Locus ID | Motif 1 | Motif 2 | Motif 3 | |||
| Os10g34540 | 64 | ANDAKAEGNKFFGAGEYERALSQYETALQIAAEL | 105 | SACHSNRAVCFLKLGKYDETIKECTKALELNPSY | 139 | LKALLRRGEAHEKLEHYDEAIADMKKIIELDPSN |
| Os12g41190 | 14 | SAALKDQGNEQFKAGNYLKAAALYTQAIKLDPDN | 48 | PTLYSNRAAAFLHLVKLNKALADADTTIKLKPQW | 82 | EKGHFRKGCVLESMEHYEEAISSFQIALQHNPQN |
| Os05g03910 | 40 | AASEKEQGNEYFKQKKFAQAIECYSRSIGLSPSA | 74 | VAFANRAMAYLKLRRFEEAENDCTEALNLDDRY | 107 | VKAYSRRITARKELGKLKEAMDDAEFAVSIDPNN |
| Os04g59394 | 181 | AEFFKSKGNEFMRSKQHLKAVELYTCAIALSRNN | 215 | AIYYCNRAAAYTLLNMFNEAVEDCLKSIEIDPNY | 249 | SKAYSRLGSAYFALGKYHDALYKGYLKASELDPS |
| Os10g36250 | 63 | AAELKGKGNACFSKREFEQALGFYSQALRYFPIS | 106 | ATLYVNRASTMHKLGLLEECLRDCDRAISVSPNY | 140 | AKAWYRRGMVNASFRNYSSSIHDLEVALSMEVTS |
| Os05g01310 | 329 | SLEAKSRGDDAFRNKDYLVAVDAYTQAIELNPND | 363 | ATLHSNRSLCWLRAGQAERALEDARACRALRPDW | 397 | AKACYREGAALRLLQRFEEAANAFYEGVQLEPEN |
| Os12g43940 | 323 | RSDMKQQGDAAFKKQDYLNASVFYTQALKVDPFD | 357 | GTLFSNRSLCWLRMGDGERALDDANACEKLRPKW | 391 | AKSYYRQGAALMFLKEYERAHRALGRALELDPES |
| Os02g29190 | 228 | ATDLKSLGNKAVEKKDYLSATGFYSKALYLYPDD | 262 | ATLFSNRSLCWHRMGDGGKALLDAHECRKLRSDW | 296 | PKAYYRLGAALMLLKDYESACEALYNGFKLDPGN |
| Os02g29210 | 247 | ATELKSLGNKAVEKKDYLSATGFYSKALDLYPDD | 281 | ATLFSNRSLCWHHMGNGGKALLDAYECRKLRPDW | 315 | PKAYYRQGAALMLLKDYESACETLYDGLKLDPGN |
| Os03g42350 | 273 | IAEFKSLGLEAVEKKDYLSAAGFYSEAMDLDPDD | 307 | ATLLSNRSLCWLYLGEGGKALVDAHKCRKMRPDW | 341 | PKACYRQGAALMLLKDYVSACEALFDGFKLDPED |
| Os03g47650 | 343 | KAQLKSLGASAVQGKDYVGASKFYSEAIQLDPTD | 377 | ATLHSNRSFCYLKSGEAREALVDAKTCIGLKPDW | 411 | PKGYYRKGAALMSLKEYKEACDAFMDGVKLDPAS |
| Os01g07640 | 318 | KDELKLQGNSSFNNEDYDAAILLYSMAMKFDNTD | 352 | AKLYSNRSACWLNLGIGDEALSDAQICSKMQPDW | 386 | AKGYYRQGMAFSLLQDYASASYVLRRALKLDPQN |
| Os09g38390 | 45 | AQELKEEGNKLFQRREHERALLNYEKAIKLLPRG | 83 | AYLHSNLAACYMQMSPPDHYRAINECNLALDASP | 119 | SKALLKRARCFEALGRLDLAYRDVAKVLAVEPNN |
| Os09g03890 | 53 | AIELKDEGTRLFQRRDYEEAAIKFGEAIKLLPKE | 91 | AFLHCNAAACYMHMNPEDLDHAIEECNLALEASP | 127 | TKALLKRARCFEALDKLDLACKDVQKVLSLEPSN |
| Os08g41390 | 406 | AGAKKEEGNALFKLGKYVRASKRYEKAAKFIEYD | 455 | VTCNLNNAACKLKLKDYKQAEKLCTKVLELDSQN | 489 | VKALYRRAQAYMQLADLELAEVDIKKALEIDPDN |
| Os04g28420 | 411 | AAKKKDEGNVWFKMGKYAKASKRYEKAAKYIEYD | 460 | VSCKLNNAACKLKLKEYREAEKLCTKVLELESTN | 494 | VKALYRRTQAYIELADLELAELDVKKALEIDPDN |
| Os02g28980 | 456 | AAKKKDEGNAWFKMEKYARASKRYGKALNFIQYD | 505 | VSCKLNNAACKLKLKDYKEAKELCTEVLELDSMN | 539 | VKAFYRRAQAHMYLVDFDLAELDIKKALEIDPDN |
| Os11g05090 | 191 | ADRRKIEGNEYFKEKKFEEAMQQYEMAIAYMGDD | 242 | NPCHLNMAACLIKLKRFDEAIAQCSIVLAEDENN | 276 | VKALFRRGKARAELGQTESAREDFLKAKKHSPED |
| Os12g05090 | 186 | ADRRKIEGNEYFKEKKFEEAMQQYEMAIAYMGDD | 237 | NPCHLNMAACLIKLKRFDEAIAQCTIVLSEDENN | 271 | VKALFRRGKARAELGQTESAREDFLKAKKYSPED |
| Os05g01460 | 68 | AELRRIEGNSCFNKARLGAAIDCYTEAIALCPDV | 102 | AVYWLNRGLCHFKRKEWAKVEEDSRRALALDDTL | 136 | VKGHYLLGCAMLEKEQCALAIKEFNKALDLLKSS |
| Os08g02140 | 14 | AELLKQEGNAFFKKDRISAAIDAYTGAIALCPKV | 48 | AVYWTNRALCYKRRNEWVRAEEDCRTAIQLDSHS | 82 | VKAHYMLGLALLNKDELAEGIKELEKSLELGRGA |
| Os06g06760 | 154 | ADHHRDRGNDFFKQKRYQEAAMHYTEAMKKNPKD | 188 | PRVFSNRAQCHIYLGALPEGLEDADKCIALDPTF | 222 | LKGYLRKAKVQLLMGNYEIALATYVEGLKCDPNN |
| Os05g11550 | 12 | SEELKLKANDAFKANKFSLAIELYSQAIELNSSN | 46 | AVYWANRAFAHTKLEEYGSAVQDASKAIEIDARY | 80 | SKGYYRRGAAYLAMGKFKEALKDFQQVKRISPND |
| Os03g50010 | 470 | AEAAKEKGNIAFKEKQWQKAINFYTEAIKLNNKV | 504 | ATYYSNRAAAFLELASYRQAEADCTSAIDIDPKI | 538 | VKAYLRRGTAREMLGYYKEAVDDFSHALVLEPMN |
| Os02g51810 | 497 | AELLKEKGNSAFKGRKWSKAVEFYSDAIKLNGTN | 531 | ATYYSNRAAAYLELGRYKQAEADCEQALLLDKKN | 565 | VKAYLRRGIAREAVLNHQEALQDIRHALALEPQN |
| Os02g51730 | 225 | PEKLKEMGNEEYREGHYAEAVALYDQAIMVDPTR | 259 | PAYWSNKAAALAALGRLIEAVGDCREAVRIDPSY | 293 | GRAHHRLGGLYLRLGEPDKAIHHFKQSANDSTGA |
| Os05g31062 | 98 | LLSHKAAGNEAFQARRYSEAVEQYSAALARNSDS | 136 | AVCFCNRAASYQALGQVTDAIADCSLAMVLDATY | 170 | LKAISRRATLYEMIRDYGQAANDLRKLISLIEKQ |
| Os05g31056 | 538 | CETWRTSGNQAYTNGHFATAEEYYTRGINSVSGH | 582 | MLCYSNRAATRMSLGRMREALQDCLIATSIDPTF | 616 | LKAKVRAANCQLALGDLEDALRSYTACLTSSKTS |
| Os01g11920 | 438 | VARARSLGNELFNSGKFSEACLAYGEGLKHHPVN | 472 | PVLYCNRAACRFKLGQWEKSIEDCNEALKIQPNY | 506 | PKALLRRAASYGKMERWAESVKDYEVLRKELPGD |
| Os05g11990 | 247 | VAQARTLGNELFHSGKFAEAFLAYGEGLKHHPAN | 281 | SVLYCNRAACMFKLGQWEKSIEDCNEALKIQPNY | 315 | WKALLRRAASYGKIEQWADSVKDYEVLRRELPGD |
| Os01g42960 | 215 | VAKARAQGNELYKAAKFSDASIAYSEGLKYEPSN | 249 | PVLYCNRAACWGKLERWEKAVDDCNEALRIQPNY | 283 | TKALLRRASSYAKLERWADCVRDYEVLHKELPAD |
| Os05g50990 | 218 | LQEVTRAGNEWYKKGHYGEALRHYDQAVALCPDS | 252 | AACRSNRAAALIGLGRLAEALRECEEAIRRDPAS | 286 | GRAHSRLAALCLRFGMVERAREHFMLAGQVNQSD |
| Os02g43020 | 2 | ADEAKAKGNAAFSAGRYEEAARHFTDAIALAPGN | 36 | HVLYSNRSAALASVHRYSEALADAEKTVELKPDW | 70 | AKGYSRLGAAHLGLGDAASAVAAYEKGLALDPTN |
| Os04g45480 | 2 | ADEAKAKGNAAFSAGRFEEAAAHFTDAIALAPDN | 36 | HVLYSNRSAAYASLHRYPEALADAERTVALRPDW | 70 | AKGCSRLGAARLGLGDAAGAVAAYEKGLALEPSN |
| Os02g29150 | 533 | ATELKSLGNKAVEKKDYLSATGFYSQAVDLYPDD | 567 | ATLFSNRSLCWHHMGDGHKALLDAYECRKLRPDW | 601 | LKAYYRQGAALMLLKDYESACETLYDGFKLDPGN |
The conserved residues are indicated in bold, and the substitutions in bold-italics. The numbers on the left of each motif refer to the amino acid positions in the sequences of proteins.
Figure 2. Phylogenetic tree showing sequence relationships between the CC-TPR proteins from rice and Arabidopsis.
Full-length protein sequences were aligned using Clustal X 2.0.10. A phylogenetic tree was then derived using the neighbor-joining method in Clustal X. The numbers indicate bootstrap values based on 1000 replicates. The ankyrin containing proteins are highlighted in red, the protein with FBD and F-box is highlighted in green, and the protein with protein kinase (STYKc) and U-box is highlighted in blue.
Expression patterns of CC-TPR proteins in Arabidopsis
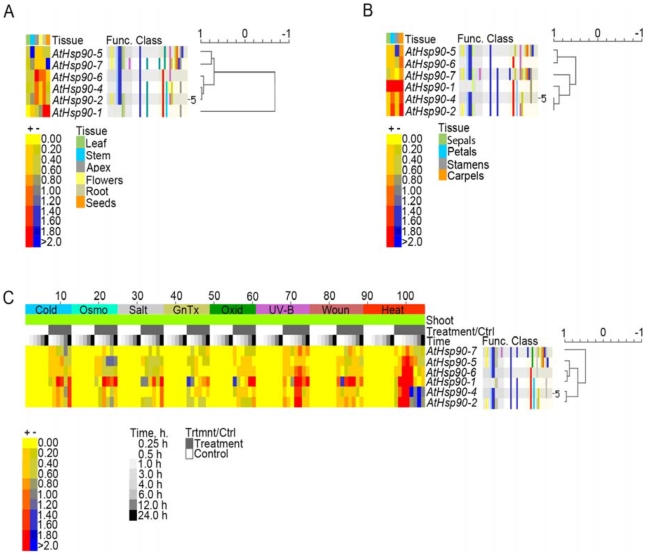
A collaborative microarray project (based on Affymetrix ATH1 arrays), dubbed AtGenExpress, has utilized 79 different Arabidopsis samples in triplicate to generate expression data [29], [30]. We rationalized that the expression patterns of Hsp90 co-chaperones must overlap to some degree with the expression patterns of Hsp90 family members. Data to this end could help formulate functional hypotheses for these proteins in the context of the Hsp90/Hsp70 chaperone machinery. We first performed e-Northern analysis with the Expression Browser at BAR using the AtGenExpress extended tissue and stress series data sets for the AtHsp90 gene family. The Arabidopsis Hsp90 gene family members are referred to as AtHsp90-1 (AT5G52640), AtHsp90-2 (AT5G56030), AtHsp90-3 (AT5G56010), AtHsp90-4 (AT5G56000), AtHsp90-5 (AT2G04030), AtHsp90-6 (AT3G07770), and AtHsp90-7 (AT4G24190). AtHsp90-1 to AtHsp90-4 are cytoplasmic, AtHsp90-5 is plastidial, AtHsp90-6 and AtHsp90-7 are localized to the mitochondria and ER, respectively [10]. Of these, AtHsp90-3 was not present on the ATH1 array and is therefore missing from our analysis.
When absolute expression values retrieved by AVT were used to compare the expression levels of AtHsp90 gene family members, AtHsp90-4 was found to have the highest level of constitutive expression, and AtHsp90-1 to be most responsive to HS (data not shown). For e-Northern data from BAR depicting tissue expression, the median level of expression across all samples displayed for a particular gene is used as control value for calculating the relative level. Data in Figure 3A shows that with the exception of AtHsp90-1, which is highly expressed in seeds, most other members have higher expression in the shoot apex, followed by roots and flowers. Further dissection of the flower into sepals, petals, stamens and carpels revealed that in general carpels and petals have higher levels of AtHsp90 transcripts as compared to the other two floral organs (Figure 3B). AtHsp90 genes are induced by various abiotic stresses, such as cold, salt, UV-B, etc., although maximum induction is in response to HS (Figure 3C). These data are in accordance with our earlier observations in Brassica napus of the relatively high expression of Hsp90 in apex, flowers and seeds, as well as of its responsiveness to cold temperatures [46].
Figure 3. Expression profiles of AtHsp90 genes.
(A) e-Northern results for expression of AtHsp90 genes in root, stem (second internode), leaf (cauline leaves), apex (shoot apex; inflorescence), flowers (stage 12), and seeds (stage 10, without siliques). (B) e-Northern results for expression of AtHsp90 genes in individual floral organs (flowers stage 12), sepals, petals, stamens and carpels. (C) e-Northern results for expression of AtHsp90 genes in response to different abiotic stresses. The control is mock treatment at each time point. The colour scale indicates the log2-level of expression above or below the median. Dark red indicates more than 4-fold above the median, while dark blue indicates 4-fold below. The clustering tree can be seen to the right of the heatmap.
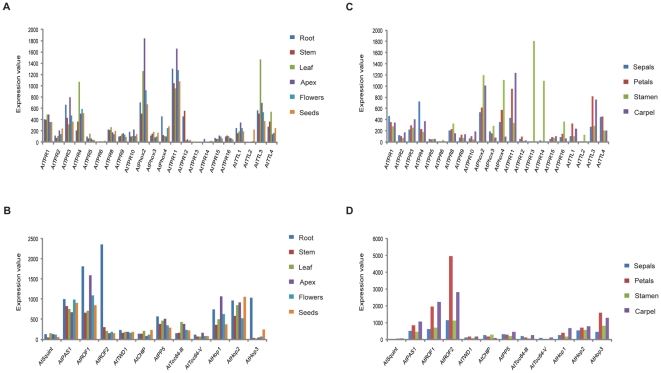
To compare the expression levels of CC-TPR genes (AtTPR7 and AtPhox1 were not present on the ATH1 array) in different plant parts, the absolute expression values retrieved by AVT were plotted (Figure 4). This was done because the low level expression of some TPR genes led to artifacts in the output of the Expression Browser. Several conclusions can be drawn from the data represented in Figure 4: 1) most genes are expressed in different plant parts, albeit at much lower levels than the highly expressed AtHsp90-4; 2) of the newly identified genes, AtTPR11 and AtPhox2 are expressed at relatively high levels in most tissues (Figure 4A), suggesting that the encoded proteins may be general co-chaperones of Hsp90/Hsp70; 3) of the known co-chaperones, immunophilins (AtPAS1, AtROF1 and 2) and Hop (AtHop1, 2 and 3), are expressed at relatively high levels (Figure 4B) and, like their animal counterparts, are abundant co-chaperones of plant Hsp90. The near to exclusive expression of AtTPR13 and AtTPR14 in stamens, and the relatively high expression of AtPhox genes, AtTPR16, AtTTL2, AtTPR6 and AtTPR8 in stamens as compared to other floral organs (Figure 4C) is noteworthy. The immunophilins and AtHop, similar to AtHsp90, have predominant expression in petals and carpels (Figure 4D). Together, these data indicate that the Hsp90/Hsp70 chaperone machinery components are expressed in all plant tissues, albeit at different levels. It is possible that some components may function as general co-chaperones, while others may have tissue-specific functions within the context of the Hsp90/Hsp70 chaperone machinery. The expression profiles of the newly identified CC-TPR proteins provide an initial working hypothesis for delineating their functions.
Figure 4. Expression profiles of Arabidopsis CC-TPR genes in different plant parts.
Absolute expression values were retrieved by AVT and plotted as such. The developmental stage with the highest expression value was used for each transcript. Expression profiles of new CC-TPR genes (A) and of known CC-TPR co-chaperones (B) for root, stem, leaf, apex, flowers and seeds. Expression profiles of new CC-TPR genes (C) and of known CC-TPR co-chaperones (D) for individual floral organs: sepals, petals, stamens and carpels.
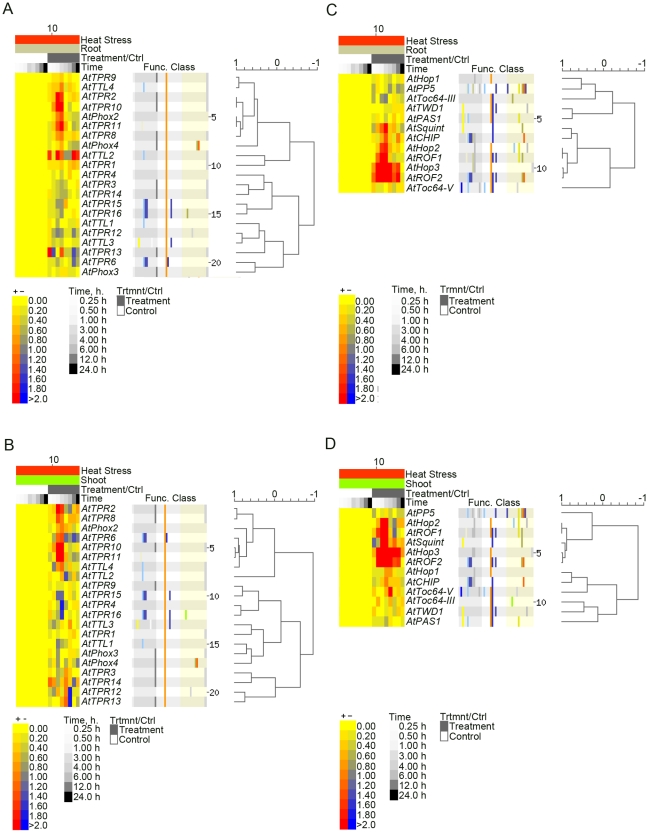
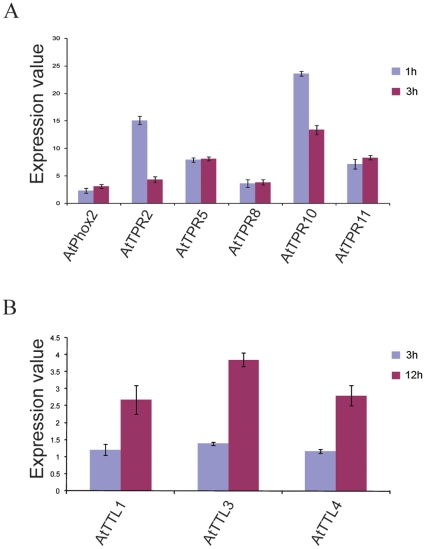
e-Northern analysis of CC-TPR genes by BAR and AVT using the stress series data set indicated that heat is the most prominent signal for induction of transcript abundance. AtTPR2, AtTPR5 (by AVT), AtTPR8, AtTPR10, AtPhox2 and AtTPR11 are heat-responsive in both root and shoot (Figure 5A, B). The fold-change values for AtTTL2, AtTPR13 and AtTPR14 in the output of Figure 5 are unreliable due to their low expression; these genes are therefore not considered as heat-responsive. AtROF1, AtROF2, AtHop2, AtHop3 and AtSquint are also heat-induced both in root and shoot (Figure 5C, D), an attribute that has been noted before [15], [19]. The heat-induced expression patterns of CC-TPR genes are similar to that of AtHsp90-1. This observation supports a co-chaperone role for the encoded proteins during HS. The co-expression of this subset of genes with AtHsp90-1 was confirmed using Expression Angler. AtHsp90-1 was used as query in AtGenExpress stress series, and the data generated by Expression Angler was searched for the presence of CC-TPR genes during HS. AtROF1, AtROF2, AtHop2, AtHop3, AtTPR2, AtTPR10 and AtTPR11 were within the top 156 proteins that are HS-induced and co-expressed with AtHsp90-1 (data not shown). As a final confirmation, the HS response of AtTPR2, AtTPR5, AtTPR8, AtPhox2, AtTPR10 and AtTPR11 in leaf tissue was checked by qRT-PCR. All of these genes showed induction of expression in response to heat (Figure 6A).
Figure 5. Expression profiles of Arabidopsis CC-TPR genes in response to HS.
e-Northern results for expression of new CC-TPR genes in roots (A) and shoots (B) of seedlings exposed to HS. e-Northern results for expression of known CC-TPR co-chaperones in roots (C) and shoots (D) of seedlings exposed to HS.
Figure 6. Transcript expression analysis by qRT-PCR in response to HS and BR treatment.
(A) For HS treatment, 10 day-old Arabidopsis seedlings were exposed to 38°C for 1 and 3 h at which time the plant tissue above the medium was collected and quick-frozen for RNA isolation. (B) BR treatment was given to 21 day-old seedlings for 3 and 12 h and the tissue was collected as in A. Transcripts were analyzed by qRT-PCR. Bars indicate mean ± SD.
Analysis of the AtGenExpress data set revealed some interesting information. One was the stress and ABA-responsive expression of the AtPhox gene family. AtPhox2 was found induced by heat (Figure 5), AtPhox3 by cold and salt stress, and AtPhox4 by cold, osmotic and salt stress, as well as pathogen infection. In accordance with the role of ABA in stress tolerance [47], AtPhox3 and 4 were found to be ABA-responsive (data can be retrieved by AVT). The second interesting observation was the brassinosteroid (BR) responsive expression of the AtTTL gene family. Three out of four TTL genes that have wide spread tissue expression, were found to be most responsive to BR as compared to other hormones. Since BR is a relatively new hormone [48], [49], the list of BR-regulated genes is still growing. To add AtTTL genes to this list we confirmed the BR-responsive expression of AtTTL1, AtTTL3 and AtTTL4 in Arabidopsis seedlings by qRT-PCR. All three genes showed 2 to 3-fold induction by BR at 12 h (Figure 6B). These results strongly suggest that AtTTL1, 3 and 4 are BR response genes.
Expression patterns of CC-TPR proteins in rice
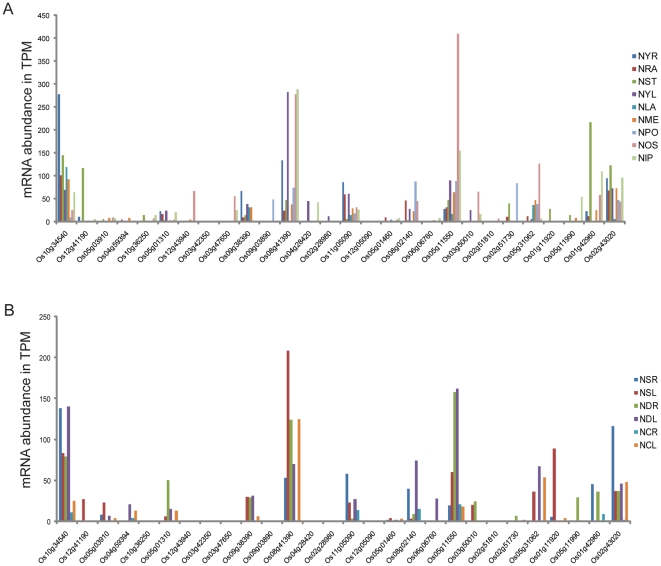
We searched tissue-specific and stress-related libraries in the rice MPSS database (http://mpss.udel.edu/rice/) using 17 nucleotides long signatures to obtain information on the relative transcript abundance of rice CC-TPR genes. Of the 35 rice CC-TPR genes being reported here, no expression was detected for seven genes (Os02g29190, Os02g29210, Os01g07640, Os05g31056, Os05g50990, Os04g45480 and Os02g29150) in the tested conditions. Genes that were widely expressed (several tissues) at relatively high levels (Figure 7A) and most strongly induced by stress (salinity, drought and cold) (Figure 7B) are Os10g34540 (AtTPR1), Os08g41390 (AtROF1), Os05g11550 (AtPP5), and Os02g43020 (AtHop). The most closely related Arabidopsis proteins are given in parentheses. Tissues that showed highest transcript abundance in general are ovary and mature stigma (NOS), stem (NST), immature panicle (NIP), young leaves (NYL), and young root (NYR). Since we had observed a subset of Arabidopsis CC-TPR genes to be expressed exclusively or at the highest level in stamens, we paid close attention to the rice genes expressed similarly in pollen. Based on maximal expression of Os02g51730 (closest to AtTPR13/AtTPR14, Figure 2) in pollen as compared to its expression in other tissues (Figure 7A), and the same of AtTPR13/AtTPR14 in stamens (Figure 4C), it is tempting to speculate that Os02g51730 is a functional ortholog of AtTPR13/AtTPR14. Similarly, salt and drought -responsive expression of Os01g11920, Os05g11990 and Os01g42960 (Figure 7B), together with their sequence and structural similarities with AtTTLs (Figure 2), suggest that these proteins are functional orthologs of each other. The protein with the unique combination of CC-TPR, protein kinase, and U-box domains (Os06g06760) was found expressed at low levels in immature panicle and to be induced by drought stress.
Figure 7. Transcript abundance of rice CC-TPR genes in tissue-specific and salinity, drought and cold -specific MPSS libraries.
(A) Transcript abundance in NYR (young roots, 14 days); NRA (mature roots, 60 days) replicate A; NST (mature stem, 60 days); NYL (young leaves, 14 days); NLA (mature leaves, 60 days) replicate A; NME (crown vegetative meristematic tissue, 60 days); NPO (mature pollen); NOS (ovary and mature stigma); and NIP (immature panicle, 90 days). (B) Transcript abundance in NSR (young roots, 14 days) and NSL (young leaves, 14 days) stressed by 250 mM NaCl for 24 h; NDR (young roots) and NDL (young leaves) stressed by drought for 5 days; NCR (young roots) and NCL (young leaves) stressed by 4°C cold for 24 h.
AtTPR1 and AtTPR2 interact with Hsp90 by the molecular ‘clamp’ mechanism
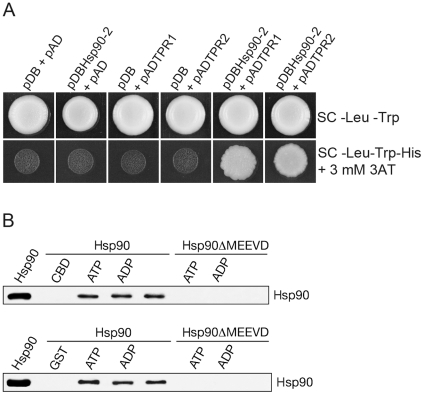
We applied yeast two-hybrid and in vitro binding assays as a proof-of-concept trial for analysis of the newly identified proteins for binding to Hsp90. AtHsp90-2 fused to the DNA-binding domain (pDBHsp90-2) and AtTPR1/TPR2 fused to the activation domain (pADTPR1/TPR2) were co-expressed in yeast. Growth of yeast on selection medium when co-transformed with pDBHsp90-2and pADTPR1/TPR2 only, but no other plasmid combination (Figure 8A), indicated positive interaction between Hsp90 and AtTPR1/TPR2. This interaction was further validated in in vitro binding assays. TPR1-CBD and TPR2-GST immobilized on chitin and Glutathione Sepharose 4B beads, respectively, were incubated with purified recombinant AtHsp90-2 in the absence or presence of ATP and ADP. Both TPR1-CBD (Figure 8B, top panel) and TPR2-GST (bottom panel) bound Hsp90 in a nucleotide independent manner, but no binding was seen to CBD or GST alone. Neither TPR1-CBD nor TPR2-GST bound to Hsp90ΔMEEVD, confirming that the interaction requires MEEVD for carboxylate clamp formation. These results provide evidence for the validity and success of our in silico strategy of identifying TPR proteins with potential to interact with Hsp90/Hsp70.
Figure 8. Interaction of AtTPR1 and AtTPR2 with Hsp90.
(A) Positive interaction between AtHsp90-2 and AtTPR1/AtTPR2 in yeast, resulting in the activation of reporter genes, was detected by growth on SC –Leu –Trp –His + 3 mM 3AT (lower panel). No growth on this medium was observed for plasmid combinations (as indicated in the figure) when either AtHsp90-2 or AtTPR1/AtTPR2 or both were absent. Yeast cells were grown on SC –Leu –Trp to select for both pDB and pAD plasmids (upper panel). (B) In vitro binding of recombinant Hsp90 or Hsp90ΔMEEVD to immobilized TPR1-CBD (upper panel) and TPR2-GST (lower panel) in the absence of any nucleotide or in the presence of 5 mM ADP or 5 mM ATP. After formation of complexes, washing of the beads and elution of proteins, Hsp90 was detected by immunoblotting. An aliquot of purified Hsp90 was run on the gel to mark the position of Hsp90 (extreme left). Controls were immobilized CBD and GST incubated with Hsp90.
Discussion
Identification of new CC-TPR proteins in Arabidopsis and rice by in silico methods
Co-chaperones are an integral part of the Hsp90/Hsp70 chaperone protein folding machinery, which has key roles in numerous cellular processes [1]–[5]. The known CC-TPR co-chaperones (Hop, immunophilins, PP5, CHIP, TOM70, Tpr2) bind to the MEEVD motif of Hsp90 and regulate the functions of Hsp90 [5], [8]. Orthologs of these are present in plants, and mutations in some have shown striking phenotypes. Due to the significance of the CC-TPR co-chaperones in the biochemical regulation of the Hsp90/Hsp70 machinery, we set out to uncover all CC-TPR proteins encoded by the model plant Arabidopsis genome based on the information available through studies in animal and yeast systems. The confirmation of two of the 24 newly identified CC-TPR candidates to bind to the MEEVD motif of Hsp90 indicates a high probability for these proteins as bona fide interactors of either Hsp90 or Hsp70 or both. This data set provides a useful framework that will accelerate studies of the Hsp90/Hsp70 chaperone machinery in plants, as well as of the newly identified CC-TPR proteins, most of which have no assigned functions.
To predict the CC-TPR co-chaperone network in Arabidopsis, sequences with the well characterized protein interaction domain called ‘TPR’ were extracted from the InterPro database, followed by careful identification of the conserved residues involved in binding with the MEEVD motif of Hsp90 [8]. Within this protein set, 12 proteins are orthologs of previously known co-chaperones of Hsp90/Hsp70, and 24 are new proteins with potential to serve as co-chaperones of Hsp90/Hsp70. A similar search in rice identified 35 CC-TPR proteins. The presence of all CC-TPR proteins (GmHop, AtROF1, AtROF2, AtTWD1, AtCHIP, AtSQUINT, LePP5 and AtTOC64) previously characterized in Arabidopsis or another plant species to bind to Hsp90/Hsp70 in our data (Table 1, Figure 2), not only validates our in silico search strategy but also lends support to the prediction of the newly identified CC-TPR proteins as co-chaperones of Hsp90/Hsp70. The data gathered in the present study provides many valuable insights: 1) several aspects of the Hsp90/Hsp70 TPR co-chaperone system are conserved in yeast, plant and human; 2) the Hsp90/Hsp70 TPR co-chaperone system appears to be the largest in plants due to the presence of gene families; 3) the identification of 24 new CC-TPR proteins in Arabidopsis with potential to act as co-chaperones indicates that the TPR domain is utilized by the Hsp90/Hsp70 machinery on a much larger scale than previously understood; 4) the presence of numerous other known functional domains in the newly identified plant CC-TPR proteins adds new functional dimensions to these proteins as well as to the Hsp90/Hsp70 chaperone machinery; 5) the plant CC-TPR co-chaperone network appears to have evolved unique features as judged by the combination of protein domains unique to plants, such as the thioredoxin domain in TTLs, and the FBD domain of unknown function in F-Box and BRCT domain containing plant proteins; and 6) the presence of a CC-TPR protein with both FBD and F-box, and another protein with protein kinase and U-box domains in rice, but not in Arabidopsis, indicates that some CC-TPR proteins may perform species-specific functions. Finally, the information provided here on the newly identified CC-TPR proteins will facilitate efficient investigation of their biological functions and significance. The in silico approach used here is the first of its kind to identify CC-TPR proteins in a genome-wide context with potential to interact with Hsp90/Hsp70 by the carboxylate clamp mechanism.
CC-TPR proteins can bind to both Hsp90 and Hsp70
The interaction of Hsp90 and Hsp70 with TPR proteins occurs via the conserved C-terminal EEVD motif in both Hsp90 and Hsp70 and the conserved carboxylate clamp residues (K5N9-N6-K2R6) in the TPR domain [8]. Additional contacts involving charged and hydrophobic amino acids determine the specificity of interaction. In case of Hop, Hsp70 and Hsp90 interact with two separate domains, viz TPR1 and TPR2a, respectively [45], but in the case of CHIP a single TPR binds either chaperone protein [50]. This is accomplished by accommodating either the methionine of Hsp90 (DDTSRMEEVD) or the isoleucine of Hsp70 (GSGPTIEEVD) into a hydrophobic pocket of CHIP, which is not present in Hop, resulting in the peptide being twisted into a conformation so that no further specific contacts are required [50]. Additional examples of TPR proteins binding to both Hsp90 and Hsp70 include the yeast CNS1 [51], and the human Tpr2 [36]. CNS1 binds to both Hsp90 and Hsp70 with comparable affinities and while it exerts no influence on the ATPase activity of Hsp90, it activates the ATPase activity of Hsp70 up to 30-fold. Tpr2 binds Hsp90 with slightly lower affinity as compared to Hsp70, but requires ATP for binding Hsp70 in the presence of Hsp90. The DnaJ homologous J domain in Tpr2 stimulates ATP hydrolysis and polypeptide binding by Hsp70 [36]. These data bring to light the importance of TPR co-chaperones and of Hsp70 regulation in the context of the Hsp90 chaperone cycle, and raise the possibility that the TPR proteins identified herein may interact with either Hsp90 or Hsp70 or both. Experimental analyses of interaction with both Hsp90 and Hsp70 are required for each of these proteins to understand their mode of action and its implications within the context of the Hsp90/Hsp70 chaperone machinery.
AtTPR1 and AtTPR2 bind Hsp90 through its MEEVD motif
AtTPR1 and AtTPR2 were verified for binding to Hsp90 as proof-of-concept of interaction of newly identified CC-TPR proteins with Hsp90 by the ‘carboxylate clamp’ mechanism. Both proteins bound Hsp90 in yeast two-hybrid and in vitro binding assays. Neither protein bound to Hsp90 lacking MEEVD, indicating that interaction occurs by the ‘carboxylate clamp’ mechanism [52], [53]. During the course of this study, a tomato TPR protein (SlTPR1) with highest similarity to AtTPR1 and Os10g34540 was demonstrated to interact with ethylene receptors, and its overexpression in tomato and Arabidopsis caused a range of developmental phenotypes [54]. Although no connections were made with Hsp90/Hsp70, we checked and found the presence of the consensus carboxylate clamp residues in the tomato protein, suggesting that it is an interactor of Hsp90/Hsp70. Based on the above information, it can be expected that important cellular functions will be unveiled in the future for the newly CC-TPR identified proteins.
Putative functions of the CC-TPR proteins
The different subcellular localization possibilities and the different combinations of protein domains (Table 1, Figures 1 and 2), suggest a diverse range of functions for the CC-TPR proteins, which likely add to and/or regulate the functional capacity of the Hsp90/Hsp70 chaperone system. Proteins that have a single TPR domain and no additional known domains may function as co-chaperones by modulating the ATPase activity of Hsp90/Hsp70. It is possible that their functional specificity is derived, at least in part, through specific temporal and spatial expression patterns. Proteins with more than one TPR domain or other domains like DnaJ, ankyrin, or Phox/PBI may function additionally through recruitment of other proteins. The presence of the SET domain in AtTPR9 and Os10g36250, together with the high probability of AtTPR9 protein to be localized in the nucleus (Table 1) suggests that it cooperates with the Hsp90/Hsp70 machinery in the process of chromatin remodelling [2]. Recent identification of the targets of thioredoxins in plants suggests that these proteins could influence nearly every major cellular process [55]. The involvement of AtTTL1 in ABA signaling pathway and stress responses [35] has set the stage for further investigation of the functions of TTLs along these lines.
Subcellular localization of a protein can provide clues to its functions. AtTPR10 with one TPR domain and ankyrin repeats has a high probability of being localized to the plastid. Since the plastidial Hsp90 and Hsp70 lack the EEVD motif at their C-terminus, we speculate that the interaction of the TPR domain is limited to cytosolic Hsp90/Hsp70, which chaperones the trafficking of precursor proteins to the chloroplast [23]. Whether the ankyrin domain recruits a client for the cytosolic Hsp90/Hsp70 system or a plastidial partner for AtTPR10 itself, are questions that need to be answered in the future. Experimental documentation of the plastidial localization of AtTPR10 is a prerequisite for addressing the functions of this protein. The identification of a protein in rice (Os06g06760) that contains both protein kinase and U-box domains is intriguing. To date, no such combination of domains has been noted in a CC-TPR protein. As a unique CC-TPR protein, functional analysis of this protein should be a priority.
In conclusion, the present study has uncovered a number of new potential interactors of Hsp90/Hsp70: future investigations of these will provide a better understanding of the Hsp90/Hsp70 chaperone machinery, its functions and its mode of action in plant cells. Due to the critical requirement of the Hsp90/Hsp70 system for cell viability and normal functioning of the cell, which has ramifications in human health, knowledge of this system is a high priority in any model organism.
Supporting Information
Amino acid sequence alignment of AtPhox1-4. The numbers on the side indicate the amino acid positions in the proteins. Alignment was performed using MEGA4 software. Black, grey and light grey shading indicates 100%, 75% and 50% conservation of amino acids, respectively.
(3.51 MB TIF)
Amino acid sequence alignment of of AtTTL1-4. The numbers on the side indicate the amino acid positions in the proteins. Alignment was performed using MEGA4 software. Black, grey and light grey shading indicates 100%, 75% and 50% conservation of amino acids, respectively.
(3.41 MB TIF)
Sequences of primers used in quantitative RT-PCR (qPCR) analysis.
(0.03 MB DOC)
Acknowledgments
We thank Dr. Mark Daley (Department of Computer Science, University of Western Ontario) for help with the HMMER program, and Ian Watt for help with analysis of rice data.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research was supported by a grant to P.K. from the Natural Sciences and Engineering Research Council of Canada. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pratt WB, Toft DO. Regulation of Signaling Protein Function and Trafficking by the hsp90/hsp70-Based Chaperone Machinery. Exp Biol Med. 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 2.Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J. 2008;410:439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- 3.Whitesell L, Lindquist SL. Hsp90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 4.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 5.Smith DF. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones. 2004;9:109–121. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caplan AJ. What is a co-chaperone? Cell Stress Chaperones. 2003;8:105–107. doi: 10.1379/1466-1268(2003)008<0105:wiac>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 8.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, et al. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 9.Cheung-Flynn J, Roberts PJ, Riggs DL, Smith DF. C-terminal sequences outside the tetratricopeptide repeat domain of FKBP51 and FKBP52 cause differential binding to Hsp90. J Biol Chem. 2003;278:17388–17394. doi: 10.1074/jbc.M300955200. [DOI] [PubMed] [Google Scholar]
- 10.Krishna P, Gloor G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:238–246. doi: 10.1379/1466-1268(2001)006<0238:thfopi>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, et al. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirasu K. The HSP90 – SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 13.Stancato LF, Hutchison KA, Krishna P, Pratt WB. Animal and plant cell lysates share a conserved chaperone system that assembles the glucocorticoid receptor into a functional heterocomplex with hsp90. Biochemistry. 1996;35:554–561. doi: 10.1021/bi9511649. [DOI] [PubMed] [Google Scholar]
- 14.Reddy RK, Kurek I, Silverstein AM, Chinkers M, Breiman A, et al. High-molecular-weight FK506-binding proteins are components of heat-shock protein 90 heterocomplexes in wheat germ lysate. Plant Physiol. 1998;118:1395–1401. doi: 10.1104/pp.118.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z, Quick MK, Kanelakis KC, Gijzen M, Krishna P. Characterization of a plant homolog of hop, a cochaperone of hsp90. Plant Physiol. 2003;131:525–535. doi: 10.1104/pp.011940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Fuente van Bentemm S, Vossen JH, de Vries KJ, van Wees S, Tameling WI, et al. Heat shock protein 90 and its co-chaperone protein phosphatase 5 interact with distinct regions of the tomato I-2 disease resistance protein. Plant J. 2005;43:284–298. doi: 10.1111/j.1365-313X.2005.02450.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Sullivan WP, Felts SJ, Prasad BD, Toft DO, et al. p23-like proteins in plants have unique characteristics but are conserved in hsp90-binding. Cell Stress Chaperones. 2010;15:703–715. doi: 10.1007/s12192-010-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith MR, Willman MR, Wu G, Berardini TZ, Moller B, et al. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:5424–5429. doi: 10.1073/pnas.0812729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aviezer-Hagai K, Skovorodnikova J, Galigniana M, Farchi-Pisanty O, Maayan E, et al. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol Biol. 2007;63:237–255. doi: 10.1007/s11103-006-9085-z. [DOI] [PubMed] [Google Scholar]
- 20.Smyczynski C, Roudier F, Gissot L, Vaillant E, Grandjean O, et al. The C terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. J Biol Chem. 2006;281:25475–25484. doi: 10.1074/jbc.M601815200. [DOI] [PubMed] [Google Scholar]
- 21.Kamphausen T, Fanghänel J, Neumann D, Schulz B, Rahfeld JU. Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J. 2002;32:263–276. doi: 10.1046/j.1365-313x.2002.01420.x. [DOI] [PubMed] [Google Scholar]
- 22.Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, et al. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem. 2006;281:30603–30612. doi: 10.1074/jbc.M604604200. [DOI] [PubMed] [Google Scholar]
- 23.Qbadou S, Becker T, Mirus O, Tews I, Soll J, et al. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 2006;25:1836–1847. doi: 10.1038/sj.emboj.7601091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen G, Yan J, Pasapula V, Luo J, He C, et al. The chloroplast protease subunit ClpP4 is a substrate of the E3 ligase AtCHIP and plays an important role in chloroplast function. Plant J. 2007;49:228–237. doi: 10.1111/j.1365-313X.2006.02963.x. [DOI] [PubMed] [Google Scholar]
- 25.Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, et al. InterPro, progress and status in 2005. Nucleic Acids Res. 2005;33:D201–205. doi: 10.1093/nar/gki106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, et al. An "electronic fluorescent pictograph" browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: the Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;35:D213–218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 30.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 31.Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:156–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Krishna P, Reddy RK, Sacco M, Frappier JR, Felsheim RF. Analysis of the native forms of the 90 kDa heat shock protein (Hsp90) in plant cytosolic extracts. Plant Mol Biol. 1997;33:457–466. doi: 10.1023/a:1005709308096. [DOI] [PubMed] [Google Scholar]
- 34.Bhangoo MK, Tzankov S, Fan AC, Dejgaard K, Thomas DY, et al. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol Biol Cell. 2007;18:3414–3428. doi: 10.1091/mbc.E07-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosado A, Schapire AL, Bressan RA, Harfouche AL, Hasegawa PM, et al. The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for the osmotic stress responses and abscisic acid sensitivity. Plant Physiol. 2006;142:1113–1126. doi: 10.1104/pp.106.085191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, et al. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22:3613–3623. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moffatt NS, Bruinsma E, Uhl C, Obermann WM, Toft D. Role of the cochaperone Tpr2 in Hsp90 chaperoning. Biochemistry. 2008;47:8203–8213. doi: 10.1021/bi800770g. [DOI] [PubMed] [Google Scholar]
- 38.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min J, Zhang X, Cheng X, Grewal SI, Xu RM. Structure of the SET domain histone lysine methyltransferase Clr4. Nat Struct Biol. 2002;9:828–832. doi: 10.1038/nsb860. [DOI] [PubMed] [Google Scholar]
- 40.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 41.Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, et al. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Ito T, Matsui Y, Ago T, Ota K, Sumimoto H. Novel modular domain PB1 recognizes PC motif to mediate functional protein-protein interactions. EMBO J. 2001;20:3938–3946. doi: 10.1093/emboj/20.15.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inaba K, Murakami S, Suzuki M, Nakagawa A, Yamashita E, et al. Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell. 2006;127:789–801. doi: 10.1016/j.cell.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 44.Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 45.Lässle M, Blatch GL, Kundra V, Takatori T, Zetter BR. Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J Biol Chem. 1997;272:1876–1884. doi: 10.1074/jbc.272.3.1876. [DOI] [PubMed] [Google Scholar]
- 46.Krishna P, Sacco M, Cherutti JF, Hill S. Cold-Induced Accumulation of hsp90 Transcripts in Brassica napus. Plant Physiol. 1995;107:915–923. doi: 10.1104/pp.107.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verslues PE, Zhu JK. Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem Soc Trans. 2005;33:375–379. doi: 10.1042/BST0330375. [DOI] [PubMed] [Google Scholar]
- 48.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 49.Krishna P. Brassinosteroid-Mediated Stress Responses. J Plant Growth Regul. 2003;22:289–297. doi: 10.1007/s00344-003-0058-z. [DOI] [PubMed] [Google Scholar]
- 50.Murata S, Chiba T, Tanaka K. CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol. 2003;35:572–578.b. doi: 10.1016/s1357-2725(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 51.Hainzl O, Wegele H, Richter K, Buchner J. Cns1 is an activator of the Ssa1 ATPase activity. J Biol Chem. 2004;279:23267–23273. doi: 10.1074/jbc.M402189200. [DOI] [PubMed] [Google Scholar]
- 52.Carrello A, Ingley E, Minchin RF, Tsai S, Ratajczak T. The common tetratricopeptide repeat acceptor site for steroid receptor-associated immunophilins and hop is located in the dimerization domain of Hsp90. J Biol Chem. 1999;274:2682–2689. doi: 10.1074/jbc.274.5.2682. [DOI] [PubMed] [Google Scholar]
- 53.Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, et al. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Z, Arciga-Reyes L, Zhong S, Alexander L, Hackett R, et al. S1TPR1, a tomato tetratricopeptide repeat protein, interacts with the ethylene receptors NR and LeETR1, modulating ethylene and auxin responses and development. J Exp Bot. 2008;59:4271–4287. doi: 10.1093/jxb/ern276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, et al. Thioredoxin targets in plants: the first 30 years. J Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment of AtPhox1-4. The numbers on the side indicate the amino acid positions in the proteins. Alignment was performed using MEGA4 software. Black, grey and light grey shading indicates 100%, 75% and 50% conservation of amino acids, respectively.
(3.51 MB TIF)
Amino acid sequence alignment of of AtTTL1-4. The numbers on the side indicate the amino acid positions in the proteins. Alignment was performed using MEGA4 software. Black, grey and light grey shading indicates 100%, 75% and 50% conservation of amino acids, respectively.
(3.41 MB TIF)
Sequences of primers used in quantitative RT-PCR (qPCR) analysis.
(0.03 MB DOC)