Abstract
The ergot alkaloids are a diverse class of fungal-derived indole alkaloid natural products with potent pharmacological activities. The biosynthetic intermediate chanoclavine-I aldehyde 1 represents a branch point in ergot biosynthesis. Ergot alkaloids festuclavine 2 and agroclavine 3 derive from alternate enzymatic pathways originating from the common biosynthetic precursor chanoclavine-I aldehyde 1. Here we show that while the Old Yellow Enzyme homolog EasA from the ergot biosynthetic gene cluster of Aspergillus fumigatus acts on chanoclavine-I aldehyde 1 to yield festuclavine 2, EasA from Neotyphodium lolii, in contrast, produces agroclavine 3. Mutational analysis suggests a mechanistic rationale for the switch in activity that controls this critical branch point of ergot alkaloid biosynthesis.
The ergot alkaloids are a diverse class of fungal-derived indole alkaloid natural products with potent pharmacological activities.1 The biosynthetic intermediate chanoclavine-I aldehyde 1 can be enzymatically converted into festuclavine 2, which is further derivatized to form alkaloids such as the fumigaclavines in Aspergillus fumigatus (Scheme 1).1 Alternatively, in certain fungal species such as Claviceps purpurea and Neotyphodium lolii, chanoclavine-I aldehyde 1 is converted to agroclavine 3, which goes on to form lysergic acid-derived ergot alkaloids such as ergopeptines.1 Festuclavine 2 and agroclavine 3 differ only by the degree of unsaturation in the D ring (Scheme 1). Although chanoclavine-I aldehyde 1 had been proposed as an ergot alkaloid biosynthetic intermediate many years ago,2 the enzyme-catalyzed mechanism of D ring formation has remained elusive. Recently, we and others have reported that a homolog of Old Yellow Enzyme from A. fumigatus, EasA_Af, reduces the alkene of the chanoclavine-I aldehyde α, β unsaturated carbonyl moiety (C8-C9, Scheme 1A).3 This reduction facilitates an intramolecular reaction between the aldehyde and the amine moieties to allow formation of the D ring of festuclavine 2 (Scheme 1A). Yet it is still unclear how chanoclavine-I aldehyde 1 is converted into agroclavine 3, which retains the C8-C9 double bond of the starting precursor, but with this alkene in the opposite geometrical configuration. Formation of agroclavine 3 from chanoclavine-I aldehyde 1 was investigated previously by Floss and colleagues in whole-cell precursor feeding studies.2, 4 Results of these studies clearly demonstrated precursor-product relationships but also raised unanswered questions about the enzymes and mechanisms involved. One particularly puzzling step implicated a cis-trans isomerization involving intermolecular transfer of a hydrogen between successive substrate molecules. Here we show that an EasA homolog from the ergot biosynthetic gene cluster of N. lolii, EasA_Nl, acts on chanoclavine-I aldehyde 1 to yield agroclavine 3 rather than festuclavine 2, therefore supporting the previously proposed cis-trans isomerization step required for the conversion of chanoclavine-I aldehyde 1 into agroclavine 3.2, 4 Mutational analysis suggests a mechanistic rationale for the switch in activity of EasA– from reductase in A. fumigatus to isomerase in N. lolii– that controls this critical branch point of ergot alkaloid biosynthesis.
Scheme 1.
A. Reduction of C8-C9 alkene by EasA_Af leads to festuclavine 2. B. EasA_Nl leads to formation of agroclavine 3.
Based on previous analyses3 and homology of EasA with Old Yellow Enzyme, a flavoenzyme that typically reduces the alkene of an α, β unsaturated carbonyl moiety,5 the role of EasA in fungal producers of festuclavine 2 is clear– reduction of the C8-C9 double bond of chanoclavine-I aldehyde 1 (Scheme 1A). However, fungal producers of agroclavine 3, which retains the C8-C9 alkene, also contain an EasA homolog in the ergot gene clusters.1 EasA therefore likely plays a different role in this biosynthetic pathway. Notably, an A. fumigatus (a festuclavine 2 producer) ΔeasA disruption strain transformed with easA from C. purpurea (an agroclavine 3 producer), yielded agroclavine 3 derived alkaloids.6 This experiment strongly suggests that EasA is the checkpoint that controls whether chanoclavine-I aldehyde 1 is converted to festuclavine 2 or agroclavine 3. To gather mechanistic data to test this hypothesis, an in vitro assay for agroclavine 3 producing EasA enzymes is required. Unfortunately, when EasA from C. purpurea was heterologously expressed in E. coli, the resulting protein was catalytically inactive with chanoclavine-I aldehyde 1 and lacked the expected flavin cofactor as evidenced by UV-vis spectroscopy. However, EasA from another agroclavine 3 producer N. lolii (EasA_Nl) could be expressed as active, holo-enzyme from E. coli (Supporting Information, SI).
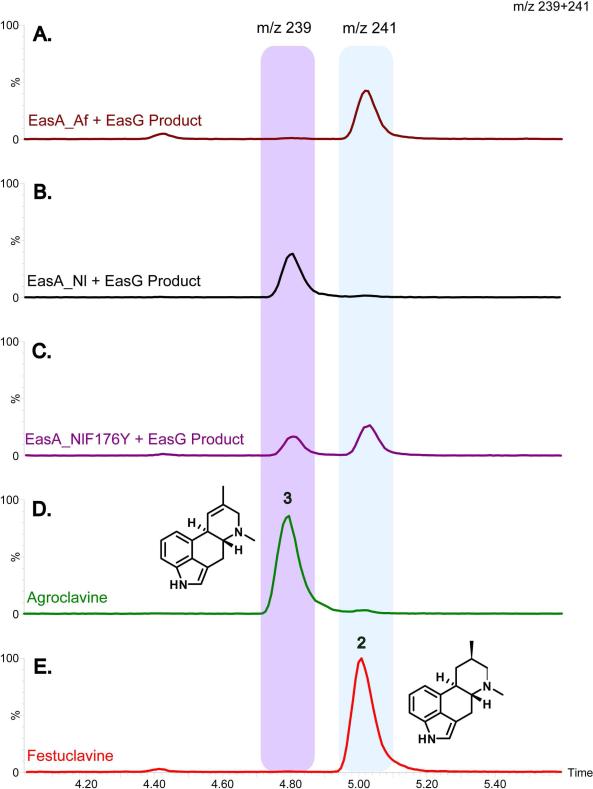
We compared the products of EasA_Af and EasA_Nl when incubated with chanoclavine-I aldehyde 1 substrate7a in the presence of NADPH-dependent oxidoreductase EasG (A. fumigatus). Recent work by Wallwey et al. has revealed that EasG reduces the cyclic iminium product generated by EasA_Af to form festuclavine 2 (Scheme 1A).3b EasG, which is present in all ergot biosynthetic gene clusters, likely plays a similar role in agroclavine 3 biosynthesis (Scheme 1A); EasG from N. lolii exhibits 65% amino acid similarity to EasG from A. fumigatus (SI). As expected, when EasA_Af and EasG were incubated with chanoclavine-I aldehyde 1 and NADPH, a product with a mass and retention time identical to an authentic standard of festuclavine 2 was observed (Figure 1A,E, SI).7b Gratifyingly, in contrast, EasA_Nl and EasG yielded a product with a mass and retention time identical to agroclavine 3 (Figure 1B,D, SI).7b Therefore, EasA_Nl exhibits the biochemical activity predicted by the alkaloid production profile of the producing organism, validating that EasA controls formation of festuclavine 2 vs. agroclavine 3 from chanoclavine-I aldehyde 1.
Figure 1.
LCMS chromatograms (selected ion monitoring at m/z 239 and 241) showing product formation in EasA assays. A. EasA_Af + EasG. B. EasA_Nl + EasG. C. EasA_Nl Phe176Tyr + EasG. D. Agroclavine 3 standard m/z 239. E. Festuclavine 2 standard m/z 241.
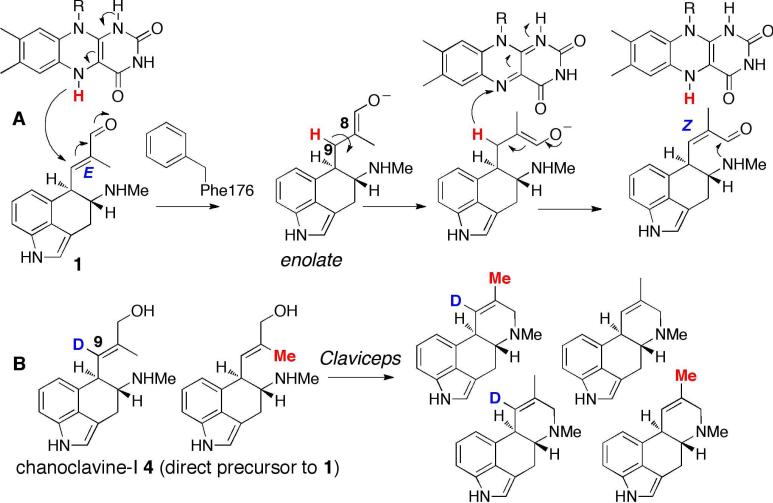
Although the C8-C9 double bond is retained in both chanoclavine-I aldehyde 1 and agroclavine 3, the alkene geometry switches from E in 1 to Z in the product 3.4 This switch from E to Z is required to allow intramolecular iminium formation (Scheme 1B). EasA_Nl therefore displays an isomerase rather than the reductase function of EasA_Af. This switch in enzymatic function clearly explains how the biosynthetic intermediate 1 with the alkene E configuration is converted into product 3 with Z configuration.
To explore the mechanistic basis for the catalytic differences between these two EasA homologs, we compared the sequences of EasA_Nl and EasA_Af (76% amino acid similarity), along with the sequences of other EasA homologs from both festuclavine 2 and agroclavine 3 producing fungal species (SI). The most intriguing difference between the two sets of producers was substitution of a phenylalanine for a tyrosine residue in the sequences of fungi that produce agroclavine and are thus expected to have EasA with isomerase activity. Previous mechanistic work on Old Yellow Enzyme from Saccharomyces carlsbergensis suggests that this tyrosine residue acts as a general acid that protonates the α carbon of the alkene substrate after a hydride from the flavin is added to the β carbon.5a We therefore speculate that in isomerase EasA_Nl, the substitution to Phe, which cannot serve as a proton donor, would allow enolization of the substrate following donation of a hydride from the flavin. Subsequent rotation around the C8-C9 bond, tautomerization, and transfer of the hydride back to the flavin leads to the Z alkene geometry to allow intramolecular iminium ion formation (Scheme 2A). Notably, our data and interpretation provide an explanation for observations documented by Floss and coworkers in labeled precursor feeding experiments. These investigators performed numerous labeling experiments supporting a mechanism that involves an intermolecular transfer of the hydrogen at C9.1c, 2, 4e The most conclusive of these experiments showed that separately labeled 2H and 13C chanoclavine-I 4 (the direct precursor to chanoclavine-I aldehyde 1) are converted by Claviceps to doubly labeled agroclavine 3 derived products (Scheme 2B) (see Table 5 of reference 2). Doubly labeled product indicates that the hydrogen at C9 undergoes an intermolecular transfer from one substrate to another. Floss proposed that this transfer may occur by addition of a hydrogen at C9, followed by removal of a hydrogen, and that one of these steps may occur in a nonstereoselective fashion.4e Our results suggest that this intermolecular H transfer likely results from hydrogen transfer between flavin and substrate as depicted in Scheme 2A. Additionally, Floss proposed that the transferred hydrogen could be either a proton or a hydride;1c, 4f the involvement of a flavin containing enzyme suggests that the hydrogen transfer at C9 likely involves a hydride. More detailed studies are required to further explore this aspect of the enzyme mechanism, and to clarify whether these hydrogen transfer steps are non-stereoselective.
Scheme 2.
A. Proposed isomerization reaction catalyzed by EasA_Nl. B. The products of whole cell labeling experiments– which derive from a mixture of unlabeled, single and double labeled 3 shown above–reported in reference 5 suggest that H of C9 undergoes intermolecular transfer.
To validate the hypothesis that the Tyr to Phe mutation is responsible for this switch in activity from a reductase (as found in A. fumigatus) to an isomerase (as found in N. lolii), we made the EasA_Nl Phe176Tyr and the EasA_Af Tyr178Phe mutations. Gratifyingly, EasA_Nl Phe176Tyr produced a compound with a mass and retention time identical to festuclavine 2 when incubated with chanoclavine-I aldehyde 1, EasG and NADPH (Figure 1C). A compound with a mass and retention time matching the wild type product, agroclavine 3, was also observed, suggesting that while the Phe176Tyr mutation is critical for reduction of the alkene substrate, other residues also contribute to a complete switch in enzymatic activity. Interestingly, the activity of EasA_Af Tyr178Phe mutant was not qualitatively different from wild type EasA_Af; both the mutant and wild type EasA_Af enzymes appeared to produce only festuclavine 2 with no agroclavine 3 observed (SI). More extensive mutations may be required to engineer an isomerase-type enzyme. The active site must rigorously exclude all other proton sources such as water that could protonate C8 after hydride addition to C9. Additionally, it is possible that the residues of EasA_Nl make the enolate shown in Scheme 2A more thermodynamically accessible than in EasA_Af homologs that yield festuclavine 3. Whereas the presence of Phe versus Tyr at the active site profoundly alters enzyme activity, more detailed mutational analysis along with structural characterization of the product stereochemistry will allow us to clarify mechanistic details of this switch in activity, and to further reconcile this proposed mechanism with previously reported whole cell labeling studies.2, 4e Amino acid sequence analysis of Old Yellow Enzymes of unknown function in the NCBI GenBank database indicates that 5% have Phe rather than Tyr at the critical active site position. Functional analyses of these homologs would be required to test their ability to isomerize rather than reduce alkene substrates. Nonetheless, the presence of the Phe for Tyr substitution suggests that isomerase functionality for this class of enzymes may extend beyond the ergot alkaloid pathway.
Supplementary Material
Acknowledgement
USDA NIFA2008-35318-04549 and NIH GM074820 for support. Scientific article number (supplied upon acceptance) from the West Virginia Agriculture Experiment Station.
Footnotes
Supporting Information Available: Experimental procedures, sequence alignments and additional experimental data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1a.Schardl C, Panaccione DG, Tudzynski P. In: The Alkaloids. Cordell G, editor. Vol. 63. Elsevier; 2006. pp. 45–86. [DOI] [PubMed] [Google Scholar]; b Panaccione DG, Coyle CM. Appl. Environ. Microbiol. 2005;71:3112–3118. doi: 10.1128/AEM.71.6.3112-3118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Floss HG, Anderson JA. In: The Biosynthesis of Mycotoxins. Steyn PS, editor. Academic Press; 1980. pp. 17–67. [Google Scholar]
- 2.Floss HG, Tcheng-Lin M, Chang C-J, Naidoo B, Blair GE, Abou-Chaar CI, Cassady JM. J. Am. Chem. Soc. 1974;96:1898–1909. doi: 10.1021/ja00813a042. [DOI] [PubMed] [Google Scholar]
- 3a.Cheng JZ, Coyle C, Panaccione D, O'Connor SE. J. Am. Chem. Soc. 2010;132:1776–1777. doi: 10.1021/ja910193p. [DOI] [PubMed] [Google Scholar]; b Wallwey C, Matuschek M, Xie X-L, Li S-M. Org. Biomol. Chem. 2010;8:3500–3508. doi: 10.1039/c003823g. [DOI] [PubMed] [Google Scholar]
- 4a.Groeger D, Erge D, Floss HG. Naturforsch., B. 1966;21:827–832. [PubMed] [Google Scholar]; b Fehr T, Acklin W, Arigoni D. Chem. Commun. 1966:801–802. [Google Scholar]; c Voigt R, Bornschein M, Rabitzsch G. Pharmazie. 1967;22:326–329. [Google Scholar]; d Floss HG, Hornemann U, Schilling N, Groeger D, Erge D. Chem. Commun. 1967:105–106. doi: 10.1021/ja01025a048. [DOI] [PubMed] [Google Scholar]; e Floss HG, Hornemann U, Schilling N, Kelley K, Groeger D, Erge D. J. Am. Chem. Soc. 1968;90:6500–6507. doi: 10.1021/ja01025a048. [DOI] [PubMed] [Google Scholar]; f Floss HG. Tetrahedron. 1976;32:873–912. [Google Scholar]
- 5a.Kohli RM, Massey V. J. Biol. Chem. 1998;273:32763–32770. doi: 10.1074/jbc.273.49.32763. [DOI] [PubMed] [Google Scholar]; b Williams RE, Bruce NC. Microbiology. 2002;148:1607–1614. doi: 10.1099/00221287-148-6-1607. [DOI] [PubMed] [Google Scholar]
- 6.Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. Appl. Environ. Microbiol. 2010;76:3898–3903. doi: 10.1128/AEM.02914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Isolation and characterization of substrate chanoclavine-I aldehyde 1 is described in (3a). (b) Characterization of authentic standards of festuclavine 2 and agroclavine 3 are reported in (3a).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.