Abstract
Studies in the field of wound healing have utilized a variety of different housekeeping genes for RT-qPCR analysis. However, nearly all of these studies assume that the selected normalization gene is stably expressed throughout the course of the repair process. The purpose of our current investigation was to identify the most stable housekeeping genes for studying gene expression in mouse wound healing using RT-qPCR. To identify which housekeeping genes are optimal for studying gene expression in wound healing, we examined all articles published in Wound Repair and Regeneration that cited RT-qPCR during the period of Jan/Feb 2008 until July/August2009. We determined that ACTIN, GAPDH, 18S and β2M were the most frequently used housekeeping genes in human, mouse, and pig studies. We also investigated nine commonly used housekeeping genes that are not generally used in wound healing models: GUS, TBP, RPLP2, ATP5B, SDHA, UBC, CANX, CYC1, and YWHAZ. We observed that wounded and unwounded tissues have contrasting housekeeping gene expression stability. The results demonstrate that commonly used housekeeping genes must be validated as accurate normalizing genes for each individual experimental condition.
Keywords: RT-qPCR, wound healing, housekeeping genes
INTRODUCTION
To investigate the molecular mechanisms of wound healing, quantitative PCR (RT-qPCR) has been widely used to analyze the expression of candidate genes. In recent years the utilization of the technique has grown, with an increasing number of published papers citing its use for the study of gene expression (1). RT-qPCR is popular mainly due to its ability to efficiently amplify small quantities of RNA in a relatively short period of time. For example, gene expression results can be obtained in under 40 minutes using reagents such as Taqman Fast Universal PCR Master Mix (Applied Biosystems).
Despite its advantages, RT-qPCR has certain drawbacks which affect the interpretation of its results: 1) reliable extraction of equal amounts of non-degraded RNA from each sample; 2) consistent reverse transcriptase efficiency resulting in equal amounts of cDNA in all samples; 3) adequate primer specificity; 4) presence of inhibitors in samples. In order to overcome these drawbacks, results are most often normalized to the expression of “housekeeping genes” that do not change their expression levels across sample or treatment groups (2). Housekeeping genes are recognized as cellular maintenance genes which regulate basic and ubiquitous cellular functions, and therefore are valid candidates to act as internal controls for gene expression. The stable expression of a housekeeping gene in any RT-qPCR reaction is mandatory since it plays a critical role in the interpretation of the final result (3).
Studies in the field of wound healing have utilized a variety of different housekeeping genes for RT-qPCR analysis. However, nearly all of these studies assume that the selected normalization gene is stably expressed throughout the course of the repair process. Generally the stability of the housekeeping gene is not assessed, leaving questions about whether normalization is accurate. The purpose of our current investigation is to identify the most stable housekeeping genes for studying gene expression in mouse wound healing using RT-qPCR. To identify which housekeeping genes are commonly used for studying gene expression in wound healing, we examined all articles published in Wound Repair and Regeneration (WRR) that cited RT-qPCR during the period ofJan/Feb 2008 until July/August2009. Twenty-six such articles were identified. From the methods, we determined ACTIN, GAPDH, 18S and β2M were the most frequently used housekeeping genes in human, mouse, and pig tissues in these studies. These genes are typically used because they are believed to be stable; however the discussion of why specific housekeeping genes were chosen for the RT-qPCR normalization was generally absent.
Several Microsoft Excel based applications are available to assess the degree of variation in the candidate housekeeping genes. In the current study, we analyzed the stability of expression of 13 different housekeeping genes using the geNorm analysis program version 3.4 developed by Vandesompele et al. in 2002 (4). We selected this specific application due to its general acceptance in the literature. The program uses an algorithm to determine the stability of the candidate housekeeping gene across samples. This measurement is denoted M, which is the average paired variation of a particular gene against all other candidate genes. Genes with the lowest M-values are considered to have the most stable expression (4–5).
We investigated nine commonly used housekeeping genes that are not generally used in wound healing models: GUS, TBP, RPLP2, ATP5B, SDHA, UBC, CANX, CYC1, and YWHAZ. Validation was carried out by normalizing the target gene, keratinocyte growth factor-2 (kgf-2), to the most stable housekeeping gene, which we determined to be TATA-box binding protein, TBP.
MATERIALS & METHODS
Animals and Wound Models
All animal procedures were approved by University of Illinois Institutional Animal Care and Use Committee. BALB/c-strain female mice, 2–3 months of age were used for the described experiments. This strain has been well-characterized in wound healing studies. Mice were anesthetized using a 100 mg/kg ketamine and 5mg/kg xylazine solution. The dorsal skin was shaved and cleansed with 70% isopropyl alcohol. Six excisional full-thickness dermal wounds were made on the dorsal surface of each mouse using a sterile 1-mm punch-biopsy instrument (Acu-Punch,Acuderm Inc., Ft. Lauderdale, FL), with three on each side of the midline. The excised skin derived at the time of wound placement was used as the normal, uninjured skin control for subsequent experiments. At 24hr, 48hr, 72hr, and 5 days after injury, five mice per time point were sacrificed and the wounds harvested; samples were placed in RNAlater (Ambion) solution for real timeRT-PCR analysis.
Real time PCR
Normal skin and wound samples stored in RNAlater solution (Ambion, USA) were homogenized in TRIzol reagent (Invitrogen, USA). Total RNA was isolated, checked for purity, and the concentration quantified spectrophotometrically. 1 μg of total RNA was isolated and treated with DNase I (Invitrogen, Carlsbad, CA, USA); and reverse transcription was performed with Omniscript Reverse Transcriptase (Qiagen, Valencia, CA, USA). cDNA concentration was assessed using a NanoDrop ND-1000 Spectrophotometer (Nanodrop), to determine 25ng of cDNA for each sample. A list of the housekeeping genes used and their source catalog numbers are shown in Table 2A, B. cDNA samples, primers, and AmpliTaq Gold® Fast PCR Master Mix (Applied Biosystems) were loaded onto MicroAmp 96-well PCR reaction plates (Applied Biosystems), and the amplification protocol was run using the StepOnePlus Real Time PCR system (Applied Biosystems). geNorm analysis was performed to determine housekeeping gene stability across samples and tissues. However, before Ct values from the quantitative real-time PCR were input into geNorm, all CT values were transformed into relative quantification data. Highest Ct values were subtracted from all other Ct values for each gene measured. This is called “the delta Ct” value, with the highest delta Ct value as 0; all other values are less than 0. For each data point, the 2^(−delta Ct) equation was applied. All data were expressed relative to the expression of the least expressed gene. Keratinocyte Growth factor 2 primers were purchased from Applied Biosystems: KGF-2- Mm01297079_m1. The results were quantified using the comparative 2−ΔΔCt method. Normal/unwounded skin and tongue tissues were normalized to 1.
Table 2A.
Candidate reference genes (Source: Applied Biosystems)
| Gene Symbol | Gene Name | Cellular Function | TaqMan Assay Number* |
|---|---|---|---|
| B2M | β-2-microglobulin | histocompatibility complex antigen | Mm00437762_m1 |
| TBP | TATA box binding protein | transcription factor | Mm01277043_m1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | dehydrogenase | 4352339E |
| GUSb | Glucurodinase,beta | glycosidase | Mm03003537_s1 |
| RPLP2 | Rribosomal protein, large p2 | translation, ribosomal protein | Mm00782638_s1 |
| ACTB | Actin,,beta | cytoskeleton | 4352341E |
| 18S | Ribosomal RNA 18S | member of ribosome RNA | 4308310 |
The primer and probe sequences for the TaqMan assays are not available.
Table 2B.
Candidate reference genes (Source: PrimerDesign Ltd*)
| Gene Symbol | Gene Name | Cellular Function |
|---|---|---|
| UBC | Ubiquitin C | ubiquitination |
| YWHAZ | Phospholipase A2 | signal transduction |
| ATP5B | ATP synthase subunit | ATP production |
| CANX | Calnexin | retain protein in ER |
| CYC1 | Cytochrome c-1 | electron transport chain |
| SDHA | Succinate dehydrogenase complex, subunit A | dehydrogenase |
The above listed genes are provided in geNorm™ Housekeeping Gene Selection Kit with PerfectProbe.
Data analysis
For analysis of gene stability we used the geNorm application, a Microsoft Excel program available at http://medgen.ugent.be/~jvdesomp/genorm/. The premise of this program is that ratios between constantly expressed, non-normalized housekeeping genes should remain regular. M, the average pair-wise variation of a single housekeeping control gene was calculated from the raw expression data. The suggested cut-off M-value is 1.5 by geNorm software. The lowest M value is the most stable, so the gene with the highest instability (highest M value) is removed at each step. A new M value for each remaining gene is calculated until only two genes remain. Since these calculations are based on ratios, the final two genes cannot be resolved from each other. Normalization factors were calculated using the geometric mean to control for changes in relative gene expression and outlying values. Pair-wise variation (Vn/nC1) was calculated by geNorm between two sequential normalization factors for all the samples (4).
Statistical analysis
GraphPad software (GraphPad Software, San Diego, CA) was used to analyze quantitative data. Comparisons were considered statistically significant when p<0.05 by one-way ANOVA and Bonferroni post-test in wounds vs normal tissues.
RESULTS
Selection of candidate housekeeping genes
Housekeeping genes are cellular maintenance genes which regulate basic and ubiquitous cellular functions. In many RT-qPCR reactions, these genes are used as internal control genes without proper validation. We researched articles, from 2008 to 2009: Volume 17, Issue 4, in Wound Repair and Regeneration to identify which housekeeping genes are commonly used (Table 1) in wound healing models. We observed that GAPDH, ACTIN, 18S, and β2M are the four most commonly used housekeeping genes in wound healing experiments. However, validation of these genes was generally not shown. Nine housekeeping genes which were not typically used in wound healing related experiments were selected for our studies: GUS, TBP, RPLP2, ATP5B, SDHA, UBC, CANX, CYC1 and YWHAZ. Pre-designed primer/probe sets for β2M, GUS, TBP, GAPDH, RPLP2, 18S, and ACTIN were purchased from Applied Biosystems (Table 2A). Primer/probe sets for ATP5B, SDHA, UBC, CANX, CYC1, and YWHAZ were obtained from geNorm™ Housekeeping Gene Selection Kit with Perfect Probe (Table 2B). Special attention was paid to selecting genes that belong to different functional classes, which significantly reduces the chance that genes might be co-regulated.
Table 1.
Typical housekeeping genes used in human, mouse, and pig wound models
| Gene | Number of articles referencing each gene* |
|---|---|
| ACTIN | 8 |
| GAPDH | 8 |
| 18S | 6 |
| β2M | 1 |
| HPRT** | 1 |
| RPS9** | 1 |
| CYCLOPHILIN A | 2 |
| YWHAZ | 1 |
26 articles in Wound Repair and Regeneration (Jan 2008-Aug 2009) were analyzed to determine the most commonly used housekeeping genes.
HPRT- hypoxanthine ribosyltransferase; RPS9- ribosomal protein S9.
Identification of the most stable housekeeping genes in normal skin
Normal skin (unwounded tissue) was harvested from the dorsum of mice and RNA was extracted. Single strand cDNA was synthesized simultaneously from each extract in order to minimize any variation during this step of the process. The expression of the transcripts of 13 potential housekeeping genes was then assayed using this cDNA.
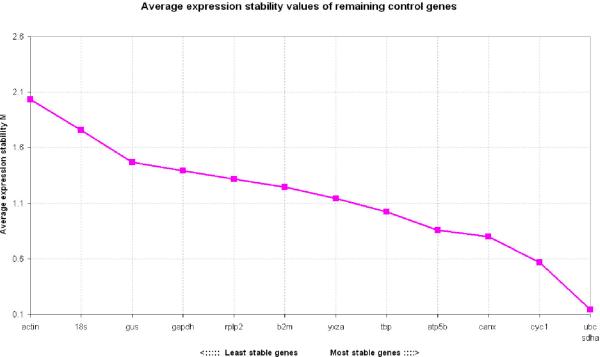
Transformed expression data were analyzed with the geNorm pairwise analysis (4) which determines a gene expression stability measure, M, for each candidate housekeeping gene based on the average pairwise variation between a particular gene and all other control genes. The expression stability was as follows from the most stable to the least stable: UBC, SDHA, CYC1, CANX, ATP5B, TBP, YWHAZ, β2M, RPLP2, GAPDH, GUS, 18S, and ACTIN (Fig. 1).
Figure 1. Gene expression stability of candidate housekeeping genes in normal skin.
Values of M with geNorm software that compares gene expression without accounting for experimental groups and proceeds to the stepwise exclusion of the genes whose relative expression levels are more variable among tissues samples. Threshold for eliminating a gene as unstable was M≥ 1.5 Lower values of M correspond to the most stable genes, thus the most suitable for normalization.
Identification of the most stable housekeeping genes in wounded skin
Six 1mm wounds were placed on the dorsum of each mouse using a biopsy punch. Wounds were harvested after 24h, 48h, 72h, and 5 days. To identify the most stable housekeeping genes for each wounded time point, RNA was extracted from samples. Single strand cDNA was synthesized simultaneously from each extract in order to minimize any variation during this step of the process. The expression of the transcripts of 13 potential housekeeping genes was then assayed using this cDNA. As described above, transformed expression data were analyzed with the geNorm software (4).
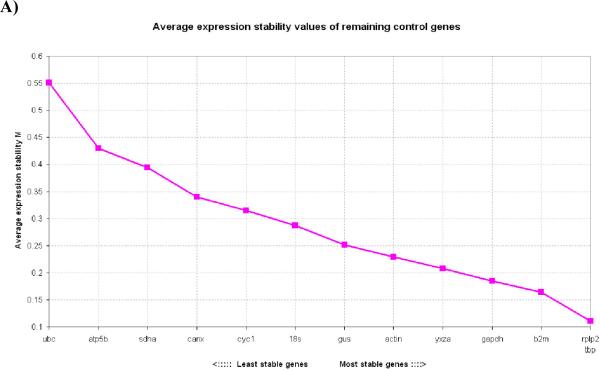
In 24h wounds, the order of expression stability from the most stable (lowest M values) to the least stable (highest M values) was: RPLP2, TBP, β2M, GAPDH, YWHAZ, ACTIN, GUS, 18S, CYC1, CANX, SDHA, ATP5B, UBC (Fig. 2A). As noted, at this time point UBC is the least stable gene which is in contrast to its superior stability in normal, uninjured skin tissue.
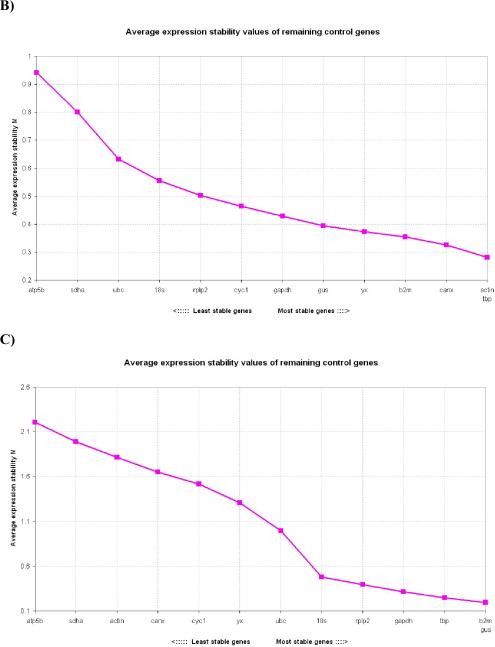
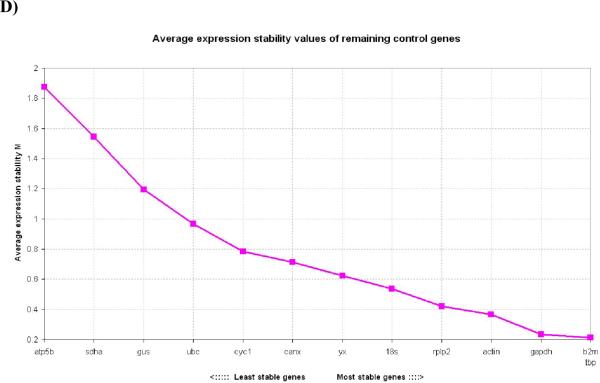
Figure 2. Gene expression stability of candidate housekeeping genes in skin wounds.
2A) Gene expression stability of candidate housekeeping genes in 1mm 24h skin wounds; 2B) Gene expression stability of candidate housekeeping genes in 1mm 48h skin wounds; 2C) Gene expression stability of candidate housekeeping genes in 1mm 72h skin wounds; 2D) Gene expression stability of candidate housekeeping genes in 1mm 5 day skin wounds. Threshold for eliminating a gene as unstable was M≥ 1.5 Lower values of M correspond to the most stable genes, thus the most suitable for normalization.
In 48h wounds, the expression stability from the most stable to the least stable was: ACTIN, TBP, CANX, β2M, YWHAZ, GUS, GAPDH, CYC1, RPLP2, 18S, UBC, SDHA, ATP5B (Fig. 2B). As opposed to normal skin in which ACTIN was the least stable gene, in 48h wounds it was one of the most stable genes.
In 72h wounds, the expression stability calculated in the genes analyzed was from the most stable to the least stable: β2M, GUS, TBP, GAPDH, RPLP2, 18S, UBC, YWHAZ, CYC1, CANX, ACTIN, SDHA, ATP5B (Fig. 2C). Once again, wounded tissues exhibited contrasting gene stability data to normal skin. Gene SDHA was one of the least stable genes in 72h wounds, as opposed to being one of the most stable in normal skin.
In 5 day wounds, the expression stability from the most stable to the least stable was: β2M, TBP, GAPDH, ACTIN, RPLP2, 18S, YWHAZ, CANX, CYC1, UBC, GUS, SDHA, ATP5B (Fig. 2D). 5 day wounds also had contrasting gene stability data to normal skin.
In all wound tissues, the gene with the most consistent stable expression was TBP with its order changing slightly in each group. We observed that wounded and unwounded tissues have contrasting housekeeping gene expression stability (Table 3). We provide a summary of the 3 most stable housekeeping genes for each tissue in Table 3. According to Vandesompele et al (2002) using the 3 best housekeeping genes is in most cases a valid normalization strategy, and results in much more accurate and reliable normalization compared to the use of only one single housekeeping gene. A normalization factor based on the expression levels of the 3 most stable housekeeping genes is calculated.
Table 3.
Summary of 3 most stable Housekeeping Genes for each tissue
| Tissue | tbp | rplp2 | β2m | gapdh | ywhaz | actin | gus | 18S | cyc1 | canx | sdha | Atp5b | ubc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normal skin | √ | √ | √ | ||||||||||
| 24h wounds | √ | √ | √ | ||||||||||
| 48h wounds | √ | √ | √ | ||||||||||
| 72h wounds | √ | √ | √ | ||||||||||
| 5 day wounds | √ | √ | √ |
Normalization of Keratinocyte Growth Factor-2 (KGF-2) in skin wound samples
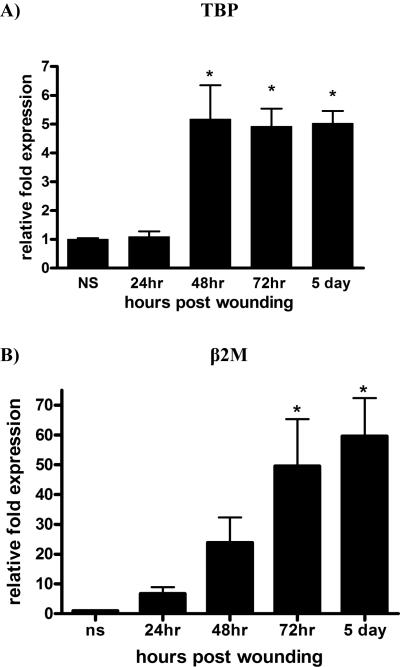
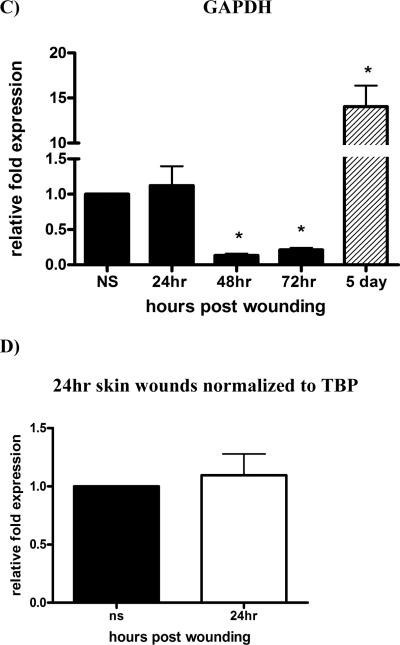
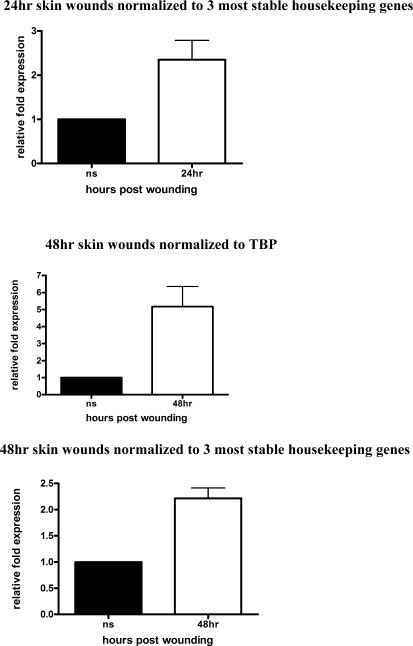
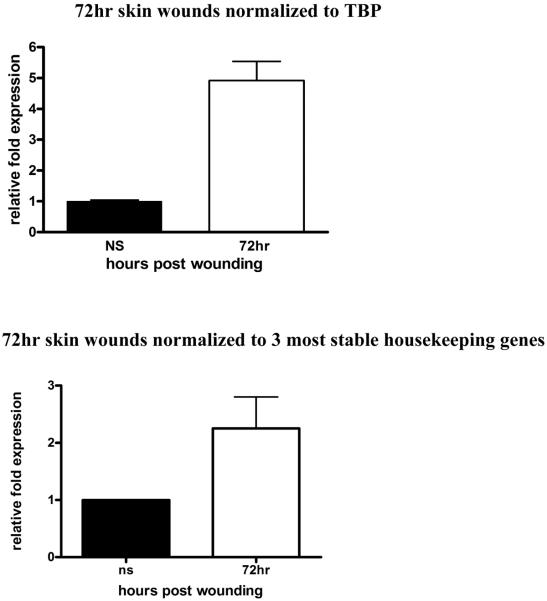
Keratinocyte Growth Factor-2 (KGF-2) is a potent mitogenic factor for keratinocytes (6) and its level of mRNA has been reported to increase by as much as 160 times upon skin injury (7). We analyzed its expression in wounds harvested from 24h, 48h, 72h and 5 days. We normalized KGF-2 to TBP (Fig. 3A), which we determined to be the most stable housekeeping gene across the spectrum of the healing stages (Table 3). β2M is the next “most suitable” candidate housekeeping gene for skin wound samples (Table 3); in Figure 3B, we show its expression profile is similar to KGF-2 normalized to TBP. In contrast, we normalized KGF-2 to GAPDH, which is generally used in wounding related RT-PCR reactions (Fig. 3C).
Figure 3. KGF-2 expression normalized in wounds.
RT-PCR was performed on RNA isolated from normal tissues or wound samples at indicated times post-injury. To determine relative changes in KGF-2 mRNA levels, results were normalized to various housekeeping genes. A) KGF-2 normalized to TBP, *P<0.0003 by One-way analysis of variance; B) KGF-2 normalized to β2M, *P<0.0004 by One-way analysis of variance C) KGF-2 normalized to GAPDH, *P<0.0001 by One-way analysis of variance; D) KGF-2 normalized to single housekeeping gene, TBP vs 3 most stable housekeeping genes for each time point, chosen according to their M values from Table 3. A normalization factor was determined for each time point.
Normal skin was normalized to 1 (A–D).
The GAPDH “M” value traced closely behind TBP in most cases (Figure 2A thru 2D). However, there is a drastic difference between the expression profiles of KGF-2 normalized to GAPDH versus TBP. The reason for this difference lies in the calculation of relative quantitation, meaning that it is best to select for a housekeeping gene that has a comparable number of copies per cell as the target gene, allowing for measurement of the genes to be performed on the same linear scale. KGF-2 is a low-to mid-range expressing gene in normal skin, thus it is best to normalize it to a housekeeping gene, such as TBP, which also is a low-to mid-range expressing gene. Alternative housekeeping genes, such as GAPDH which is a high-range expressing in normal skin tissue and in skin wounds, are best used as normalizing genes for target genes at comparable expression levels.
In a multiple housekeeping gene approach, the three most stable housekeeping genes are chosen according to their “M” value. Normalization factor is determined for these 3 genes by using the geometric mean of housekeeping gene expression as a normalization factor instead of the arithmetic mean, because it helps to control for possible outlying values and abundance differences among the different genes. In Figure 3D, we demonstrate how single vs multiple housekeeping gene approach affects KGF-2 expression at each wound healing time point. For each time point, the 3 most stable housekeeping genes were chosen according to their “M” values shown in Table 3 and a normalization factor was determined for each time point.
This data demonstrates importance of selecting appropriate housekeeping genes, and the effect of inappropriate selection on the data output.
DISCUSSION
In this study, we investigated for the first time the variability in the expression of a panel of housekeeping genes used for the normalization of real-time quantitative RT-qPCR in a murine wound healing model. When the results from the normal skin were compared to those of wounded tissue, we identified unique housekeeping genes that were most stable in each tissue and therefore could be validated as the most suitable internal controls for these RT-qPCR reactions. Our results indicate that there is a difference in housekeeping gene expression stability between wounded and unwounded tissues.
The candidate housekeeping genes that we have analyzed are generally used as internal controls, because they are believed to have relatively stable expression. We paid special attention to select genes from different functional families. This is required for geNorm analysis, the currently preferred software program for selection of the best housekeeping gene (4). The software calculates the internal control gene-stability measure “M”, which is defined as the average pairwise variation of a particular gene with all other control genes (4). This measure relies on the principle that the expression ratio of two ideal internal control genes is identical in all samples, regardless of the experimental condition or cell type. Variation of the expression ratios of two housekeeping genes reflects the fact that one of them is not constantly expressed. An increasing variation in ratios corresponds to decreasing expression stability. The 1.5 cut-off for “M” is optimally determined by the geNorm program. It measures deviation from standard, so 1.5 means deviation of 50%. If the deviation is higher than 50%, we consider them statistically insignificant for a scientifically valuable correlation to emerge. In essence, it is a way to quickly sort out those genes which deviate too far for futher analysis. Genes with a M value < 1.5 are not automatically considered to exhibit statistically important correlation, and they are further analyzed using stepwise exclusion method. Stepwise exclusion of the gene with the highest M value results in a combination of two constitutively expressed housekeeping genes that have the most stable expression in the tested samples.
We must also mention that the significance of testing the quality of RNA must not be overlooked. From previous experiments in our laboratory, we realize that RNA quality has a profound impact on the results, in terms of the significance of differential expression and variability of reference genes. Thus, we routinely test the integrity (18S/28S) and concentration of RNA by using an Experion instrument per the manufacturer's instruction. Besides testing the purity and integrity of RNA, we also the purity of input RNA material; it should be free of contaminating DNA. We performed a proper DNase treatment on all of our RNA samples to eliminate the presence of DNA.
Our results indicate that the TBP can be used as a reference gene for relative gene quantification and normalization in 1mm 24h, 48h, 72h and 5 day wounds as determined by geNorm program. However, in normal skin UBC, SDHA, and CYC1 were found to be the most stable housekeeping genes. Although ubiquitin is not commonly used as a normalization gene, in our normal skin studies it was found to be quite suitable. Previously, Czechowski et al. (8) pointed out that genes with fairly low levels of expression, such as genes of the ubiquitin complex, may be of particular interest for normalizing expression levels of genes which have moderate to low levels of expression.
Previous studies have also mentioned paying special attention to housekeeping genes in diseased skin, such as an increase in expression of GAPDH in all areas of epidermis of psoriatic plaque compared to normal skin (14). Interestingly, GAPDH and ACTIN genes are generally the most widely used housekeeping genes. ACTIN has been described as a high expression housekeeping gene in skin tissues (15). However, expression stability of ACTIN and GAPDH has been shown to vary significantly across tissues, cell types, and during cell proliferation and development (9, 10, 11, 12). De Jonge (13) et al. identified candidate housekeeping genes in human gene array samples, which had enhanced stability among different cells types and experimental conditions, with none of the commonly used housekeeping genes found in the top 50 stably expressed genes. The current study supports the idea that GAPDH and ACTIN may not be highly suitable as internal controls for wound healing studies that utilize RT-qPCR. Thus, caution should be applied in the selection of an appropriate housekeeping gene for such investigations. As we observed in our study, the selection of an appropriate internal control is essential for obtaining meaningful results in RT-qPCR studies. We tested the need for careful housekeeping gene selection for normalization in two additional wound healing models including a 3mm skin wound and a 1mm tongue wound (unpublished data). These studies have confirmed that best housekeeping gene for normalization must be independently selected for each model. For example, in 3mm skin wounds, we performed analysis of several target genes that are significant in wound healing. These genes are: Epidermal Growth Factor (EGF), Transforming Growth Factor beta 1 and 3 (TGFβ1 and 3), Vascular Endothelial Growth Factor (VEGF), and Keratinocyte Growth Factor 1 (KGF 1). We normalized these target genes to 3 different housekeeping genes, such as GAPDH, TBP, and β2M. This unpublished data also verified our conclusion of the importance of selecting appropriate housekeeping gene, and the affect of inappropriate selection on the data output.
Furthermore, although our results have identified suitable control genes for wound healing, this analysis is likely specific to the particular model that we utilized. Therefore, control gene selection must be validated for each experimental model, cell and tissue type or in healing vs non-healing diabetic wounds. In the latter case, the above discussed normalization analysis might not be sufficient due to the absence of set time course and might require additional/alternative methods.
ACKNOWLEDGMENTS
This publication was supported by Grants RO1-GM50875 (LAD), P20-GM078426 (LAD) and F31 (add your grant number) (AT). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS, NIDCR, or NIH.
ABBREVIATIONS
- KGF-2
Kerationcyte Growth Factor-2
- TBP
TATA-box binding protein
- B2M
β-2-microglobulin
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GUSb
Glucurodinase, beta
- RPLP2
Rribosomal protein, large p2
- ACTB
Actin, beta
- 18S
Ribosomal RNA 18S
- UBC
Ubiquitin C
- YWHAZ
Phospholipase A2
- ATP5B
ATP synthase subunit
- CANX
Calnexin
- CYC1
Cytochrome c-1
- SDHA
Succinate dehydrogenase complex, subunit A
REFERENCES
- 1.VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–26. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- 2.Nygard A, Jorgensen CB, Cirera S, Fredholm M. Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol Biol. 2007;8:67. doi: 10.1186/1471-2199-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–37. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 4.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepa A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002 Jun 18;3(7) doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. Epub 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minner F, Poumay Y. Candidate Housekeeping Genes Require Evalutation before the Selection for Studies of Human Epidermal Keratinocytes. J Investigative Dermatol. 2009;129:770–773. doi: 10.1038/jid.2008.247. [DOI] [PubMed] [Google Scholar]
- 6.Finch PW, Rubin JS, Miki T, Ron D, Aaronson SA. Human KGF is FGF-related with properties of a paracrine effector of epithelial cell growth. Science. 1989;245:752–55. doi: 10.1126/science.2475908. [DOI] [PubMed] [Google Scholar]
- 7.Werner S, Peters KG, Longaker MT, Fuller-Pace F, Banda MJ, Williams LT. Large induction of keratinocyte growth factor expression in the dermis during wound healing. Proc Natl Acad Sci. 1992;89:6896–900. doi: 10.1073/pnas.89.15.6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative realtime PCR. Biochem Biophys Res Commun. 2004;313:856–62. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 10.Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR. Beta-actin--an unsuitable internal control for RT-PCR. Mol Cell Probes. 2001;15:307–11. doi: 10.1006/mcpr.2001.0376. [DOI] [PubMed] [Google Scholar]
- 11.Glare EM, Divjak M, Bailey MJ, Walters EH. Beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57:765–770.12. doi: 10.1136/thorax.57.9.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deindl E, Boengler K, Van Royen N, Schaper W. Differential expression of GAPDH and beta3-actin in growing collateral arteries. Mol Cell Biochem. 2002;236:139–146. doi: 10.1023/a:1016166127465. [DOI] [PubMed] [Google Scholar]
- 13.de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PloS One. 2007;2:e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Rees J. Variation in epidermal housekeeping gene expression in different pathological states. Acta Derm Venereol. 2000;80:2–3. doi: 10.1080/000155500750012397. [DOI] [PubMed] [Google Scholar]
- 15.Bär M, Bär D, Lehmann B. Selection and validation of candidate housekeeping genes for studies of human keratinocytes - review and recommendations. J Invest Dermatol. 2009;129:770–3. doi: 10.1038/jid.2008.428. [DOI] [PubMed] [Google Scholar]