Abstract
Heme proteins are ubiquitous in biology and are commonly involved in critical processes such as electron transfer, oxidative phosphorylation, and signal transduction. Both the protein environment and the heme cofactor contribute to generate the range of chemical properties needed for these diverse functions. Using the H-NOX (Heme Nitric oxide/OXygen binding) protein from the thermophilic bacterium Thermoanerobacter tengcongensis, we have shown that heme electronic properties can be modulated by porphyrin distortion within the same protein scaffold without changing the heme ligation state or coordination environment. The degree of heme distortion was found to be directly correlated to the electron density at the heme iron, as evidenced by dramatic changes in the heme redox potential and pKa of the distal ligand (−OH vs. H2O). Protein-induced porphyrin distortion represents a new strategy to rationally tune the electronic properties of protein-bound porphyrins and could be used to engineer proteins with desired reactivity or functionality.
Hemoproteins are involved in diverse and complex redox reactions, including electron transfer and oxygen activation.1,2 A unifying feature of hemoproteins is the redox-active heme cofactor. However, the redox potentials of these proteins span an unusually wide range, from +450 to −550 mV (vs. SHE).2–4 Such a large variation of redox potentials has been attributed to various aspects of protein structure. For example, in human myoglobin and cytochromes, the hydrophobic residues in the heme-binding pocket are an important determinant of redox potential,5–11 and changes in this pocket alter the redox potential by more than 400 mV.3,12,13 Changes in the ligation and spin state of the heme iron also exert great influence upon the redox properties of hemoproteins, including those involved in electron transfer.14–16 Nevertheless, despite decades of study, those structural factors responsible for this variance in redox potentials remain to be fully elucidated. A complete understanding of the relationship between the structure and redox chemistry of hemoproteins would facilitate the rational design of proteins with desirable chemical properties.
The heme cofactor is an aromatic, highly conjugated macrocycle that adopts a planar conformation when free in solution (Figure S1a). However, evidence from high-resolution crystal structures and spectroscopic studies has shown that the heme-porphyrin structure need not be planar when protein-bound.17–19 Importantly, modes of porphyrin distortion, while energetically unfavorable,20 are conserved within classes of hemoproteins,19–21 suggesting that such distortions are likely to be important for protein function. The effect of porphyrin distortion on heme redox properties has been investigated by the use of synthetic porphyrins that adopt various conformations.18,22–25 Unfortunately, the information obtained through studies of synthetic heme analogues is inherently limited by the need to alter the porphyrin structure. An ideal model system would allow for the degree of porphyrin-distortion to be varied without altering the native heme.
It has been shown previously that the Heme Nitric oxide/OXygen binding (H-NOX) protein from T. tengcongensis (Tt H-NOX) contains a highly distorted heme cofactor. Moreover, when the distortion from planarity was lessened, the redox potential decreased by 171 mV.17,26 These earlier studies did not address whether a direct correlation exists between the degree of heme distortion and the redox potential. In the present study, site-directed mutagenesis was utilized to systematically modulate the porphyrin conformation in Tt H-NOX, while maintaining the hydrophobic character of the heme-binding pocket, and native ligands for Fe present in the wild type protein.
The I5L and I5L/P115A Tt H-NOX mutants were selected as candidates for altering the porphyrin conformation. These mutants disrupt the van der Waals contacts with the heme (Figure S1b–c).17,27 Notably, neither hydrophilic nor aromatic groups were introduced, thereby minimizing changes to the hydrophobicity and electrostatics in the heme pocket. The net change in partition coefficients were relatively small at values of 0.11, −0.38, and −0.49 for the I5L, I5L/P115A, and P115A mutants, respectively.28 These mutants were expected to have very similar heme environments while maintaining the coordination state of the wild-type protein.
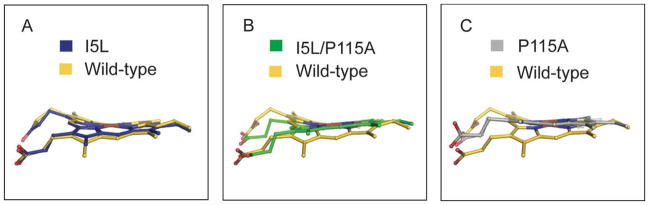
High-resolution crystal structures of the I5L and I5L/P115A mutants (2.15 and 2.04 Å, respectively) were solved in order to establish changes in the porphyrin conformation (Table S1). Structural analysis revealed that the I5L and I5L/P115A mutants have significantly flatter heme cofactors than wild-type Tt H-NOX (rmsd from planarity of 0.46 Å) (Figure 1a–b). The rmsd from planarity of both I5L heme molecules in the asymmetric unit (AU.) is 0.34 Å, whereas the rmsd from planarity for the heme molecules in the AU of I5L/P115A is 0.20 Å and 0.16 Å, respectively.
Figure 1.
Comparison of A) I5L (blue) B) I5L/P115A (green), and C) P115A17 (silver) overlaid with wild-type Tt H-NOX (gold).
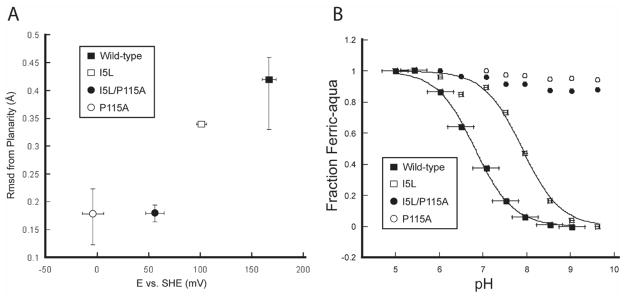
To establish whether changes in the heme conformation correlated with heme electronic properties, the reduction potentials of the I5L and I5L/P115A mutants were measured using previously described methods (Figure S2).17,29 The midpoint potentials for the I5L and I5L/P115A mutants were 101.5 ± 5.0 and 56.0 ± 8.9 mV, respectively. These midpoint potentials, in addition to those of the wild-type protein and the P115A mutant, were plotted against mean heme rmsd from planarity (Figure 2A) showing a clear trend between the degree of heme distortion and reduction potential. Taken together, the data show that as the planarity of the heme cofactor increases, the redox potential decreases.
Figure 2.
(A) Heme distortion versus reduction potential and (B) pH titration curves in wild-type Tt H-NOX and mutants.
UV-vis spectra of the ferric complexes of the wild-type and mutant proteins provide evidence for electron deficiency at the heme iron in the more distorted porphyrins. The absorption data indicate that distorted hemes bind an axial water ligand, in contrast to wild-type Tt H-NOX which binds a hydroxide ligand. The UV-vis spectrum of wild-type Tt H-NOX is characteristic of a 6-coordinate hydroxide-bound heme with a Soret maximum at 415 nm and α/β bands at 590/550 nm, respectively (Table 1 and Figure S3).30 In contrast, the UV-vis spectra of both the I5L/P115A and P115A mutants are characteristic of 6-coordinate aqua-bound heme groups,30 with Soret maxima at 404 and 405 nm and α/β bands at 615/530 and 610/533 nm, respectively (Figure S3). The spectrum of the ferric I5L mutant, which displays an intermediate degree of heme distortion, is characteristic of a mixture of both aqua and hydroxide-bound heme with a Soret maximum of 408 nm and α/β bands at 588/542 nm. These data suggest that the porphyrin distortion influences the pKa of the bound aqua ligand in H-NOX.
Table 1.
UV-vis spectral features and pKa values of ferric wild-type Tt H-NOX and mutants
| Tt H-NOX | Soret (nm) | α/β (nm) | pKa |
|---|---|---|---|
| Wild-type | 415 | 590/550 | 6.8 |
| I5L | 408 | 588/542 | 7.9 |
| I5L/P115A | 404 | 615/530 | >10 |
| P115A | 405 | 610/533 | >10 |
To test this hypothesis, pH titrations of the ferric wild-type and mutant Tt H-NOX proteins were carried out and the spectral changes recorded. The pKa values for the distal bound water for wild-type and I5L Tt H-NOX were 6.8 and 7.9, respectively (Figure 2B and Table 1). The pKa values of the I5L/P115A and P115A mutants could not be determined, due to protein instability at high pH, but no change in the absorption spectra of these mutants was observed up to a pH of 10, establishing a lower limit for their pKa values.
Results here show that heme distortion in wild-type Tt H-NOX leads to less electron density at the heme iron, as evidenced by the decreased pKa of the bound water ligand in the ferric oxidation state. The higher reduction potential is also consistent with less electron density localized at the iron in wild-type. High-resolution crystallographic data indicate that porphyrin distortion perturbs the metal site in Tt H-NOX away from an optimal octahedral geometry (Table S2–S3). Studies with saddled, synthetic porphyrins have shown that deviations from octahedral geometry favor an increase in the reduction potential of the Fe(III)/Fe(II) couple.31 Deviations of a similar nature are seen in the protein-bound porphyrins here as are similar changes in reduction potential.
Although it has been established for some time that heme pocket hydrophobicity and axial ligands contribute to heme redox potential, the data here provide direct evidence that protein-induced porphyrin distortion may alter the heme electronic properties. Modulating the redox potentials of porphyrins through this methodology represents a novel strategy to alter porphyrin chemistry and could be used, in concert with ligand and hydrophobic effects, to tailor proteins to have desired redox potentials and functionalities.
Supplementary Material
Acknowledgments
We thank S. Y. Reece for critical discussions and assistance in redox potentiometry. We thank E. Boon, M. Volgraf, and K. Connell for initial studies with the I5L and I5L/P115A mutants. We thank J. Yoon, T. Agapie, J. J. Woodward, J. J. Curley, M. B. Winter and members of the Marletta and Kuriyan Labs for critical discussion and review of the manuscript. We thank staff at the ALS beamline 8.2.1 for assistance during data collection. This work was supported by the NIH Grant GM070671.
Footnotes
Supporting Information Available: Supplemental figures, experimental details, X-ray diffraction data, and quantitative heme distortion analysis. The material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Sykes AG. Heme-Fe Proteins. Vol. 51 Academic Press; San Diego: 2001. [Google Scholar]
- 2.Reedy CJ, Elvekrog MM, Gibney BR. Nucleic Acids Res. 2008;36:D307–313. doi: 10.1093/nar/gkm814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shifman JM, Gibney BR, Sharp RE, Dutton PL. Biochemistry. 2000;39:14813–14821. doi: 10.1021/bi000927b. [DOI] [PubMed] [Google Scholar]
- 4.Paoli M, Marles-Wright J, Smith A. DNA Cell Biol. 2002;21:271–280. doi: 10.1089/104454902753759690. [DOI] [PubMed] [Google Scholar]
- 5.Varadarajan R, Zewert TE, Gray HB, Boxer SG. Science. 1989;243:69–72. doi: 10.1126/science.2563171. [DOI] [PubMed] [Google Scholar]
- 6.Mauk AG, Moore GR. J Biol Inorg Chem. 1997;2:119–125. [Google Scholar]
- 7.Rivera M, Seetharaman R, Girdhar D, Wirtz M, Zhang X, Wang X, White S. Biochemistry. 1998;37:1485–1494. doi: 10.1021/bi972390g. [DOI] [PubMed] [Google Scholar]
- 8.Gunner MR, Honig B. Proc Natl Acad Sci USA. 1991;88:9151–9155. doi: 10.1073/pnas.88.20.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertrand P, Mbarki O, Asso M, Blanchard L, Guerlesquin F, Tegoni M. Biochemistry. 1995;34:11071–11079. doi: 10.1021/bi00035a012. [DOI] [PubMed] [Google Scholar]
- 10.Pascher T, Chesick JP, Winkler JR, Gray HB. Science. 1996;271:1558–1560. doi: 10.1126/science.271.5255.1558. [DOI] [PubMed] [Google Scholar]
- 11.Mao JJ, Hauser K, Gunner MR. Biochemistry. 2003;42:9829–9840. doi: 10.1021/bi027288k. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy ML, Gibney BR. Curr Opin Struct Biol. 2001;11:485–490. doi: 10.1016/s0959-440x(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 13.Cowley AB, Kennedy ML, Silchenko S, Lukat-Rodgers GS, Rodgers KR, Benson DR. Inorg Chem. 2006;45:9985–10001. doi: 10.1021/ic052205k. [DOI] [PubMed] [Google Scholar]
- 14.Fisher MT, Sligar SG. J Am Chem Soc. 1985;107:5018–5019. [Google Scholar]
- 15.Tezcan FA, Winkler JR, Gray HB. J Am Chem Soc. 1998;120:13383–13388. [Google Scholar]
- 16.Battistuzzi G, Borsari M, Cowan JA, Ranieri A, Sola M. J Am Chem Soc. 2002;124:5315–5324. doi: 10.1021/ja017479v. [DOI] [PubMed] [Google Scholar]
- 17.Olea C, Jr, Boon EM, Pellicena P, Kuriyan J, Marletta MA. ACS Chem Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Chem Soc Rev. 1998;27:31–41. [Google Scholar]
- 19.Jentzen W, Ma JG, Shelnutt JA. Biophys J. 1998;74:753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson KK, Hobbs JD, Luo LA, Stanley KD, Quirke JME, Shelnutt JA. J Am Chem Soc. 1993;115:12346–12352. [Google Scholar]
- 21.Ma JG, Laberge M, Song XZ, Jentzen W, Jia SL, Zhang J, Vanderkooi JM, Shelnutt JA. Biochemistry. 1998;37:5118–5128. doi: 10.1021/bi972375b. [DOI] [PubMed] [Google Scholar]
- 22.Patra R, Chaudhary A, Ghosh SK, Rath SP. Inorg Chem. 2008;47:8324–8335. doi: 10.1021/ic800944q. [DOI] [PubMed] [Google Scholar]
- 23.Barkigia KM, Chantranupong L, Smith KM, Fajer J. J Am Chem Soc. 1988;110:7566–7567. [Google Scholar]
- 24.Ravikanth M, Chandrashekar TK. Coordination Chem. 1995;82:105–188. [Google Scholar]
- 25.Kadish KM, Lin M, Van Caemelbecke E, De Stefano G, Medforth CJ, Nurco DJ, Nelson NY, Krattinger B, Muzzi CM, Jaquinod L, Xu Y, Shyr DC, Smith KM, Shelnutt JA. Inorg Chem. 2002;41:6673–6687. doi: 10.1021/ic0200702. [DOI] [PubMed] [Google Scholar]
- 26.Tran R, Boon EM, Marletta MA, Mathies RA. Biochemistry. 2009;48:8568–8577. doi: 10.1021/bi900563g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Proc Natl Acad Sci USA. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abraham DJ, Leo AJ. Proteins. 1987;2:130–152. doi: 10.1002/prot.340020207. [DOI] [PubMed] [Google Scholar]
- 29.Dutton PL. Methods Enzymol. 1978;54:411–435. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 30.Antonini E, Brunori M. Hemoglobin and Myoglobin in Their Reactions with Ligands. 1. Vol. 21 North-Holland Publishing Company; Amsterdam: 1971. [Google Scholar]
- 31.Grinstaff MW, Hill MG, Birnbaum ER, Schaefer WP, Labinger JA, Gray HB. Inorg Chem. 1995;34:4896–4902. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.