Abstract
Development of the mouse central nervous system was reported to be normal in the absence of either Sox4 or its close relative Sox11 despite strong and widespread expression of both transcription factors. Here we show that combined absence of both Sox proteins in the mouse leads to severe hypoplasia of the developing spinal cord. Proliferation of neuroepithelial precursor cells in the ventricular zone was unaffected. These cells also acquired their correct positional identity. Both glial and neuronal progenitors were generated and neurons appeared in a similar spatiotemporal pattern as in the wildtype. Rates of cell death were however dramatically increased throughout embryogenesis in the double deficient spinal cord arguing that Sox4 and Sox11 are jointly and redundantly required for cell survival. The absence of pronounced proliferation, patterning, specification, and maturation defects furthermore indicates that the decreased cell survival is not a secondary effect of one of these events. We therefore conclude that the two Sox proteins directly function as pro-survival factors during spinal cord development in neural cell types.
Keywords: spinal cord, neurogenesis, gliogenesis, apoptosis, proliferation, cell survival
INTRODUCTION
Several Sox proteins are known transcriptional regulators of vertebrate nervous system development (Wegner and Stolt 2005). Many more are broadly expressed throughout this tissue. These include the three closely related SoxC factors Sox4, Sox11 and Sox12 (Uwanogho et al. 1995; Hargrave et al. 1997; Kuhlbrodt et al. 1998; Bergsland et al. 2006; Dy et al. 2008; Hoser et al. 2008). In the sympathetic nervous system (SNS) SoxC protein expression is restricted to neuronal progenitor cells (Potzner et al. 2010), whereas in the developing central nervous system (CNS) SoxC proteins occur in uncommitted precursors as well as neuronal and glial cells (Uwanogho et al. 1995; Hargrave et al. 1997; Kuhlbrodt et al. 1998; Bergsland et al. 2006; Potzner et al. 2007; Dy et al. 2008; Hoser et al. 2008).
Although many developmental disturbances occur in Sox4-deficient and Sox11-deficient mice and lead to pre- or perinatal lethality, nervous system defects were not apparent (Schilham et al. 1996; Cheung et al. 2000; Sock et al. 2004; Hoser et al. 2008). The absence of overt neural phenotypes has led to the assumption that SoxC proteins may function redundantly during nervous system development.
Functional redundancy has indeed been recently shown in mice with multiple SoxC gene deficiencies during early organogenesis (Bhattaram et al. 2010) and in the SNS (Potzner et al. 2010). During SNS development, SoxC proteins first promote the proliferation of sympathetic progenitors and later enhance their survival so that the SNS is severely hypoplastic in mice with combined Sox4 and Sox11 deficiencies. Because of slightly different expression patterns, Sox11 is more important for the early effect on proliferation, whereas Sox4 is the predominant survival factor.
Further evidence for a role in nervous system development comes from overexpression studies. Overexpression of a Sox4 transgene in the mouse had shown that prolonged Sox4 expression in glial cells of the CNS interferes with their terminal differentiation (Hoser et al. 2007; Potzner et al. 2007). SoxC overexpression in the developing chicken neural tube, in contrast, led to the premature expression of panneuronal markers and precocious establishment of neuronal properties and thus implicated SoxC proteins in neuronal maturation (Bergsland et al. 2006).
Here, we have used mice with combined deficiencies to address the function of Sox4 and Sox11 in the developing spinal cord (SC) as a model region for the CNS.
MATERIALS AND METHODS
Mouse husbandry, genotyping, BrdU labelling, and dissections
Mice used in this study carried Sox4loxP (Penzo-Mendez et al. 2007) and Sox11lacZ (Sock et al. 2004) alleles on a mixed 129SvJ × C57Bl/6J × NMRI background. For conditional deletion of the Sox4loxP allele, a Brn4∷Cre transgene (bcre-32 line) was used (Ahn et al. 2001). Genotyping was performed by PCR. Primer sequences are available upon request. For BrdU labelling, pregnant mice were injected intraperitoneally with 100 μg BrdU (Sigma) per gram body weight 1 h or 24 h before dissection (Stolt et al. 2003). Embryos were obtained from 9.5 days post coitum (dpc) to 16.5 dpc from staged pregnancies, underwent fixation in 4% paraformaldehyde, and were frozen at −80°C in Jung Tissue Freezing Medium (Leica, Nussloch, Germany) after cryoprotection (Stolt et al. 2003; Potzner et al. 2010).
Immunohistochemistry, in situ hybridization and TUNEL
Immunohistochemistry and TUNEL were performed on 10 μm cryotome sections, in situ hybridization on 14 μm sections. Each experiment was carried out at least three times per genotype on transverse thoracic level sections from different embryos. For immunohistochemistry, the following primary antibodies were used in various combinations: guinea pig antisera against Lbx1 (1:2000 dilution, gift of C. Birchmeier, MDC, Berlin), Lmx1b (1:1000 dilution, gift of C. Birchmeier, MDC, Berlin), Sox4 (1:1500 dilution, Hoser et al. 2008), Sox10 (1:1000 dilution, Maka et al. 2005), Sox11 (1:1000 dilution, Hoser et al. 2008), Tlx1 (1:1000 dilution, gift of C. Birchmeier, MDC, Berlin), rabbit antisera against Ki67 (1:500 dilution, NeoMarkers), Olig2 (1:5000, gift of D. Rowitch, UCSF, San Francisco), phospho-Histone H3 (1:1000 dilution, Upstate Biotechnology), Sox2 (1:500 dilution, generated against a peptide spanning amino acids 10–38 of mouse Sox2), and mouse monoclonals against Tuj1 (1:4000 dilution, Covance), NeuN (1:600 dilution, Chemicon), Hb9 (1:50 dilution, Developmental Studies Hybridoma Bank), Islet-1 (1:2000 dilution, Developmental Studies Hybridoma Bank), Nkx2.2 (1:50 dilution, Developmental Studies Hybridoma Bank). Secondary antibodies conjugated to Alexa 488, Cy2 or Cy3 immunofluorescent dyes (Dianova and Molecular Probes) were used for detection. Incorporated BrdU was visualized by Alexa-488 coupled mouse monoclonals against BrdU (1:20 dilution, Molecular Probes). TUNEL was performed according to the manufacturer`s protocol (Chemicon).
In situ hybridizations were performed using DIG-labelled antisense riboprobes for Irx3, Nkx6.1 and Olig2 as described (Stolt et al. 2003).
Quantifications
Numbers of immunoreactive cells or DAPI-positive nuclei in one half of the SC were counted per section. Data were obtained from 12 or more sections from at least three different embryos for each genotype and embryonic age. Diagrams show mean values ± SEM. Statistical significance was determined by unpaired two-tailed Student's t test using Microsoft Excel software and P values are given in the figure legends.
RESULTS
SoxC gene expression and deletion in the developing SC
The occurrence and distribution of Sox4 and Sox11 in the developing SC has been extensively characterized at the transcript level (Uwanogho et al. 1995; Hargrave et al. 1997; Kuhlbrodt et al. 1998; Bergsland et al. 2006; Dy et al. 2008; Hoser et al. 2008). At the protein level, only few data are available. Therefore we analyzed this by immunohistochemistry. At 10.5 dpc, both SoxC proteins were uniformly present at high levels throughout the SC with its homogeneous precursor cell population (Suppl. Fig. S1A,K). At 11.5 and 12.5 dpc, SoxC proteins were still present throughout the SC, but levels in the ventricular zone (VZ) had started to decrease (Suppl. Fig. S1B,C,L,M) arguing that SoxC proteins are gradually lost from the precursor cell population. From 14.5 dpc onwards, Sox4 staining additionally decreased in the mantle zone, particularly in the more lateral and ventral regions (Suppl. Fig. S1D,E). A similar decrease was also observed with a slight temporal delay for Sox11 (Suppl. Fig. S1N,O). Considering that the ventrolateral regions generally contain the most mature neurons in the developing SC, Sox4 and Sox11 expression is likely lost during neuronal maturation. Sox4 and Sox11 were also present in glial cells, although usually at relatively low levels as judged by co-immunohistochemistry with NeuN as a neuronal and Sox9 as a glial marker (Suppl. Fig. S2A–F and data not shown). At the time of birth, Sox4 and Sox11 had almost disappeared from the SC (data not shown). These results show congruence between the occurrence of SoxC proteins and transcripts. They confirm that Sox4 and Sox11 are expressed in neuroepithelial precursors, committed glial and neuronal progenitors and in maturing, but not fully mature neurons.
Double heterozygous animals with a Sox11lacZ allele and a constitutive Sox4 null allele were not viable (data not shown). Therefore we combined the Sox11lacZ allele with a conditional Sox4 allele (Penzo-Mendez et al. 2007) and a Brn4∷Cre transgene (Ahn et al. 2001). The Brn4∷Cre transgene is already expressed at 8.5 dpc in the developing CNS and present throughout the SC at 9.5 dpc. Outside the CNS, it is only expressed in the developing limb ectoderm. Consistent with the early and restricted expression of the Brn4∷Cre transgene, Sox4 protein had already disappeared from the SC at 9.5 dpc but remained detectable in the surrounding mesodermal tissues (Suppl. Fig. S1U–X). The recombined Sox4 allele is henceforth referred to as Sox4Δ.
In the absence of Sox11, we failed to detect a compensatory upregulation of Sox4 expression. The Sox4-specific staining was very similar in its pattern and intensity in Sox11lacZ/lacZ and wildtype embryos from 10.5 dpc to 16.5 dpc (compare Suppl. Fig. S1A–E to Suppl. Fig. S1F–J). Likewise, there was no indication of Sox11 upregulation in Sox4Δ/Δ SC (compare Suppl. Fig. S1K–O to Suppl. Fig. S1P–T) arguing that any potential compensatory activity between these two SoxC proteins in the SC is not dependent on increased expression of one protein in the absence of the other.
Cell number reductions in the Sox4 and Sox11 double deficient SC
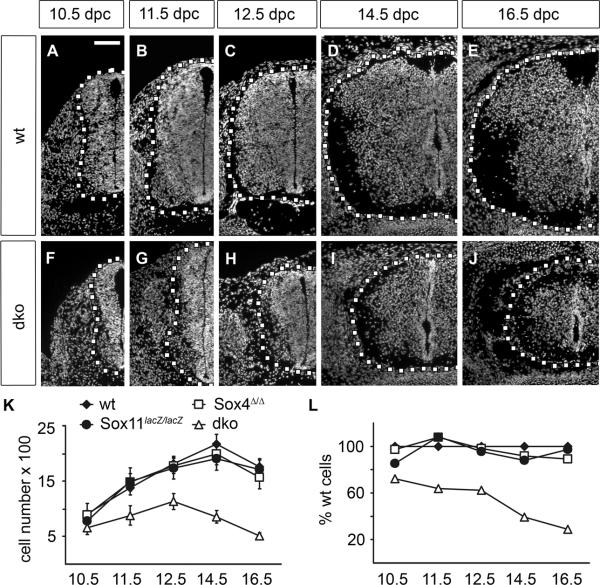
Sox4Δ/Δ Sox11lacZ/lacZ mice did not survive after birth thus limiting our analysis to embryogenesis. First we studied SC appearance at several developmental stages. It became immediately obvious that the SC of Sox4Δ/Δ Sox11lacZ/lacZ embryos was dramatically smaller than that of wildtype controls. The size reduction was already obvious at 10.5 dpc (Fig. 1A,F) and became even more pronounced later (compare Fig. 1B–E to Fig. 1G–J). While the dorsal horn had started to form in the wildtype SC at 14.5dpc (Fig. 1D), no such structure was discernible in the double mutant (Fig. 1I,J). Counting the number of DAPI-positive nuclei in the SC per cross section revealed the presence of approximately 660 cells in one half of the Sox4Δ/Δ Sox11lacZ/lacZ SC at 10.5 dpc as opposed to 910 in the wildtype (Fig. 1K). By 12.5 dpc, the number of cells reached its maximum in Sox4Δ/Δ Sox11lacZ/lacZ embryos with 1100 per SC half compared to 1800 cells in the wildtype. Whereas the cell number in the wildtype SC continued to increase to a maximum of 2200 per half at 14.5 dpc, cell numbers had already started to decline in the Sox4Δ/Δ Sox11lacZ/lacZ SC by this age (Fig. 1K). At 16.5 dpc, 510 cells were counted in the Sox4Δ/Δ Sox11lacZ/lacZ embryos per SC half compared to 1800 cells in the wildtype. When set into relation to the wildtype SC cell number, double-deficient SC contained 70% of the wildtype SC cells at 10.5 dpc (Fig. 1L). From this value cell numbers continuously decreased to 30% at 16.5 dpc.
Fig. 1. Size comparison of embryonic SC from wildtype mice and mice with SoxC deficiencies.
(A–J) DAPI staining was performed on transverse thoracic level sections of wildtype (wt) (A–E) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) (F–J) embryos at 10.5 dpc (A,F), 11.5 dpc (B,G), 12.5 dpc (C,H), 14.5 dpc (D,I) and 16.5 dpc (E,J). Only one half of the SC is shown with its circumference indicated by a dotted line. Scale bar in A is valid for all panels and corresponds to 100 μm. (K,L) Cell numbers in SC were determined in wildtype (wt, filled diamond), Sox4loxP/loxP, Brn4::Cre (Sox4Δ/Δ, open square), Sox11lacZ/lacZ (filled circle) and Sox4Δ/Δ Sox11lacZ/lacZ (dko, open triangle) embryos by counting DAPI-positive nuclei in one SC half per section, and are presented as absolute numbers (K) and relative to wildtype numbers in percent (L). Reductions in cell number relative to the age-matched wildtype were statistically significant only for Sox4Δ/Δ Sox11lacZ/lacZ SC (P <0.001 at 10.5 dpc, 12.5 dpc, 14.5 dpc, 16.5 dpc; P <0.005 at 11.5 dpc) as determined by Student's t test.
We also included Sox4Δ/Δ and Sox11lacZ/lacZ single mutant SC in our analysis. By gross inspection, single mutant SC were morphologically indistinguishable from the wildtype and cell numbers were not significantly different (Fig. 1K,L and data not shown). As defects were only apparent in the double mutant, we restricted all further analyses to double mutant SC.
Normal dorsoventral patterning in the Sox4 and Sox11 double deficient SC
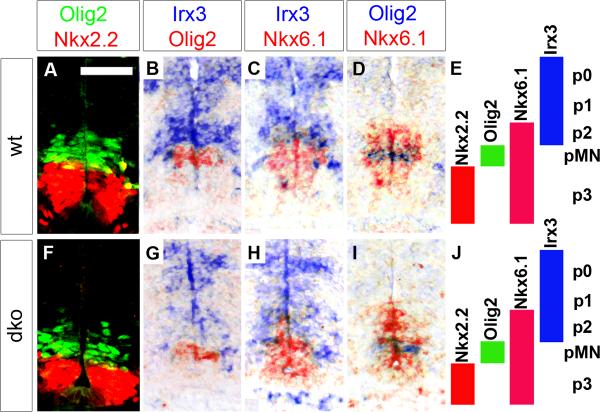
Several ways could be envisaged in which SoxC proteins function. Their occurrence in neuroepithelial precursor cells and committed progenitors is, for instance, compatible with a role in SC patterning. To address this issue, we compared the localization and boundaries of ventral domains in the VZ of wildtype and Sox4Δ/Δ Sox11lacZ/lacZ SC at 11.5 dpc (Fig. 2E,J). In both genotypes, the ventralmost p3 domain was marked by Nkx2.2 expression and was immediately adjacent to the pMN domain which followed dorsally and expressed Olig2 (Fig. 2A,F). Irx3 in contrast was expressed in the more dorsal p2, p1 and p0 domains (Fig. 2B,G). The p2 domain was similarly defined in Sox4Δ/Δ Sox11lacZ/lacZ and wildtype SC as the VZ part with overlapping Irx3 and Nkx6.1 expression (Fig. 2C,H). The pMN domain was furthermore embedded in both genotypes in a larger Nkx6.1-positive region that consists of p3, pMN and p2 domain (Fig. 2D,I). Ventral domain markers are thus similarly expressed in the VZ of wildtype and Sox4Δ/Δ Sox11lacZ/lacZ SC arguing that SoxC proteins do not influence dorsoventral patterning processes in the developing SC. This conclusion was also supported by studies with the dorsal domain markers Pax6 and Pax7 (data not shown).
Fig. 2. VZ patterning in embryonic SC from wildtype mice and mice with SoxC deficiencies.
Immunohistochemistry with antibodies specific for Nkx2.2 and Olig2 (A,F) and in situ hybridizations with antisense riboprobes for Olig2, Nkx6.1, and Irx3 (B–D,G–I) were performed on transverse thoracic level sections of wildtype (wt) (A–D) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) (F–I) embryos at 11.5 dpc to determine the integrity of the ventral VZ domains as summarized in E and J. In situ hybridization signals obtained with different probes on immediately adjacent sections were superimposed using Adobe Photoshop with the color of one of the signals being converted to red. A magnification of the signal displaying part of the VZ is shown. Scale bar in A is valid for all panels and corresponds to 50 μm.
Mild reduction of neuroepithelial precursors in the Sox4 and Sox11 double deficient SC
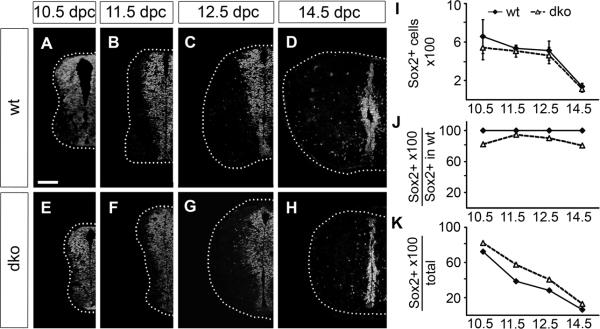
The presence of transcription factors responsible for positional identity of neuroepithelial precursors in the VZ of Sox4Δ/Δ Sox11lacZ/lacZ SC already indicates that these cells exist. Neuroepithelial precursors are furthermore characterized by Sox2 expression independent of their position along the dorsoventral axis. With Sox2 as a marker, we indeed detected neuroepithelial precursor cells at all dorsoventral levels of the Sox4Δ/Δ Sox11lacZ/lacZ SC from 10.5 dpc until 14.5 dpc but the VZ appeared slightly thinner than in the wildtype (compare Fig. 3A–D to Fig. 3E–H). From 14.5 dpc onwards, neuroepithelial precursors and VZ vanished similarly in wildtype and Sox4Δ/Δ Sox11lacZ/lacZ SC (Fig. 3D,H).
Fig. 3. Neuroepithelial precursor cells in the embryonic SC of wildtype mice and mice with SoxC deficiencies.
(A–H) Immunohistochemistry was performed with antibodies directed against Sox2 on transverse thoracic level sections of wildtype (wt) (A–D) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) (E–H) embryos at 10.5 dpc (A,E), 11.5 dpc (B,F), 12.5 dpc (C,G), and 14.5 dpc (D,H). Only one half of the SC is shown placed on a black background with its circumference indicated by a dotted line. Scale bar in A is valid for all panels and corresponds to 100 μm. (I–K) Sox2-positive cell numbers were determined in one SC half per section for wildtype (wt, filled diamond) and Sox4Δ/Δ Sox11lacZ/lacZ (dko, open triangle) embryos at various times and are presented as absolute numbers (I), relative to wildtype Sox2-positive cell numbers (J) and relative to overall cell numbers in the respective genotype (K). Reductions in cell number relative to the age-matched wildtype were not statistically significant as determined by Student's t test.
Quantification of Sox2-positive cells in both genotypes confirmed their reduction in Sox4Δ/Δ Sox11lacZ/lacZ SC (Fig. 3I). Compared to wildtype, between 10% and 25% fewer Sox2-positive VZ cells were detected in Sox4Δ/Δ Sox11lacZ/lacZ SC at any given time (Fig. 3J). The difference, however, did not reach statistical significance. In effect, Sox2-positive VZ cells contributed a higher proportion to the overall cell number in Sox4Δ/Δ Sox11lacZ/lacZ than in wildtype SC (Fig. 3K) arguing that neuroepithelial precursors do not represent the cell population most severely affected by loss of Sox4 and Sox11.
Mild decrease of glial populations in the Sox4 and Sox11 double deficient SC
Both glia and neurons are specified from neuroepithelial precursors of the VZ. To study glial development in Sox4Δ/Δ Sox11lacZ/lacZ SC, we first characterized the oligodendroglia using Sox10 as a lineage-specific marker. Similar to the wildtype, Sox10-positive cells began to appear in Sox4Δ/Δ Sox11lacZ/lacZ SC around 12.0–12.5 dpc in the immediate vicinity of the pMN domain (Suppl. Fig. S3A,D). Two days later, Sox10-positive oligodendrocyte progenitors had started to colonize the Sox4Δ/Δ Sox11lacZ/lacZ SC. At 14.5 dpc, the density of oligodendrocyte progenitors was still higher in ventral regions of the SC similar to the wildtype and consistent with their ventral origin (Suppl. Fig. S3B,E). By 16.5 dpc, oligodendrocyte progenitors were more uniformly distributed throughout the SC. The behaviour of oligodendrocyte progenitors in Sox4Δ/Δ Sox11lacZ/lacZ and wildtype SC thus resembled each other (Suppl. Fig. S3C,F).
Although approximately equal numbers of oligodendrocyte progenitors were specified at 12.5 dpc in both genotypes, their numbers increased less strongly in Sox4Δ/Δ Sox11lacZ/lacZ SC (Suppl. Fig. S3G). We counted only 50% as many oligodendrocyte progenitors in the mutant as in the wildtype SC at 14.5 dpc and an even lower amount of 40% at 16.5 dpc (Suppl. Fig. S3H). However, when set into relation to the total cell number, Sox10-positive cells accounted for a higher percentage of cells in the mutant SC than in the wildtype from 14.5 dpc onwards (Suppl. Fig. S3I). We thus conclude that oligodendrocyte progenitors are affected in Sox4Δ/Δ Sox11lacZ/lacZ SC but less than other cell populations.
In addition to oligodendroglia, the embryonic SC also contains astrocyte progenitors. Unfortunately, no single marker exists that exclusively labels these cells during early development. To follow these cells and count their number, we had to resort to a marker combination. From 12.5 dpc onwards, Sox2 labels glial cells in the mantle zone in addition to neuroepithelial precursors in the VZ (Fig. 3C,D,G,H). Subtraction of the Sox10-positive oligodendroglial fraction from these Sox2-positive cells in the mantle zone identifies the astrocyte progenitors.
Their number was lower in Sox4Δ/Δ Sox11lacZ/lacZ than in wildtype SC at 14.5 dpc to 16.5 dpc (Suppl. Fig. S3J). Expressed in relative terms, Sox4Δ/Δ Sox11lacZ/lacZ SC contained only 75% as many astrocyte progenitors as the wildtype at 14.5 dpc (Suppl. Fig. S3K). This level decreased further to 40% at 16.5 dpc. The reduction of astrocyte progenitors thus nearly equals the one of oligodendrocyte progenitors. Similar to oligodendrocyte progenitors, astrocyte progenitors nevertheless constitute a higher fraction of cells in the mutant than in the wildtype SC from 14.5 dpc onwards (Suppl. Fig. S3L).
Strong decrease of neuronal populations in the Sox4 and Sox11 double deficient SC
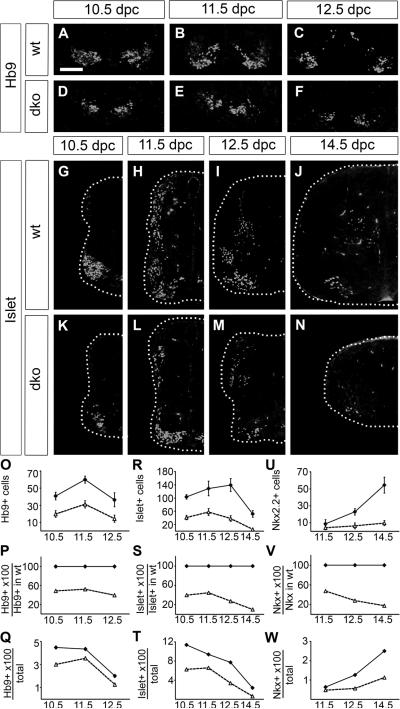
Since glia were not predominantly responsible for the observed loss of SC cells, we turned our analysis to neuronal populations. Early born motoneurons were studied using antibodies against Hb9 which is first expressed in all motoneuron progenitors and stays on in the medial column during motoneuron maturation. Hb9-positive cells appeared at the same time in wildtype and Sox4Δ/Δ Sox11lacZ/lacZ SC (Fig. 4A,D) and occupied comparable positions in the ventral horn area (compare Fig. 4A–C to Fig. 4D–F). Their number was, however, severely reduced in Sox4Δ/Δ Sox11lacZ/lacZ SC (Fig. 4O) and amounted to 50% of wildtype values at 10.5 dpc and 11.5 dpc (Fig. 4P). At 12.5 dpc, Hb9-positive cell numbers even dropped to 40% of wildtype levels. Relative to the total cell number, Hb9-positive cells were furthermore underrepresented at all time points in the mutant SC (Fig. 4Q).
Fig. 4. Neurons in the embryonic SC of wildtype mice and mice with SoxC deficiencies.
(A–N) Immunohistochemistry was performed with antibodies directed against Hb9 (A–F) and against Islet-1 (G–N) on transverse thoracic level sections of wildtype (wt) (A–C,G–J) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) (D–F,K–N) embryos at 10.5 dpc (A,D,G,K), 11.5 dpc (B,E,H,L), 12.5 dpc (C,F,I,M) and 14.5 dpc (J,N). In (A–F), the ventral most part of the SC is shown. In (G–N), one half of the SC is shown placed on a black background with its circumference indicated by a dotted line. Scale bar in A is valid for all panels and corresponds to 100 μm. (O–W) Numbers of Hb9-positive motoneurons (O–Q), Islet-1-positive moto- and interneurons (R–T) and Nkx2.2-positive ventral interneurons (U–W) were determined in one SC half per section for wildtype (wt, filled diamond) and Sox4Δ/Δ Sox11lacZ/lacZ (dko, open triangle) embryos at various times and are presented as absolute numbers (O,R,U), relative to wildtype marker-positive cell numbers (P,S,V) and relative to overall cell numbers in the respective genotype (Q,T,W). Reductions in cell number relative to the age-matched wildtype were statistically significant (P <0.001) for all markers and all analyzed embryonic ages in Sox4Δ/Δ Sox11lacZ/lacZ SC as determined by Student's t test.
When antibodies against Islet-1 were employed, similar observations were made. The antibody labels all motoneurons from the progenitor state throughout maturation as well as interneurons that arise dorsally and eventually take up their final position in the medial SC. Islet-1-positive cells were present and correctly localized in Sox4Δ/Δ Sox11lacZ/lacZ SC (compare Fig. 4G–J to Fig. 4K–N) but severely reduced in number (Fig. 4R) relative to their number in the wildtype (Fig. 4S) and compared to other cell types in the mutant SC (Fig. 4T). Similar results were also obtained for the Nkx2.2-positive ventral interneurons (Fig. 4U–W).
As the ventral neuronal populations were overproportionately affected by the loss of Sox4 and Sox11, we investigated whether this was also true for different populations of dorsal horn interneurons which are marked during their development by Lbx1, Tlx3 and Lmx1b (Müller et al. 2002). Lbx1, Tlx3 and Lmx1b were all detected in Sox4Δ/Δ Sox11lacZ/lacZ SC and their localization was comparable to the wildtype (compare Suppl. Fig. S4A–F to Suppl. Fig. S4G–L). Their numbers were, however, so strongly reduced that mutant SC failed to develop a typical dorsal horn. As exemplified for Lmx1b, quantifications confirmed the dramatic reduction in the number of marker-positive cells relative to the wildtype and relative to other cell types in the mutant SC (Suppl. Fig. S4M–O). We conclude that the loss of Sox4 and Sox11 exerts its strongest effect on neuronal populations.
Mild proliferation changes in the Sox4 and Sox11 double deficient SC
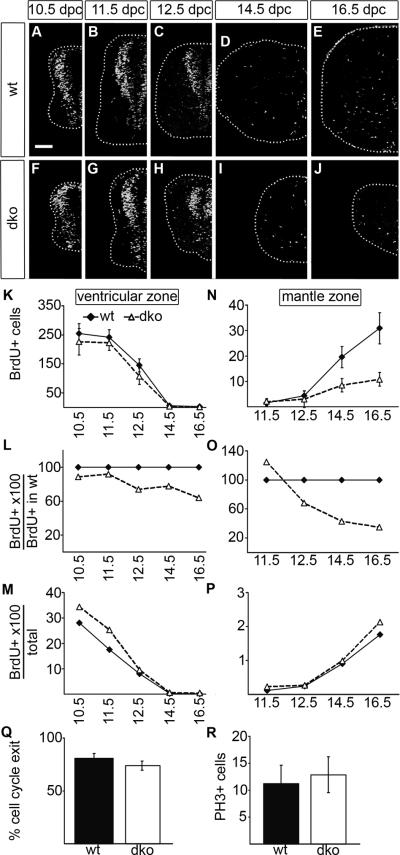
To determine the cause for the reduced cell numbers in the Sox4Δ/Δ Sox11lacZ/lacZ SC, we first studied proliferation and analyzed how many SC cells were able to incorporate BrdU in a one hour period at various times of development. In the wildtype proliferating cells were situated either in the VZ or the mantle zone (Fig. 5A–E). When localized in the VZ they correspond to neuroepithelial precursors, whereas proliferating mantle zone cells represent glial progenitors. As a consequence, proliferation in the VZ decreased at late phases of embryogenesis with depletion of the neuroepithelial precursor cell population, whereas glial proliferation in the mantle zone increased. This overall pattern was similarly observed for wildtype and Sox4Δ/Δ Sox11lacZ/lacZ SC (compare Fig. 5A–E to Fig. 5F–J).
Fig. 5. Proliferation and cell cycle parameters in the embryonic SC of wildtype mice and mice with SoxC deficiencies.
(A–J) BrdU integrated into the cellular DNA within a 1 hour period prior to embryo preparation was detected by anti-BrdU antibodies on transverse thoracic level sections of wildtype (wt) (A–E) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) (F–J) embryos at 10.5 dpc (A,F), 11.5 dpc (B,G), 12.5 dpc (C,H), 14.5 dpc (D,I) and 16.5 dpc (E,J). Only one half of the SC is shown placed on a black background with its circumference indicated by a dotted line. Scale bar in A is valid for all panels and corresponds to 100 μm. (K–P) The numbers of BrdU-labelled cells were separately determined for VZ (K,L,M) and mantle zone (N,O,P) in one half of the SC per section for wildtype (wt, filled diamond) and Sox4Δ/Δ Sox11lacZ/lacZ (dko, open triangle) embryos at various times and are presented as absolute numbers (K,N), relative to wildtype numbers of BrdU-labelled cells in the respective zone (L,O) and relative to overall cell numbers in VZ (M) or mantle zone (P) in the respective genotype. Reductions in cell number relative to the age-matched wildtype were statistically significant (P <0.001) for BrdU-positive cells in the mantle zone of Sox4Δ/Δ Sox11lacZ/lacZ SC at 14.5 and 16.5 dpc as determined by Student's t test. (Q) BrdU integrated into the cellular DNA 24 h prior to embryo preparation was detected by anti-BrdU antibodies on transverse thoracic level sections of wildtype (wt) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) embryos at 14.5 dpc and compared to Ki67-immunohistochemistry. The number of cells that were BrdU-positive but Ki67-negative were determined to quantify cell cycle exit. (R) Mitotic cells were detected with antibodies against phospho-Histone H3 (PH3) in wildtype (wt) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) embryos at 12.5 dpc. Differences between genotypes are not statistically significant in (Q,R).
We separately quantified proliferating cells in VZ and mantle zone to differentiate between neuroepithelial and glial proliferation. The number of BrdU-positive cells within the VZ was only slightly lower in the mutant than in the wildtype (Fig. 5K) and corresponded to around 90% of wildtype values at 10.5 dpc and 11.5 dpc and 70–80% at 12.5 and 14.5 dpc (Fig. 5L). As a consequence, proliferation rates within the VZ were at early times higher in Sox4Δ/Δ Sox11lacZ/lacZ SC and normalized to wildtype levels at later times (Fig. 5M). Therefore proliferation of neuroepithelial precursors is not compromised in the absence of Sox4 and Sox11.
Compared to the wildtype, BrdU-labelled cells were much less frequent in the mantle zone of Sox4Δ/Δ Sox11lacZ/lacZ SC from 14.5 dpc onwards (Fig. 5N). The relative reduction of BrdU-labelled cells in the mantle zone was even higher than the reduction in glial cell numbers (compare Fig. 5O to Suppl. Fig. S3H,K). Taking the lower total cell number into account, proliferation rates were, however, similar to the wildtype in the mantle zone (Fig. 5P).
We also analyzed cell cycle length and exit by performing Ki67 immunohistochemistry on SC that received the BrdU-label 24h before. Under these conditions, BrdU-labelled cells that are no longer Ki67-positive represent those cells that have exited the cell cycle within the last 24 hours. At 14.5 dpc we found no significant difference in this fraction of BrdU-labelled cells between Sox4Δ/Δ Sox11lacZ/lacZ and wildtype SC (Fig. 5Q) suggesting that cell cycle length and exit are unchanged. The number of cells in mitosis was also similar as indicated by comparable numbers of cells labelled with phospho-Histone H3 antibody in both genotypes (Fig. 5R).
Dramatically increased cell death in the Sox4 and Sox11 double deficient SC
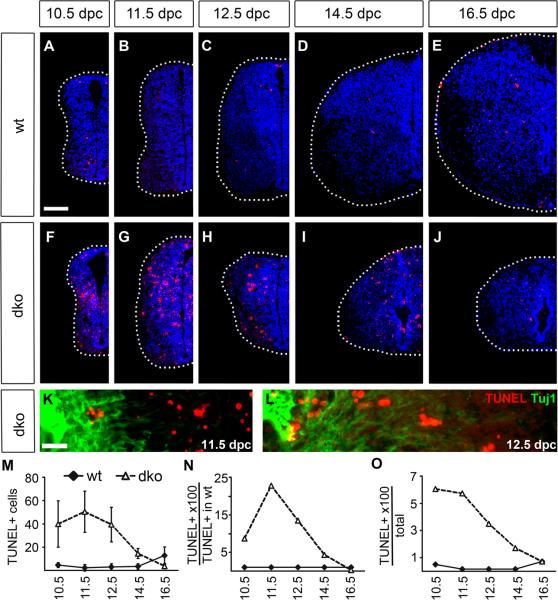
Next we studied the occurrence of apoptotic cell death by TUNEL. Already visual inspection revealed a dramatically increased rate of apoptosis in Sox4Δ/Δ Sox11lacZ/lacZ SC (compare Fig. 6A–E to Fig. 6F–J). This increase was visible at all embryonic ages until 16.5 dpc, the latest time point analyzed in this study. Apoptotic cells were found in all parts of the SC including VZ, subventricular zone and mantle zone at all dorsoventral levels and only a fraction of apoptotic cells were positive for the neuronal marker Tuj-1 (Fig. 6K,L) arguing that apoptosis is not restricted to a particular cell type in Sox4Δ/Δ Sox11lacZ/lacZ SC.
Fig. 6. Cell death in the embryonic SC of wildtype mice and mice with SoxC deficiencies.
(A–L) TUNEL (in red) was performed on transverse thoracic level sections of wildtype (wt) (A–E) and Sox4Δ/Δ Sox11lacZ/lacZ (dko) (F–L) embryos at 10.5 dpc (A,F), 11.5 dpc (B,G,K), 12.5 dpc (C,H,L), 14.5 dpc (D,I) and 16.5 dpc (E,J). In (A–J), one half of the SC is shown placed on a black background with its circumference indicated by a dotted line. Nuclei are counterstained by DAPI (blue). In (K,L), Tuj-1 immunohistochemistry (green) was additionally performed. The VZ is at the right margin of each panel. Scale bars correspond to 100 μm in A (valid for A–J) and to 25 μm in K (valid for K,L). (M–O) The numbers of TUNEL-positive cells were determined in one SC half per section for wildtype (wt, filled diamond) and Sox4Δ/Δ Sox11lacZ/lacZ (dko, open triangle) embryos at various times and are presented as absolute numbers (M), relative to wildtype numbers of TUNEL-positive cells (N) and relative to overall cell numbers in the respective genotype (O). Increases in TUNEL-positive cell number relative to the age-matched wildtype were statistically significant (P <0.001) for Sox4Δ/Δ Sox11lacZ/lacZ SC until 14.5 dpc as determined by Student's t test.
Absolute numbers of TUNEL-positive cells remained significantly higher than in the wildtype until 14.5 dpc (Fig. 6M). Compared to the wildtype SC, apoptotic cell numbers were 9-fold higher in the mutant at 10.5 dpc, 23-fold higher at 11.5 dpc, 13-fold higher at 12.5 dpc and still 4-fold higher at 14.5 dpc (Fig. 6N). Between 3.5% to 6% of all cells in the mutant SC underwent apoptosis at any given time from 10.5 dpc to 12.5 dpc compared to less than 0.5% in the wildtype (Fig. 6O). Sox4 and Sox11 are thus essentially required for cell survival in the embryonic SC over an extended period of time.
DISCUSSION
In this manuscript we show that the combined deletion of the two closely related SoxC proteins Sox4 and Sox11 causes a dramatic increase in apoptosis in the developing SC. We therefore conclude that SoxC transcription factors are essential for cell survival. Preliminary results indicate that this is also the case in other parts of the CNS. Previous analyses in single Sox4- and Sox11-deficient mice had failed to report comparable SC defects (Cheung et al. 2000; Sock et al. 2004). With knowledge of the phenotype in double deficient mice, we reassessed the single deficient mice. These analyses revealed a barely increased rate of apoptosis in Sox11-deficient SC. Apoptosis was more prominent in Sox4-deficient SC, but still much lower than in the double deficient tissue (data not shown). Cell loss in single deficient SC was furthermore not statistically significant indicating that the observed increases in apoptosis were compensated. Our results thus confirm the hypothesis that Sox4 and Sox11 function during SC development in a largely redundant manner.
Proliferation of VZ cells was not dramatically affected. Rates even appeared slightly increased, probably representing a compensatory response to the decreased survival. VZ cells also acquired their proper positional identity in the absence of Sox4 and Sox11 indicating that these factors do not have a major impact on patterning processes in the CNS. Additionally, we found no evidence for neuronal specification or maturation defects, as all the markers for neuronal progenitors and neurons were detectable in mutant SC at times and in patterns comparable to the wildtype. Oligodendrocyte and astrocyte progenitors were likewise present in double deficient SC. We conclude from these results that Sox4 and Sox11 primarily function in cell survival and that cell death in the mutant is not a secondary consequence of a role in proliferation, patterning, specification or maturation processes.
A cell survival function for Sox4 and/or Sox11 has also been postulated in several tumor entities (Frierson et al. 2002; Liu et al. 2006; Pramoonjago et al. 2006; Vanaja et al. 2006; Haram et al. 2008). In most tumors SoxC proteins appear to promote survival and inhibit apoptosis. However, there are also examples for pro-apoptotic functions, including one for Sox4 in bladder carcinoma (Aaboe et al. 2006) and for Sox11 in mantle cell lymphoma (Ek et al. 2008; Wang et al. 2008). Examples for a survival function of SoxC proteins in contrast are scarce from embryonic development, but start to become apparent and have recently been reported during early postgastrulation organogenesis and in the developing SNS (Bhattaram et al. 2010; Potzner et al. 2010). From the localization of apoptotic figures in the SC at later stages of development the survival function of SoxC proteins appears to be broad and not restricted to neuroepithelial precursors. Nevertheless, the impact of such an increased apoptosis varied between cell types. Proliferating cell types appeared to compensate for increased cell loss by hyperproliferation. This may explain why postmitotic neuronal progenitors and neurons are most strongly affected and most severely underrepresented in the absence of Sox4 and Sox11. Alternatively, Sox4 and Sox11 may be more protective for neuronal cells in accord with their higher levels in these as compared to other SC cells. Sox4 and Sox11 are expressed in neuroepithelial precursors of the VZ, neuronal and glial progenitors, and even in maturing neurons (Uwanogho et al. 1995; Hargrave et al. 1997; Kuhlbrodt et al. 1998; Bergsland et al. 2006; Dy et al. 2008; Hoser et al. 2008). It is possible that the two SoxC proteins affect survival of all these cells in a cell-intrinsic manner. However, the different cell types clearly influence each other so that it is equally plausible that loss of SoxC proteins in one cell type influences survival of another cell type. Oligodendrocyte progenitors, for instance, rely on neuronal growth factors such as PDGF for their survival and proliferation (Noble et al. 1988; Richardson et al. 1988). The dramatic reduction of the neuronal population will thus almost certainly decrease oligodendroglial survival and proliferation in the SoxC-deficient SC and may in fact explain why their proliferation is not as much upregulated as neuroepithelial precursor proliferation.
Recently, we have analyzed the role of Sox4 and Sox11 in the developing SNS and have found SoxC proteins to influence proliferation during early periods and survival at late ones (Potzner et al. 2010). Thus, there are similarities but also substantial differences in the role of SoxC proteins in SNS and SC development, further supporting the context-dependence of their function.
The role of SoxC proteins in SC development had previously been studied by electroporation in the neural tube of chicken embryos (Bergsland et al. 2006). Similar to our study, Bergsland and colleagues failed to detect any influence of SoxC proteins on proliferation and cell cycle parameters. However, Bergsland et al. found evidence for a role of SoxC proteins in activation of neuronal gene expression and neuronal maturation downstream of proneural bHLH proteins both by ectopic overexpression and siRNA-dependent downregulation. In contrast, effects on survival were very minor with less than a 2-fold increase of apoptosis after treatment with both Sox4- and Sox11-specific siRNA. On face value, these results are difficult to reconcile with ours, even if it has to be kept in mind that the studies were not only performed with different methods but also in different species and that species-specific differences in SoxC functions could exist. SoxC proteins may positively influence expression of panneuronal and subtype-specific markers and thereby exert an influence on neuronal maturation. However, it seems unlikely that Sox4 and Sox11 represent an absolute requirement for neuronal maturation as we have observed maturation of several types of neurons in the absence of both SoxC proteins.
The different effects of SoxC proteins on cell survival between our study and the work by Bergsland et al. are particularly striking. Considering that we only detected the dramatic increase in cell death in the double and not in the single mutant SC, it is possible that the siRNA-dependent knockdown had failed to reduce the combined amount of Sox4 and Sox11 in the chicken neural tube to levels low enough to detect the survival function.
Essential roles for cell survival in the developing CNS have previously been reported for a number of signalling molecules and transcription factors including Pax3, Rb, Sal proteins, JNK kinases and K-Ras (Macleod et al. 1996; Koera et al. 1997; Kuan et al. 1999; Pani et al. 2002; Bohm et al. 2008). In the corresponding mouse mutants, apoptosis was, however, restricted to specific cell types or thought to be secondary to alterations in cell cycle and proliferation characteristics. Sox4 has been reported to interact with and to stabilize p53 thereby leading to increased cell death in a lung carcinoma cell line (Pan et al. 2009). Although this proapoptotic function is the exact opposite of the antiapoptotic role that we detected in the developing SC, it may point to a more general link between SoxC proteins, p53 and the apoptotic machinery.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Rowitch, C. Birchmeier and T. Müller for the gift of antibodies and plasmids. This work was supported by grants from the DFG to E.S. (So251/3-1) and the NIH to V.L. (R01-AR54153).
REFERENCES
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- Ahn K, Mishina Y, Hanks M, Behringer R, Crenshaw E. BMPR-IA signaling is required for the formation of the apical ectodermal ridge and dorsalventral patterning of the limb. Development. 2001;128:4449–4461. doi: 10.1242/dev.128.22.4449. [DOI] [PubMed] [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattaram P, Penzo-Méndez A, Sock E, Colmenares C, Kaneko KJ, DePamphilis ML, Wegner M, Lefebvre V. Organogenesis relies on Sox4, Sox11, and Sox12 for survival of neural and mesenchymal progenitor cells. Nature Communications. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm J, Buck A, Borozdin W, Mannan AU, Matysiak-Scholze U, Adham I, Schulz-Schaeffer W, Floss T, Wurst W, Kohlhase J, Barrionuevo F. Sall1, sall2, and sall4 are required for neural tube closure in mice. Am J Pathol. 2008;173:1455–1463. doi: 10.2353/ajpath.2008.071039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Mol. Brain Res. 2000;79:180–191. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- Dy P, Penzo-Mendez A, Wang H, Pedraza CE, Macklin WB, Lefebvre V. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36:3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek S, Dictor M, Jerkeman M, Jirström K, Borrebaeck C. Nuclear expression of the non B-cell lineage Sox11 transcription factor identifies mantle cell lymphoma. Blood. 2008;111:800–805. doi: 10.1182/blood-2007-06-093401. [DOI] [PubMed] [Google Scholar]
- Frierson HF, Jr., El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM. Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol. 2002;161:1315–1323. doi: 10.1016/S0002-9440(10)64408-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haram KM, Peltier HJ, Lu B, Bhasin M, Otu HH, Choy B, Regan M, Libermann TA, Latham GJ, Sanda MG, Arredouani MS. Gene expression profile of mouse prostate tumors reveals dysregulations in major biological processes and identifies potential murine targets for preclinical development of human prostate cancer therapy. Prostate. 2008;68:1517–1530. doi: 10.1002/pros.20803. [DOI] [PubMed] [Google Scholar]
- Hargrave M, Wright E, Kun J, Emery J, Cooper L, Koopman P. Expression of the Sox11 gene in mouse embryos suggests roles in neuronal maturation and epithelio-mesenchymal induction. Dev. Dyn. 1997;210:79–86. doi: 10.1002/(SICI)1097-0177(199710)210:2<79::AID-AJA1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hoser M, Baader SL, Bosl MR, Ihmer A, Wegner M, Sock E. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J Neurosci. 2007;27:5495–5505. doi: 10.1523/JNEUROSCI.1384-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoser M, Potzner MR, Koch JMC, Bösl MR, Wegner M, Sock E. Sox12 deletion in the mouse reveals non-reciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol. Cell. Biol. 2008;28:4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koera K, Nakamura K, Nakao K, Miyoshi J, Toyoshima K, Hatta T, Otani H, Aiba A, Katsuki M. K-ras is essential for the development of the mouse embryo. Oncogene. 1997;15:1151–1159. doi: 10.1038/sj.onc.1201284. [DOI] [PubMed] [Google Scholar]
- Kuan CY, Yang DD, Samanta Roy DR, Davis RJ, Rakic P, Flavell RA. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 1999;22:667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Enderich J, Hermans-Borgmeyer I, Wegner M. Cooperative function of POU proteins and Sox proteins in glial cells. J. Biol. Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66:4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- Macleod KF, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. Embo J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- Maka M, Stolt CC, Wegner M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005;277:155–169. doi: 10.1016/j.ydbio.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Müller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature. 1988;333:560–565. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- Pan X, Zhao J, Zhang WN, Li HY, Mu R, Zhou T, Zhang HY, Gong WL, Yu M, Man JH, Zhang PJ, Li AL, Zhang XM. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci U S A. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani L, Horal M, Loeken MR. Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function: implications for Pax-3- dependent development and tumorigenesis. Genes Dev. 2002;16:676–680. doi: 10.1101/gad.969302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo-Mendez A, Dy P, Pallavi B, Lefebvre V. Generation of mice harboring a Sox4 conditional null allele. Genesis. 2007;45:776–780. doi: 10.1002/dvg.20358. [DOI] [PubMed] [Google Scholar]
- Potzner M, Griffel C, Lütjen-Drecoll E, Bösl MR, Wegner M, Sock E. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol. Cell. Biol. 2007;27:5316–5326. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potzner MR, Tsarovina K, Binder E, Penzo-Méndez A, Lefebvre V, Rohrer H, Wegner M, Sock E. Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Development. 2010;137:775–784. doi: 10.1242/dev.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Pringle N, Mosley MJ, Westermark B, Dubois-Dalcq M. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53:309–319. doi: 10.1016/0092-8674(88)90392-3. [DOI] [PubMed] [Google Scholar]
- Schilham MW, Oosterwegel MA, Moerer P, Ya J, Deboer PAJ, Vandewetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- Sock E, Rettig SD, Enderich J, Bösl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol. 2004;24:6635–6644. doi: 10.1128/MCB.24.15.6635-6644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier M-C, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic Expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Vanaja DK, Ballman KV, Morlan BW, Cheville JC, Neumann RM, Lieber MM, Tindall DJ, Young CY. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin Cancer Res. 2006;12:1128–1136. doi: 10.1158/1078-0432.CCR-05-2072. [DOI] [PubMed] [Google Scholar]
- Wang X, Asplund A, Porwit A, Flygare J, Smith C, Christensson B, Sander B. The subcellular Sox11 distribution pattern identifies subsets of mantle cell lymphoma: correlation to overall survival. Br J Haematol. 2008;143:248–252. doi: 10.1111/j.1365-2141.2008.07329.x. [DOI] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.