Abstract
Site-specific cross-linking can generate homogeneous multimeric proteins of defined valency. Pancreatic-type ribonucleases are an especially attractive target, as their natural dimers can enter mammalian cells, evade the cytosolic ribonuclease inhibitor (RI), and exert their toxic ribonucleolytic activity. Here, we report on the use of eight distinct thiol-reactive cross-linking reagents to produce dimeric and trimeric conjugates of four pancreatic-type ribonucleases. Both the site of conjugation and, to a lesser extent, the propinquity of the monomers within the conjugate modulate affinity for RI, and hence cytotoxicity. Still, the cytotoxicity of the multimers is confounded in vitro by their increased hydrodynamic radius, which attenuates cytosolic entry. A monomeric RI-evasive variant of bovine pancreatic ribonuclease (RNase A) inhibits the growth of human prostate and lung tumors in mice. An RI-evasive trimeric conjugate inhibits tumor growth at a lower dose and with a less frequent administration than does the monomer. This effect is attributable to an enhanced persistence of the trimers in circulation. On a molecular basis, the trimer is ~300-fold more efficacious and as well tolerated as erlotinib, which is in clinical use for the treatment of lung cancer. These data encourage the development of mammalian ribonucleases for the treatment of human cancers.
INTRODUCTION
Bovine seminal ribonuclease (BS-RNase1) and several amphibian ribonucleases, including ranpirnase (Onconase®; ONC), exhibit innate antitumoral activity (1-3). ONC has demonstrated responses in patients with breast, non-small cell lung, and esophageal cancer, and unresectable malignant mesothelioma (4-6). Although these two homologues of bovine pancreatic ribonuclease (RNase A (7)) differ dramatically in both primary and quaternary structure (8, 9), they both evade inhibition by ribonuclease inhibitor (RI (10)), a protein that resides in the cytosol of mammalian cells. BS-RNase is a natural dimer that evades RI by steric repulsion (11); whereas ONC is a small monomer whose truncated surface loops cannot interact intimately with RI (12). The ability to retain ribonucleolytic activity in the presence of RI is a primary determinant of the cytotoxicity of BS-RNase, ONC, and engineered ribonucleases (13).
Prior to the discovery of BS-RNase (14), lyophilization of solutions of RNase A from aqueous acetic acid was found to yield dimers and higher order oligomers of RNase A (15). These oligomers of RNase A—as large as nonameric (16)—formed by domain swapping of the N- or C-termini, or both (17), and displayed antitumoral activity concomitant with their degree of oligomerization: tetramers > trimers > dimers (18). Tandem dimers of RNase A are likewise toxic to cancer cells (19).
In contrast to domain-swapping, covalent cross-linking does not allow for monomer dissociation (20, 21). Dimers of RNase A formed by cross-linking with amine-reactive imido esters, inhibit tumor cell proliferation in vitro (22), exhibit antitumoral activity in mice (23, 24), and have enhanced persistence in the circulation of rats (25). Cross-linked trimers of RNase A generated with a dimethyl suberimidate linker displayed higher toxicity towads a cervical carcinoma cell line than did cross-linked dimers (26).
Lysine is a common residue. RNase A contains 10 lysine residues, as well as an N-terminal amino group. Accordingly, a disadvantage of using amine-reactive reagents to cross-link RNase A is the heterogeneity of the ensuing products. Moreover, the amino group in the side-chain of Lys41 has a low pKa of ~8.8 (27) and is thus especially reactive. Its modification can reduce catalytic proficiency by 105-fold (28) and obviate cytotoxicity (29).
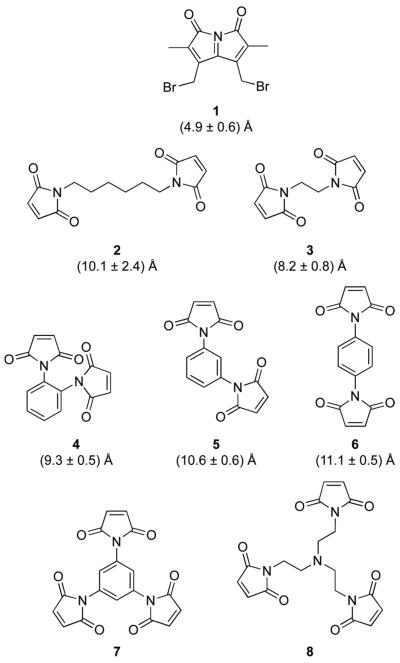
Previously, we used dibromobimane (1) (Figure 1) to prepare a BS-RNase dimer by forming thioether bonds at a particular cysteine residue (31). We found this dimer to be toxic to cancer cells in vitro, but did not determine its affinity for RI. Guided by the structure of the RI·RNase A complex (Figure 2), Youle and coworkers generated homogeneous homodimeric conjugates of both human pancreatic ribonuclease (RNase 1) and human eosinophil-derived neurotoxin (RNase 2) by installing a cysteine at a residue that contacts RI and reacting its thiol with 1,6-bis(maleimido)hexane (2) (Figure 1) (33). The resulting conjugates were less sensitive to inhibition by RI, and were cytotoxic. The thiol-reactive linker used by Youle and coworkers was, however, long and flexible, inserting 12 atoms and 9 rotatable bonds between the sulfur atoms of the nascent cysteine residues. We reasoned that greater RI-evasion, and thus greater cytotoxicity, could be achieved by using linkers that introduced fewer degrees of freedom between the ribonuclease moieties. In addition, we were inspired by the increased cytotoxicity of RNase A trimers, formed by either noncovalent, domain-swapping (18) or non-specific cross-linking (26).
Figure 1.
Structures of thiol-reactive linkers used to tether ribonucleases into dimers and trimers. The distances are between sulfur atoms in dimeric conjugates as calculated with stochastic molecular dynamics simulations (30).
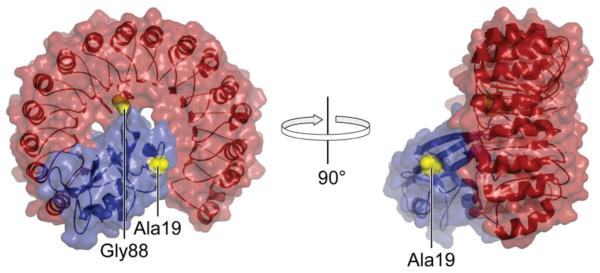
Figure 2.
Structure of the porcine RI·RNase A complex showing the location of residues 19 and 88 (yellow), which were replaced with cysteine in variants of RNase A (blue). The image was created with the program PyMOL (DeLano Scientific, South San Francisco, CA) using protein data bank entry 1dfj (32).
Here, we describe the semisynthesis and characterization of 13 dimeric and trimeric ribonuclease conjugates, derived from cross-linkers 1–8 and RNase A, BS-RNase, RNase 1, and ONC. We show that the choice of cross-linker as well as the site and valency of cross-linking affect the toxic activity toward cancer cells. The results have important implications for the development of ribonucleases as cancer chemotherapeutic agents.
MATERIALS AND METHODS
Materials
Protein production and characterization
E. coli BL21(DE3) cells, and pET22b(+) and pET27b(+) plasmids were from Novagen (Madison, WI). K-562 (derived from a continuous human chronic myelogenous leukemia line), DU 145 (human prostate carcinoma), and A549 (human non-small cell lung carcinoma) cells and were from the American Type Culture Collection (ATCC; Manassas, VA). Cell culture medium, Dulbecco's phosphate-buffered saline (DPBS), antibiotics, trypsin, and fetal bovine serum (FBS) were from Invitrogen (Carlsbad, CA). [methyl-3H]Thymidine (6.7 Ci/mmol) was from PerkinElmer (Boston, MA). Oligonucleotides for site-directed mutagenesis and sequencing, and the ribonuclease substrates, 6-FAM–dArUdAdA–6-TAMRA and 6-FAM–dArUdGdA–6-TAMRA, were from Integrated DNA Technologies (Coralville, IA).
1,6-Bis(maleimido)hexane (2), 1,2-bis(maleimido)ethane (3), tris(N-maleimidoethyl)amine (8), N-ethylmaleimide were from Thermo Fisher Scientific (Rockford, IL). 1,2-Bis(maleimido)benzene (4), 1,3-bis(maleimido)benzene (5), and 1,4-bis(maleimido)benzene (6) were from Sigma–Aldrich. Dibromobimane (1) was from Molecular Probes (Eugene, OR).
FPLC HiLoad 26/60 Superdex G75 and G200 gel-filtration columns, HiTrap Desalting, SP, and Q columns were from GE Healthcare (Uppsala, Sweden). Gel-filtration standards, SDS–PAGE molecular-weight standards, and pre-cast gels for poly(acrylamide) gel electrophoresis were from BioRad (Hercules, CA). Flat-bottom, black polystyrene (PS), Costar assay plates (96-well) with non-binding surface (NBS) were from Corning Life Sciences (Acton, MA). Molecular biology-grade bovine serum albumin (BSA) was obtained as a 20 mg/mL solution from Sigma–Aldrich (St. Louis, MO). Fluorometric enzymatic activity measurements were made using 4.5-mL methacrylate cuvettes from VWR (West Chester, PA). All other chemicals used were of commercial reagent grade or better, and were used without further purification.
Chemical synthesis
DMF was drawn from a Baker CYCLE-TAINER solvent delivery system. All other reagents were from Sigma–Aldrich or Fisher Scientific (Hanover Park, IL), and used without further purification. Thin-layer chromatography was performed using aluminum-backed plates coated with silica gel containing F254 phosphor and visualized by UV illumination or staining with I2, ceric ammonium molybdate, or phosphomolybdic acid. NMR spectra were obtained with a Bruker DMX-400 Avance spectrometer at the National Magnetic Resonance Facility at Madison (NMRFAM).
Xenograft studies
Erlotinib hydrochloride (Tarceva®) was from Genentech (South San Francisco, CA); docetaxel and cisplatin were from Sigma–Aldrich. Homozygous (nu/nu) male Balb/c (−/−) mice were from Harlan Sprague Dawley (Indianapolis, IN).
Instrumentation
[methyl-3H]Thymidine incorporation into K-562 and A549 genomic DNA was quantitated by scintillation counting using a Microbeta TriLux liquid scintillation and luminescence counter (PerkinElmer, Wellesley, MA). The molecular mass of ribonuclease variants and multimeric conjugates was confirmed by MALDI–TOF mass spectrometry using 3,5-dimethoxy-4-hydroxycinnamic acid as a matrix and a Voyager-DE-PRO Biospectrometry Workstation (Applied Biosystems, Foster City, CA) in the Biophysics Instrumentation Facility at the University of Wisconsin–Madison. Cuvette-scale fluorescence measurements were made with a QuantaMaster1 photon-counting fluorometer equipped with sample stirring (Photon Technology International, South Brunswick, NJ). Fluorescence-based activity inhibition assays, competition binding assays performed in 96-well plate format, and cell-viability assays utilizing CellTiter-Blue were read with an EnVison 2100 microplate reader (Perkin–Elmer, Waltham, MA) in the Keck Center for Chemical Genomics at the University of Wisconsin-Madison. Thermal denaturation data were acquired using a Cary 400 Bio double-beam spectrophotometer equipped with a Cary temperature controller (Varian, Palo Alto, CA) in the Biophysics Instrumentation Facility.
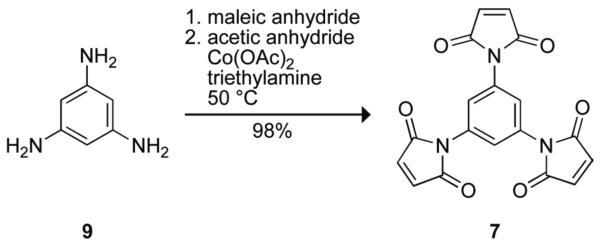
Synthesis of 1,3,5-tris(maleimido)benzene (7)
Linker 7 was designed to be a trimeric version of phenylene linkers 4–6, and to be more compact than known linker 1,3,5-tris(N-maleimidomethyl)benzene (34). Linker 7 was synthesized by the reaction shown in Scheme 1. First, 1,3,5-triaminobenzene (9) was synthesized by hydrogenation of phloroglucinol trioxime with Raney nickel, as described previously (35). Then, maleic anhydride (271 mg, 2.76 mmol) was dissolved in DMF (0.6 mL) under Ar(g). Compound 9 (98 mg, 0.78 mmol) in DMF (0.5 mL) was added dropwise, giving rise to a white precipitate. Acetic anhydride (0.7 mL, 7.41 mmol), a catalytic amount of Co(OAc)2, and triethylamine (40 μL, 0.29 mmol) were added, and the resulting brown solution was stirred for 20 h at 50 °C, protected from light. The reaction mixture was poured into ice water, and the resulting dark brown precipitate was collected by filtration. The solid was washed with water and Et2O, and dried under high vacuum (≤1 mm Hg). Linker 7 was isolated as a dark brown powder and used without further purification (286 mg, 98% yield). 1H NMR (400 MHz, DMSO-d6) β (ppm): 7.43 (s, 3H), 7.21 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ (ppm): 169.50, 134.82, 132.26, 124.12.
SCHEME 1.
Design of Cysteine-Containing Ribonuclease Variants
RNase A
We sought to test the hypothesis that greater evasion of RI would be attainable by tethering multiple ribonuclease monomers intimately via a residue within the RI–ribonuclease interface (36). In RNase A, residue 88 was chosen for conjugation (Figure 2), as single amino-acid substitutions there were known to be highly destabilizing to the RI·RNase A complex (37, 38). Residue 88 is a glycine, and its substitution with cysteine does not alter the net charge of the protein. Multimers of G88C were made with linkers 1–8 (Figure 1). Residue 19 of RNase A was chosen as a representative site outside of the RI–RNase A interface (Figure 2). Small molecules had been attached there without any detectable reduction in affinity for RI (39, 40). A trimer of A19C was made with linker 8. Trimeric conjugates were also prepared with D38R/R39D/N67R/G88C RNase A, which is a variant of RI-evasive D38R/R39D/N67R/G88R RNase A (38), using linker 8.
BS-RNase
BS-RNase And RNase A have high sequence identity (83%) and structural similarity (41). Accordingly, amino-acid substitutions in BS-RNase analogous to those that enabled RNase A to evade RI were also effective in endowing monomeric (that is, C31A/C32A) variants of BS-RNase with the ability to evade RI (42). Accordingly, we reasoned that tethering at residue 88 with linker 8 would enable BS-RNase to evade RI.
RNase 1
Although no single amino-acid substitution in the β4–β5 loop of RNase 1 has been shown to be as effective as the G88R substitution in RNase A in effecting RI-evasion, the importance of this loop in mediating contacts with RI is well established (43-45). Youle and coworkers demonstrated the utility of the G89C substitution to create RNase 1 dimers that evade RI (33). We choose that same substitution and made trimers using linker 8.
ONC
Linker 8 was used to tether ONC via residue 72 in its β4–β5 loop, which is truncated relative to that in mammalian homologues (9). The ensuing (S72C ONC)3–8 trimer, like the D38R/R39D/N67R/G88C RNase A trimer, was composed of RI-evasive monomers and thus revealed the effect of molecular size, apart from RI evasion, on cytotoxicity.
Production of Ribonucleases and RI
cDNA encoding ribonuclease variants was generated by Quickchange site-directed mutagenesis (Stratagene, La Jolla, CA) using plasmid pBXR (RNase A) (46), pSR1 (BS-RNase) (47), pHP-RNase (RNase 1) (44), pONC (ONC) (37), or pET27b(+) containing cDNA encoding D38R/R39D/N67R/G88R RNase A (38). Plasmid pHP-RNase was modified to install the missing codon for the C-terminal threonine residue (Thr128), which establishes native RNase 1 (48). RNase A, RNase 1, ONC, and G88R RNase A were produced as described previously (37, 38, 44). Free cysteine-containing variants of RNase A (A19C, G88C, and D38R/R39D/N67R/G88C), BS-RNase (C31A/C32A/G88C), RNase 1 (G89C), and ONC (S72C), were prepared in a similar manner, but with several alterations. The protein solution containing dissolved inclusion bodies was diluted by 10-fold with a thoroughly degassed solution of acetic acid (20 mM), subjected to centrifugation to remove precipitate, and dialyzed overnight against aqueous acetic acid (20 mM) that had been purged with N2(g) or Ar(g). Ribonucleases were allowed to refold for ≥3 days at 4 °C following slow dilution into 0.10 M Tris–HCl buffer at pH 8.0, containing NaCl (0.10 M), l-arginine (0.5 M), ethylenediamine tetraacetic acid (EDTA; 10 mM), reduced glutathione (GSH; 1.0 mM), and oxidized glutathione (GSSG; 0.2 mM). For free-cysteine containing variants of RNase 1, the concentrations of GSH and GSSG used were 3.0 mM and 0.6 mM, respectively. The refolding solution was purged with N2(g) or Ar(g) prior to the addition of denatured protein to prevent the oxidation of the free cysteines. Following purification by gel-filtration chromatography, the free thiol groups of the unpaired cysteine residues were protected from inadvertent air oxidation by reaction with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) as described previously (28). The resulting NTB-protected ribonucleases were applied to a HiTrap SP cation-exchange column, eluted with a linear gradient of NaCl (0.15–0.40 M) in 50 mM NaOAc buffer at pH 5.0, and stored at 4 °C until subsequent modification with thiol-reactive compounds. Protein concentration (excluding NTB-protected ribonucleases) was determined either by using a bicinchoninic acid-based kit (Thermo Fisher Scientific) (49) and measuring absorbance at 570 nm in a 96-well plate using an ELx800 microplate reader (Bio-Tek Instruments, Winooski, VT) or by UV spectroscopy using an extinction coefficient of ε278 = 0.72 (mg/mL)−1cm−1 for RNase A and its variants (50), ε278 = 0.465 (mg/mL)−1cm−1 for C31A/C32A/G88C BS-RNase (51), ε280 = 0.53 (mg/mL)−1cm−1 for RNase 1 and its variants (44), and ε280 = 0.87 (mg/mL)−1cm−1 for ONC and its variants (37).
Human RI was produced and purified as described previously (38, 45, 52) and stored in DPBS containing dithiothreitol (DTT; 10 mM). Freshly prepared RI was assumed to be 100% active (i.e., fully reduced) for up to one month following purification and was not used beyond this time without re-purification.
After their purification, ribonucleases and RI migrated as single bands during SDS–PAGE (e.g., Figure S1). The identity and integrity of purified ribonuclease variants was confirmed by MALDI–TOF mass spectrometry.
Semisynthesis of Ribonuclease Dimers and Trimers
The pH of the solution containing NTB-protected ribonucleases in HiTrap SP elution buffer was adjusted from 5.0 to ~8.0 by the addition of either 10× DPBS (to 10% v/v) or 1.0 M Tris–HCl buffer at pH 8.0 (to 8–10% v/v). Deprotection by the addition of DTT (5–10 equiv) resulted in the generation of the yellow 2-nitro-5-thiobenzoate anion (53). Ribonucleases were separated from small molecules using a HiTrap desalting column that had been equilibrated with DPBS containing EDTA (1 mM).
Semisynthetic dimers
Stock solutions (20–100 mM) of cross-linkers 3–6 and 1 were prepared in DMF, and 0.6 equiv were added to a solution containing a deprotected ribonuclease (50–250 μM). This sub-stoichiometric amount of linker was found to produce the desired dimeric (or trimeric) conjugates in high yield, as reported previously for the dimerization of C31S BS-RNase (54). A 0.6–5.0-fold molar excess of linker favored the production of a bis(maleimide)-derivatized ribonuclease monomer; a >10-fold molar excess of linker resulted in the undesirable nonspecific modification of the ε-amino group of lysine residues, despite the maleimido group being >103-fold more reactive with thiols than amines at pH 7 (55). Conjugation reaction mixtures were protected from light, and reactions were allowed to proceed at room temperature for 2 h or overnight at 4 °C. Reactions were terminated by the addition of DTT (5 equiv versus ribonuclease). The resulting crude reaction mixture was applied to a HiLoad 26/60 G75 Superdex gel-filtration column, and eluted with 50 mM NaOAc buffer at pH 5.0 containing NaCl (0.10 M) and NaN3 (0.05% w/v). Fractions were analyzed by SDS–PAGE to distinguish monomeric and multimeric species, and then combined to yield the final desired conjugate at ≥95% homogeneity. Typical yields for the production of homodimers and homotrimers were 70% and 50%, respectively. Semisynthetic variants were dialyzed exhaustively against DPBS prior to further biochemical characterization.
Semisynthetic trimers
Trimeric ribonuclease conjugates were synthesized in the manner described above for dimeric conjugates, except that the tris(maleimide)-containing linkers in DMF were added to DPBS containing ribonucleases (50–250 μM) at 0.4 molar equivalents (8) or 2.0 molar equivalents (7).
Modification of Ribonucleases with N-Ethylmaleimide
To enable comparisons of G88C RNase A and G89C RNase 1 to the wild-type enzymes, these variants were modified with N-ethylmaleimide (NEM) under conditions similar to those used for the semisynthesis of dimers and trimers, except that NEM (5 equiv from a 100 mM stock solution in DMF) was added to the deprotected protein that had been desalted into DPBS. The reaction mixture was protected from light, and the reaction was allowed to proceed overnight at 4 °C and then quenched by the addition of DTT (final concentration of 1 mM). The reaction mixture was then diluted 10-fold with NaOAc buffer (50 mM) at pH 5.0, applied to a HiTrap SP cation-exchange column, and eluted with a linear gradient of NaCl (0–0.4 M). The masses of the resulting ribonuclease–NEM conjugates were confirmed by MALDI–TOF mass spectrometry.
Biochemical Characterization of Ribonuclease Dimers and Trimers
The succinimidyl thioether formed by conjugation of a thiol and maleimide is susceptible to a low rate of spontaneous hydrolysis (56). The products are isomeric succinamic acid thioethers that bear an additional carboxylate group, which could obfuscate our analyses. Accordingly, ribonuclease conjugates were used within 3 months of their semisynthesis.
Ribonucleolytic activity
The enzymatic activity of ribonucleases was determined by assaying their ability to catalyze the cleavage of the hypersensitive fluorogenic substrate 6-FAM–dArUdAdA–6-TAMRA (20 nM) (λex = 493 nm; λem = 515 nm) (57). Assays were carried out at ambient temperature in DPBS (2.0 mL) containing BSA (0.1 mg/mL) to prevent the non-specific binding of ribonuclease to the cuvette. Values of kcat/KM were obtained with the equation:
| (1) |
where ΔI/Δt is the initial reaction velocity for cleavage of 6-FAM–dArUdAdA–6-TAMRA upon addition of ribonuclease (E) to the cuvette, and I0 and Imax are, respectively, the fluorescence intensities prior to enzyme addition and following the complete cleavage of substrate by addition of excess RNase A. Activity values for ONC and its conjugates were determined likewise using the substrate 6-FAM–dArUdGdA–6-TAMRA (50 nM) (58).
Binding by RI
The affinity of ribonucleases for RI was determined directly by using a fluorescence-based competition assay, as described previously (52). For the unlabeled competing multimeric ribonuclease conjugates, the molar concentration used was that of the constituent active sites and not that of the entire conjugate.
Inhibition of ribonucleolytic activity by RI
Twofold serial dilutions of RI (2 μM–1 pM) were prepared in 10 mM MOPS buffer at pH 7.1 containing NaCl (0.138 M), BSA (0.1 mg/mL), and DTT (5 mM) using Eppendorf Protein LoBind tubes. These RI-containing serial dilutions were delivered (50 μL/well) to a 96-well plate. Ribonucleases and their conjugates were diluted in the same buffer and added to the RI-containing wells (50 μL/well; typical final working concentration: 15 pM active site). Plates were incubated for at least 30 min at ambient temperature prior to the addition of fluorogenic substrate (6-FAM–dArUdAdA–6-TAMRA; final concentration: 50 nM) to initiate the assay. The plate was shaken briefly (30 s), and the fluorescence intensity was measured at 30-s intervals for 5 min on a microplate reader using a FITC filter set (excitation at 485 nm with a 14-nm bandwidth; emission at 535 nm with a 25-nm bandwidth; dichroic mirror cutoff at 505 nm). An identical quantity of 6-FAM–dArUdAdA–6-TAMRA was cleaved with an excess amount of ribonuclease (>10 nM), and the final fluorescence was measured to ensure that <10% of the total substrate was degraded over the course of the activity assays. Additionally, an aliquot of the untreated substrate was assayed at the end of the experiment to confirm that it had not degraded appreciably during the course of the experiment, which would be indicative of ribonuclease contamination. Initial slope values were obtained from the data points by linear regression. Relative activities were determined by comparison to a control containing no RI. Relative activity plots were fitted by nonlinear regression to a sigmoidal variable-slope curve using the equation:
| (2) |
In eq 2, h is the slope of the curve, and FT and FB correspond to the maximum relative activities at both low and high concentrations of RI, respectively.
Gel-filtration analysis
Ribonucleases (1 mL of a 1 mg/mL solution in gel-filtration buffer) were applied to a HiLoad 26/60 Superdex G200 gel-filtration column and eluted with NaOAc buffer (50 mM) at pH 5.0 containing NaCl (0.10 M) and NaN3 (0.05% w/v) at a flow rate of 4 mL/min. Gel-filtration standards were prepared and separated by chromatography using the same column.
Conformational stability
The conformational stability of dimers and trimers of RNase A was determined by measuring their Tm value, which is the temperature at the midpoint of the transition between the folded and unfolded states. Protein was dialyzed exhaustively against DPBS, and diluted to a concentration of ~25 μM in DPBS. Assays were performed by slow incremental heating (0.15 °C/min from 25–80 °C), and monitoring of the absorbance at 287 nm, which decreases upon RNase A denaturation (59, 60). Data were collected and analyzed with the program THERMAL from Varian Analytical Instruments (Walnut Creek, CA), which fitted the data to a two-state process and enabled the determination of the value of Tm (61).
Inhibition of Tumor-cell Proliferation in Vitro
Pancreatic-type ribonucleases can inhibit tumor-cell proliferation by evoking apoptosis (62). This cytotoxic activity was assessed by measuring the effect of ribonucleases on the incorporation of [methyl-3H]thymidine into the nascent DNA of K-562 cells, as described previously (37). Cells were grown in Roswell Park Memorial Institute 1640 medium (Mediatech, Herndon, VA). Cell-proliferation assays were performed at least three times in triplicate. Each data point represents the mean (± SE) of three or more experimental values. Values of IC50, which is the concentration of ribonucleases that decreases cell proliferation to 50%, were calculated by fitting the data using nonlinear regression to a sigmoidal dose–response curve, eq 3.
| (3) |
In eq 3, y is the DNA synthesis following a 4-h [methyl-3H]thymidine pulse, and h is the slope of the curve.
Inhibition of Tumor-cell Proliferation in Vivo
Trimeric ribonucleases were tested for their ability to suppress the growth of human tumors implanted into the flanks of nude mice. DU 145 cells (prostate cancer) and A549 cells (non-small cell lung cancer) were selected for both their ability to proliferate in mice and their low rate of spontaneous regression. Moreover, each line represents a clinically relevant target that is used often in the testing of new chemotherapeutic agents.
DU 145 cells were grown in Dulbecco's modified Eagle's medium (ATCC) containing FBS (10% v/v); A549 cells were grown in F12K medium (ATCC) containing FBS (10% v/v). Cells were implanted into the right rear flank of 5–6 week old male homozygous (nu/nu) nude mice. Tumors were allowed to grow to a volume of ≥75 mm3 before the initiation of treatment. All the test compounds were diluted in vehicle (DPBS), and one set of animals was treated with vehicle alone on the dosing schedule with greatest frequency. All treatments were administered either by intraperitoneal injection (i.p.) or orally with a gavage needle (p.o.); the volume of administered solution was based upon the body weight of the animal (10 μL/g). Treatment with all agents was ongoing throughout the entire experiment.
(G88C RNase A)3–8 was used to treat the DU 145 and A549 xenograft models. Mice in the A549 xenograft model were each implanted with 4 × 106 cells; treatment was administered at a dose of 25 mg/kg (i.p., 1×wk). Mice in the DU 145 xenograft model each received 3 × 106 cells; again, treatment was administered at a dose of 25 mg/kg (i.p., 1×wk). Mice in the A549 xenograft model were also treated with (G89C RNase 1)3–8 at doses of 7.5 and 75 mg/kg (i.p., 1×wk).
Mice treated with trimeric RNase A or RNase 1 were compared with mice treated with monomeric K7A/D38R/R39D/G88R RNase A at doses of 5, 15, and 45 mg/kg (i.p., qd×5) and 75 mg/kg (i.p., 1×wk), and monomeric ONC at a dose of 5 mg/kg (i.p., 1×wk), as well as the approved chemotherapeutic agents docetaxel (15 mg/kg; i.p., 1×wk), cisplatin (6 mg/kg; i.p., 1×wk), and erlotinib (100 mg/kg; p.o., 2×wk).
Tumor size was measured twice weekly using calipers, and tumor volume (mm3) was estimated by using the formula for a spheroid (eq 4):
| (4) |
The percent tumor growth inhibition (% TGI) was calculated with eq 5:
| (5) |
RESULTS
Conformational Stability
To maintain its biological activity, a ribonuclease must maintain its native tertiary structure (63). To assess whether the covalent tethering of ribonucleases into dimers or trimers compromises their conformational stability, we determined the Tm values of G88C RNase A–NEM, (G88C RNase A)2–3 and (G88C RNase A)3–8 (Table 1). All three had Tm values nearly indistinguishable from that of RNase A (Figure S2; Table 1). Thus, multimerization did not appear to be deleterious to the conformational stability of these conjugates. No evidence of cooperative unfolding was observed for dimeric or trimeric conjugates.
Table 1.
In vitro attributes of ribonucleases and their conjugates
| Ribonuclease |
Tma (°C) |
kcat/KMb (106 M−1s−1) |
Kd′c (nM) |
IC50d (μM) |
IC50e (μM) |
Z |
|---|---|---|---|---|---|---|
| RNase A | 64 | 6.4 ± 0.3 | 4.4 × 10−5 f | >25 | >25 | +4 |
| G88R RNase A | 63 | 8.2 ± 0.2 | 1.8 ± 0.3 | 3.3 ± 0.1 | 3.4 | +5 |
| G88C RNase A–NEM | 64 | 6.8 ± 0.1 | 1.4 ± 0.4 | 28 ± 2 | 28 | +4 |
| (G88C RNase A)2–1 | ND | 12 ± 1 | 8.2 ± 0.2 | 22 ± 3 | 44 | +8 |
| (G88C RNase A)2–2 | ND | 11 ± 1 | 2.6 ± 0.2 | 25 ± 1 | 50 | +8 |
| (G88C RNase A)2–3 | 64 | 6.6 ± 0.6 | 9.6 ± 0.4 | 11 ± 1 | 21 | +8 |
| (G88C RNase A)2–4 | ND | 7.4 ± 0.5 | 18 ± 2 | 10 ± 1 | 20 | +8 |
| (G88C RNase A)2–5 | ND | 8.6 ± 0.1 | 8.6 ± 1.6 | 14 ± 1 | 27 | +8 |
| (G88C RNase A)2–6 | ND | 5.5 ± 0.03 | 14 ± 3 | 27 ± 2 | 55 | +8 |
| (G88C RNase A)3–7 | ND | 12 ± 1 | 39 ± 3 | 16 ± 1 | 48 | +12 |
| (G88C RNase A)3–8 | 64 | 16 ± 0.4 | 17 ± 1 | 8.6 ± 0.5 | 27 | +13 |
| (A19C RNase A)3–8 | ND | 7.5 ± 0.4 | <1.4 | >25 | >25 | +13 |
| (D38R/R39D/N67R/G88C RNase A)3–8 | ND | 8.4 ± 0.3 | ND | 1.0 ± 0.1 | 3.3 | +16 |
|
| ||||||
| (C31A/C32A/G88C BS-RNase)3–8 | ND | 5.8 ± 0.3 | 84 ± 12 | 0.78 ± 0.05 | 2.1 | +28 |
|
| ||||||
| RNase 1 | 58 | 29 ± 1 | 2.9 × 10−7 g | >25 | >25 | +6 |
| (G89C RNase 1)3–8 | ND | 16 ± 3 | <1.4 | 1.5 ± 0.1 | 4.6 | +19 |
|
| ||||||
| ONC | 90 h | 0.0018 ± 0.00005 | >150 i | 0.22 ± 0.01 | 0.22 | +5 |
| (S72C ONC)3–8 | ND | 0.012 ± 0.0005 | ND | 0.79 ± 0.05 | 2.8 | +16 |
ND, not determined.
Values Tm (± 2 °C) in 1×DPBS and determined by UV spectroscopy.
Per active-site values of kcat/KM (± SE) for catalysis of 6-FAM–dArU(dA)2–6-TAMRA cleavage in 1×DPBS containing BSA (0.1 mg/mL) at (20 ± 2) °C. 6-FAM–dArUdGdA–6-TAMRA was the substrate for ONC and (S72C ONC)3–8.
Per active-site values of Kd′ (± SE) for the complex with RI at (20 ± 2) °C.
Per molecule values of IC50 (± SE) for incorporation of [methyl-3H]thymidine into the DNA of K-562 cells exposed to a ribonuclease and calculated with eq 3.
Per active-site values of IC50 calculated by multiplying the IC50 value by the number of monomers in the conjugate.
From ref (64).
From ref (45).
From ref (37) and determined by circular dichroism spectroscopy.
Value of Ki from ref (12).
Ribonucleolytic Activity
Site-specific covalent tethering of mammalian ribonucleases had a negligible effect on their ribonucleolytic activity (Table 1). Most multimeric conjugates as well as G88C RNase A–NEM exhibited kcat/KM values (per active site) that were within error of that of RNase A. Exceptions were (G88C RNase A)2–1, (G88C RNase A)2–2, and trimeric conjugates of G88C RNase A prepared with either of the homotrifunctional linkers (7 or 8), which displayed ~2-fold increased activity per active site toward the 6-FAM–dArUdAdA–6-TAMRA substrate. The S72C ONC trimer displayed the largest change in activity upon trimerization, as its kcat/KM value per active site was nearly 7-fold higher than that of ONC.
Affinity for Ribonuclease Inhibitor
Dimeric and trimeric ribonuclease conjugates could conceivably be bound by more than one RI molecule. In addition, the initial binding of one RI molecule to the conjugate is likely to influence the likelihood of subsequent binding events or preclude subsequent RI-binding entirely. The fluorescence-based competition assay used herein did not report on the stoichiometry of RI-binding or determine microscopic values of Kd. Hence, only values of apparent Kd (Kd′) are reported for the various conjugates.
The Kd = 1.8 nM obtained for G88R RNase A (Table 1) was in gratifying agreement with values obtained previously (38, 52). G88C RNase A–NEM exhibited a nearly equivalent affinity for RI, having a Kd = 1.4 nM. Among the dimeric conjugates, (G88C RNase A)2–4 had the lowest affinity for RI, with Kd′ = 18 nM. With the exception of (G88C RNase A)2–2 (Kd′ = 2.6 nM), the remaining dimeric conjugates of G88C RNase A all had similar affinity for RI, with Kd′ = 8.2–14 nM. Each trimeric conjugate of G88C RNase A displayed a significantly different affinity for RI. Those prepared with the novel tris(maleimide) linker 7 were substantially more evasive than those prepared with 8, with Kd′ = 39 and 17 nM, respectively. In general, trimeric conjugates of G88C RNase A had a lower affinity for RI than did dimers of G88C RNase A. (BS-RNase)3–8 trimeric conjugates were the most RI-evasive of the conjugates assayed, having Kd′ = 84 nM. Trimers of A19C RNase A, which are tethered outside of the RI interface, possessed an affinity for RI that was sufficiently high to preclude quantitation in our assay (Kd′ <1.4 nM).
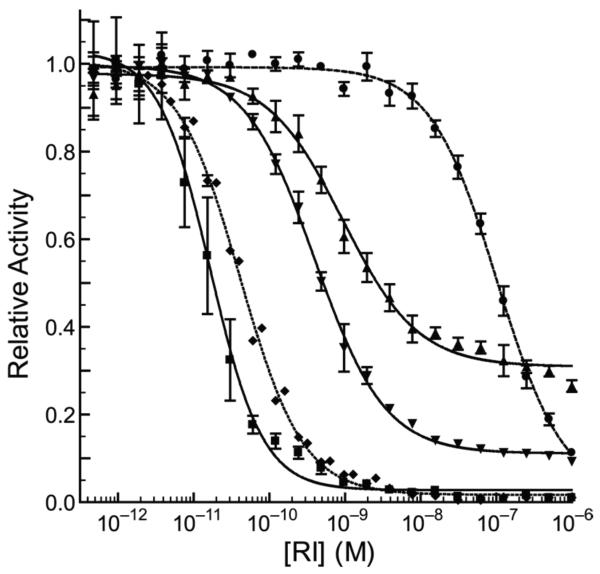
A novel assay was developed to determine the stoichiometry of RI-binding. The ability of the ribonuclease conjugates to cleave a fluorogenic substrate following pre-incubation with varying concentrations of RI was assayed. Apparent Ki′ values were obtained by applying eq 2 to the data in Figure 3.
Figure 3.
Relative enzymatic activity of ribonucleases after pre-incubation with increasing concentrations of RI. Data points are the mean (± SE) from three separate experiments, and were fitted with eq 2. G88R RNase A (◆), K31A/D38R/R39D/N67R/G88R RNase A (●), (G88C RNase A)2–3 (▼), (G88C RNase A)3–8 (▲), and (A19C RNase A)3–8 (■).
The G88R and K31A/D38R/R39D/N67R/G88R variants of RNase A had Ki′ = 41.4 pM and 103 nM, respectively. (G88C RNase A)3–8 exhibited Ki′ = 0.94 nM, which is 23-fold higher than that of G88R RNase A. Near physiological concentrations of RI, the fractional activity of the trimeric conjugate reached a plateau at ~0.31. This value is consistent with one of the three active sites in the trimer remaining inaccessible to micromolar concentrations of RI. (G88C RNase A)3–7 behaved in a nearly identical manner to (G88C RNase A)3–8 (data not shown). Conversely, (A19C RNase A)3–8, which differs from (G88C RNase A)3–8 only in that it is restrictively tethered through a residue outside of the interface with RI, is inhibited tightly and completely by RI with Ki′ = 17 pM. This value is close to the concentration of enzymic active sites present in the assay solution (15 pM), and thus merely represents an upper limit for the Ki′ value (65).
(G88C RNase A)2–3 was more readily inhibited by RI than was (G88C RNase A)3–8, with Ki′ = 0.40 nM. Interestingly, the fractional activity of this dimer also reached a plateau at micromolar concentrations of RI, though the significance of its relative activity (~0.11) is obvious. Nonetheless, we conclude that trimers in the cytosol would retain a larger amount of unfettered ribonuclease activity than would dimers.
Cytotoxicity in Vitro
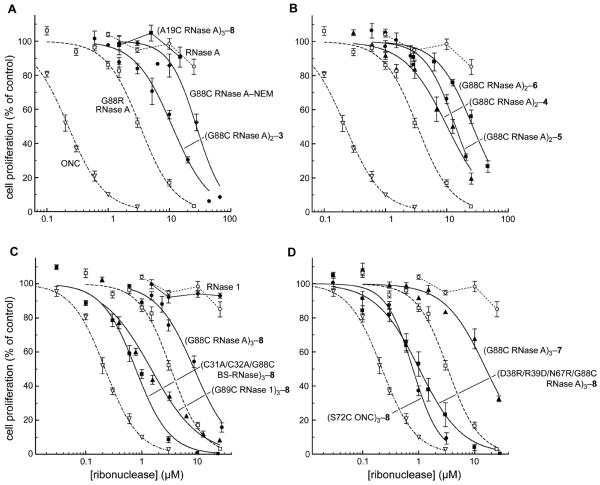
The toxicity of each ribonuclease was measured toward the K-562 human leukemia cell line. IC50 values are given in Table 1 on both a conjugate and an active-site basis, and were derived by applying eq 3 to the data in Figure 4. The results and fitted curves in Figure 4 are based upon the concentration of the entire conjugate, not just the active sites, and those will be the IC50 values discussed herein unless indicated otherwise. ONC and G88R RNase A displayed IC50 values similar to those reported previously (37, 38). (A19C RNase)3–8 exhibited no cytotoxic activity, even at concentrations of 20 μM (Figure 4A). This lack of cytotoxicity is consistent with its preserved sensitivity to RI, and underscores the importance of constraining the ribonucleases by tethering within the RI–RNase A interface. G88C RNase A–NEM (IC50 = 28 μM; Figure 4A) was nearly 10-fold less cytotoxic than G88R RNase A. This substantial decrease in cytotoxic activity was surprising because of their nearly identical affinities for RI. (G88C RNase A)2–3 (IC50 = 11 μM) was 3-fold less toxic to K-562 cells than was G88R RNase A (Figure 4A) despite its substantially greater resistance to inhibition by RI in vitro (Figure 3).
Figure 4.
Effect of ribonucleases on the proliferation of K-562 cells. The incorporation of [methyl-3H]thymidine into cellular DNA was used to monitor the proliferation of K-562 cells in the presence of ribonucleases. Data points are the mean (± SE) from at least three separate experiments carried out in triplicate, and were fitted with eq 3. Values of IC50 are listed in Table 1. For dimeric and trimeric conjugates, the concentration is of the molecule, not the constitutive active sites. Each panel depicts a different theme; data for RNase A (○), G88R RNase A (□), and ONC (▽) are repeated for comparison. (A) Tethering within or outside of the RI-ribonuclease interface. (B) Analogous dimeric conjugates made with bis(maleimido)benzene linkers 4–6. (C) Analogous trimeric conjugates of RNase A (Z = +13), RNase 1 (Z = +19), and BS-RNase (Z = +28). (D) Trimeric conjugates of evasive monomers, or made with linker 7.
The differential effects on cytotoxicity of linking two molecules of G88C RNase A either ortho-, meta-, or para- to each other about a phenyl ring is shown in Figure 4B. Surprisingly, the relatively subtle chemical differences led to detectable and reproducible differences in cytotoxicity. The IC50 values exhibited by (G88C RNase A)2–4, (G88C RNase A)2–5, and (G88C RNase A)2–6 were 10, 14, and 27 μM, respectively, consistent with our hypothesis that more restrictive tethering endows greater cytotoxicity.
Three homologous mammalian ribonucleases, RNase A, RNase 1, and BS-RNase, were tethered with linker 8 through analogous residues within their β4–β5 surface loop. The resulting trimers were tested for their ability to inhibit the proliferation of K-562 cells (Figure 4C). As reported previously (44), RNase 1 had no effect on the proliferation of K-562 cells. Per conjugate molecule, the (G88C RNase A)3–8 conjugate (IC50 = 8.6 μM) was more cytotoxic than any of the dimeric conjugates of RNase A linked through the same residue. (G89C RNase 1)3–8 (IC50 = 1.5 μM) and (C31A/C32A/G88C BS-RNase)3–8 (IC50 = 0.78 μM) were even more cytotoxic than the analogous RNase A trimer.
ONC and D38R/R39D/N67R/G88R RNase A are monomeric ribonucleases that are highly cytotoxic (IC50 = 0.27 and 0.19 μM, respectively) because they are insensitive to RI (38). The cytotoxic activities of trimeric conjugates of these two ribonucleases are shown in Figure 4D. Both (S72C ONC)3–8 (IC50 = 0.79 μM) and (D38R/R39D/N67R/G88C RNase A)3–8 (IC50 = 1.0 μM) are less cytotoxic than their monomers. On a per active-site basis, the cytotoxic activity of ONC and D38R/R39D/N67R/G88R RNase A is reduced by 10- and 17-fold, respectively, upon trimer formation.
Xenograft Mouse Studies
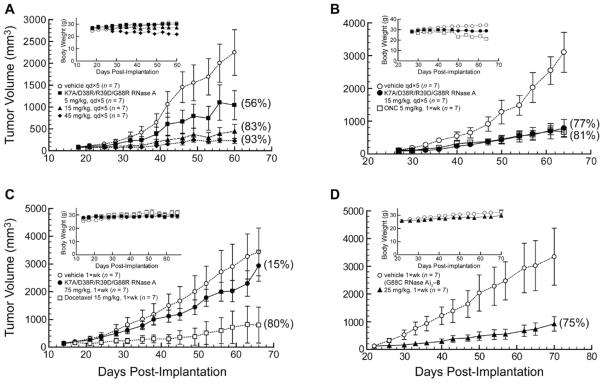
DU 145 prostate carcinoma cells are known to be sensitive to RI-evasive variants of RNase A (38). To establish an effective dose and administration schedule, mice bearing DU 145 prostate carcinoma tumors were treated with K7A/D38R/R39D/G88R RNase A and ONC. A dose of 15 mg/kg (i.p., qd×5) was identified as being optimal for eliciting the maximum inhibition of tumor growth (TGI = 83%; Figure 5A) while displaying minimal off-target effects as monitored by change in body weight (Figure 5A inset). Indeed, this dose of K7A/D38R/R39D/G88R RNase A inhibits the growth of DU 145 tumors in mice as well as the effective dose of ONC (5 mg/kg; i.p., 1×wk) (Figure 5B). Although the mice treated with the RNase A variant received 15-fold more ribonuclease (by mass) on a weekly basis than did those treated with ONC, the mammalian ribonuclease seems to be better tolerated, as indicated by the large decrease in body weight of the animals in the ONC treatment group during the course of the experiment (Figure 5B inset).
Figure 5.
Effect of K7A/D38R/R39D/G88R RNase A and (G88C RNase A)3–8 on the tumor volume and body weight (insets) of Balb c(−/−) mice in xenograft models bearing human DU 145 prostate tumors. Data points are the mean (± SE) for n mice; % TGI values in parentheses were calculated with eq 5. Vehicle control (○; n = 7). (A) K7A/D38R/R39D/G88R RNase A (■, 5 mg/kg; ▲, 15 mg/kg; ◆, 45 mg/kg; i.p., qd×5; n = 7). (B) K7A/D38R/R39D/G88R RNase A (●, 15 mg/kg; i.p., qd×5; n = 7), ONC (□, 5 mg/kg; i.p., 1×wk; n = 7). (C) K7A/D38R/R39D/G88R RNase A (●, 75 mg/kg; i.p., 1×wk; n = 7) and docetaxel (□, 15 mg/kg; i.p., 1×wk, n = 7, though 4 mice died during the course of the experiment such that n = 6 on day 28, n = 5 on day 35, n = 4 on day 49, and n = 3 on day 63). (D) (G88C RNase A)3–8 (▲, 25 mg/kg; i.p., 1×wk, n = 7).
The importance of frequent administration of K7A/D38R/R39D/G88R RNase A to achieve maximal tumor growth inhibition is shown in Figure 5C. These mice received 75 mg/kg (1×wk) instead of the previous 15 mg/kg (qd×5). Although equivalent on a mass/week basis, once-weekly administration of K7A/D38R/R39D/G88R RNase A was dramatically less effective at inhibiting tumor growth (TGI = 15% versus 83%).
Next, mice were treated with a trimeric ribonuclease: (G88C RNase A)3–8. A dose of 25 mg/kg (1×wk) of (G88C RNase A)3–8, which is a third of the ribonuclease by mass that was used in the experiments depicted in Figure 5C, was administered to mice bearing DU 145 tumors. This dose led to a dramatic inhibition of tumor growth (TGI = 75%; Figure 5D), despite the infrequent administration.
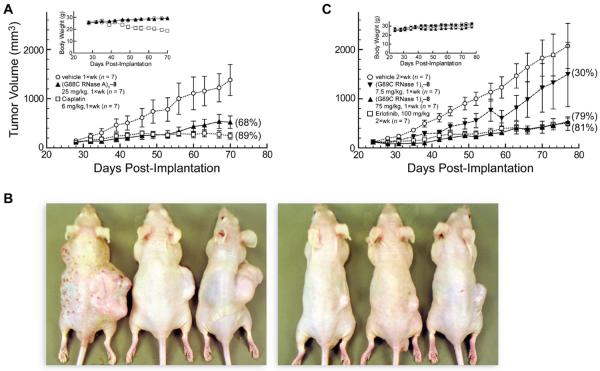
Xenograft studies were continued with A549 lung adenocarcinoma cells, which are less sensitive than DU 145 cells to RI-evasive variants of RNase A (38). A dose of 25 mg/kg (1×wk) of (G88C RNase A)3–8 inhibited the growth of these tumors substantially (TGI = 68%; Figure 6A). Photographs of mice from the vehicle and treatment groups upon completion of the experiment show the large reduction in the size of the tumors in treated animals (Figure 6B).
Figure 6.
Effect of trimeric ribonuclease conjugates on tumor volume and body weight (insets) of Balb c(−/−) mice in xenograft models bearing human A549 lung tumors. Data points are the mean (± SE) for n mice; % TGI values in parentheses were calculated with eq 5. Vehicle control (○; n = 7). (A) (G88C RNase A)3–8 (▲, 25 mg/kg; i.p., 1×wk, n = 7) and cisplatin (□, 6 mg/kg; i.p., 1×wk, n = 7). (B) Photographs of representative mice treated with vehicle (left) or (G88C RNase A)3–8 (right) on day 70 of the experiment in panel A. (C) (G89C RNase 1)3–8 (▼, 7.5 mg/kg; ▲, 75 mg/kg; i.p., 1×wk, n = 7) and erlotinib (□, 100 mg/kg; p.o., 2×wk, n = 7).
The human conjugate, (G89C RNase 1)3–8, was also administered to mice bearing A549 tumors. Although a dose of 7.5 mg/kg (1×wk) inhibited tumor growth only slightly (TGI = 30%) (Figure 6C), a 10-fold higher dose of 75 mg/kg (1×wk) inhibited the growth of these tumors at a high level (TGI = 79%) that was indistinguishable from that of a 100 mg/kg (2×wk) dose of erlotinib hydrochloride. This tyrosine kinase inhibitor was approved in 2004 for the treatment of non-small cell lung cancer (66). It is noteworthy that a single molecule of the human ribonuclease conjugate achieved the same efficacy as ~300 molecules of erlotinib.
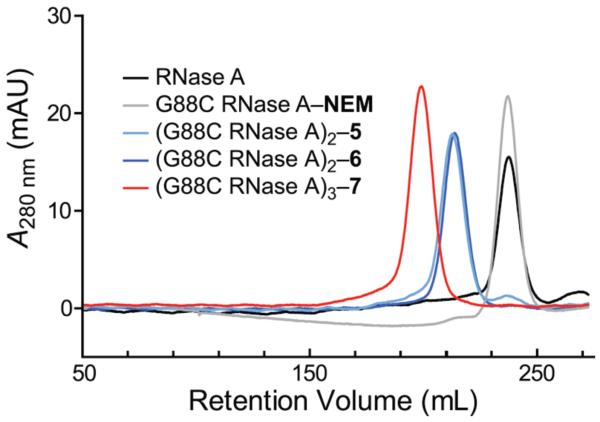
Hydrodynamic Radius
The hydrodynamic radius of RNase A conjugates was assessed by gel-filtration chromatography. As expected, the radii decreased in the order trimer > dimers > monomers (Figure 7), suggesting that trimers persist longest in plasma circulation.
Figure 7.
Elution profile of monomeric, dimeric, and trimeric ribonucleases from gel-filtration chromatography.
DISCUSSION
Thiol-reactive cross-linking reagents have found broad utility in a wide range of applications (55, 67, 68). For example, these reagents have been used as “molecular rulers” to probe the structure of large macromolecular complexes (69-74). Inspired by dimeric linkers 4–6, we synthesized the novel trimeric linker 7 (Scheme 1), expecting its phenyl scaffold to afford an even more constrained linkage than tren linker 8.
Preservation of Ribonucleolytic Activity
The enzymatic activity of RNase A is known to diminish upon non-specific cross-linking with diimido esters (21). In contrast, site-specific tethering did not have a deleterious effect on the catalytic activity of any of the ribonucleases herein (Table 1). Indeed, (S72C ONC)3–8 exhibited increased activity. The low catalytic activity of ONC is thought to originate in its low affinity for substrate (58). The increased charge density could accelerate the rate of substrate association.
Linkage Position
Site-specific tethering can allow or disallow a biomolecular interaction. We prepared two trimeric conjugates of RNase A, (A19C RNase A)3–8 and (G88C RNase A)3–8, that differ only at their site of linkage (Figure 2). Residue 19 lies within the α1–α2 loop of RNase A. Tethering three ribonuclease moieties there can readily accommodate the binding of three molecules of RI with high affinity (Ki′ ≤ 17 pM; Figure 3). This preservation of sensitivity to RI is corroborated by the results of the competition binding assay (Kd′ ≤ 1.4 nM; Table 1). In marked contrast, one active site of (G88C RNase A)3–8 remains free to degrade cellular RNA in the presence of micromolar RI (Figure 3). This evasion of RI endows (G88C RNase A)3–8 with toxicity for K-562 cells (Figure 4). This result is consistent with the report of Yamada and coworkers, who spliced human basic fibroblast growth factor (bFGF) into human pancreatic ribonuclease (RNase 1) as a separate domain through insertional fusion (75). RI-evasion, and thus efficacy as an immunotoxin, was greater when the bFGF graft was between residues 89 and 90, as opposed to residues 19 and 20, which are structurally analogous to the linkage positions explored in this work (H. Tada, personal communication).
Linker Conformation
A motivation of our work was to characterize ribonuclease multimers that were tethered more restrictively than those prepared with linker 2. All of the bifunctional reagents tested yielded dimeric ribonuclease conjugates that were indeed more RI-evasive than (G88C RNase A)2–2 (Table 1). Moreover, all but one of the reagents (6) yielded ribonuclease dimers that were more effective at inhibiting the proliferation of K-562 cells than was (G88C RNase A)2–2. The novel linker 7 likewise yielded more RI-evasive trimers (~2-fold) than those prepared with its more flexible congener, 8. Surprisingly, the greater ability of (G88C RNase A)3–7 to evade RI did not translate into greater cytotoxicity. The amino group of linker 8 is likely to be protonated at physiological pH values, giving this trimeric conjugate a net charge of +13 compared to +12 for (G88C RNase A)3–7. The cationicity of ribonucleases has been correlated with their efficiency of internalization and therefore cytotoxicity (76-79), and could contribute to the greater than expected cytotoxicity of (G88C RNase A)3–8 relative to (G88C RNase A)3–7.
Net Molecular Charge and Cytotoxicity
RNase A (Z = +4), RNase 1 (Z = +6), and BS-RNase (Z = +9) are highly cationic proteins with >80% amino-acid sequence identity and comparable ribonucleolytic activity. Yet, (G88C RNase A)3–8, (G89C RNase 1)3–8, and (C31A/C32A/G88C BS-RNase)3–8 possess significantly different net charges: Z = +13, +19, and +28, respectively. A correlation of net charge with cytotoxicity (76-79) is apparent in the trimeric ribonuclease conjugates, which have IC50 values of 8.6, 1.5, and 0.78 μM, respectively (Table 1; Figure 4C).
Molecular Size and Cytotoxicity
The hydrodynamic radius of the ribonuclease conjugates changes in a predictable manner (Figure 7). Previously, we showed that the quaternary structure of BS-RNase was a liability to its cytotoxic activity, as monomeric variants of BS-RNase that evade RI are more cytotoxic than are dimers of the same variants (42). Likewise, trimerization of D38R/R39D/N67R/G88R RNase A and ONC, which evade RI as monomers, reduces their cytotoxicity by 10- and 17-fold, respectively (Table 1). What is the basis for this decrease? The cellular uptake of fluorescent latex beads of discrete size was found to correlate inversely with size (80). Although these beads were 50–500 nm in diameter and thus larger than our conjugates, our data suggest that the correlation between cellular uptake and molecular size extends to smaller entities as well.
Molecular Size and Circulation
The glomerular capillary wall acts as a molecular sieve, removing small proteins (e.g., proteohormones) from plasma circulation but retaining large ones (e.g., albumin and IgG antibodies). In both mice and rats, the half-life of intravenously administered monomeric ribonucleases is 2–5 min (23, 25, 81), which is similar to the glomerular filtration rate (81, 82). For a particular molecule, this rate is determined largely by molecular size; only when molecular size approaches that of the filtration pore do other attributes, such as molecular charge, have an appreciable influence (83-85). Given the small size of monomeric ribonucleases (Figure 7), their rapid clearance is not surprising (86, 87). Dimeric ribonucleases exhibit a longer half-life in circulating plasma (23, 25, 81). Trimeric ribonuclease conjugates such as (G88C RNase A)3–8 are likely to persist even longer, as they are similar in size to albumin. Such greater plasma retention could contribute to the superior ability of trimeric ribonuclease conjugates to reduce tumor growth in vivo (Figures 5 and 6).
Conclusions
Using eight different thiol-reactive homobifunctional linkers (six dimeric and two trimeric), we have demonstrated that the cytotoxic activity of ribonuclease multimers correlates with how well they evade RI. Importantly, the catalytic activity and conformational stability are preserved in tethered ribonucleases. Finally, trimeric conjugates of G88C RNase A and G89C RNase 1 prepared with linker 8 were as effective as highly evasive monomeric variants of RNase A at inhibiting tumor growth in tumor-bearing mouse models, but could be administered less frequently and at a lower dose. Thus, this work demonstrates the potential of non-reducible, covalent conjugates of mammalian ribonucleases as human cancer therapeutics.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grant R01 CA073808 (NIH). We are grateful to L. D. Lavis for advice on the synthesis of linker 7, G. A. Ellis for the production and purification of RI, and J. Kalia, B. D. Smith, R. J. Johnson, G. A. Ellis, L. D. Lavis, and R. F. Turcotte for contributive discussions. T.J.R. was supported by Biotechnology Training Grant 08349 (NIH) and a William R. & Dorothy E. Sullivan Wisconsin Distinguished Graduate Fellowship. The Biophysics Instrumentation Facility was established with grants BIR-9512577 (NSF) and S10 RR13790 (NIH). The Keck Center for Chemical Genomics was established with a grant from the W.M. Keck Foundation. NMRFAM was supported by grant P41 RR02301 (NIH).
Footnotes
Supporting Information Available: Figures S1 and S2. This material is available free of charge via the internet at http://pubs.acs.org.
Abbreviations: BSA, bovine serum albumin; BS-RNase, bovine seminal ribonuclease; DMF, dimethylformamide; DPBS, Dulbecco's phosphate-buffered saline; DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); DTT, dithiothreitol; EDTA, ethylenediamine tetraacetic acid; 6-FAM, 6-carboxyfluorescein; FBS, fetal bovine serum; GSH, reduced glutathione; GSSG, oxidized glutathione; i.p., intraperitoneal; MALDI–TOF, matrix-assisted laser desorption ionization–time-of-flight; MOPS, 4-morpholinepropanesulfonic acid; NEM, N-ethylmaleimide; NTB, 2-nitro-5-thiobenzoic acid; ONC, ranpirnase or Onconase® (a registered trademark of Tamir Biotechnology); p.o., per os or by mouth; qd, quaque die or once daily; RI, ribonuclease inhibitor; RNase A, bovine pancreatic ribonuclease; PAGE, poly(acrylamide) gel electrophoresis; SDS, sodium dodecyl sulfate; 6-TAMRA, 6-carboxytetramethylrhodamine; TGI, tumor growth inhibition; Tris, tris(hydroxymethyl)aminomethane; Z, net molecular charge: arginine + lysine − aspartate − glutamate − pyroglutamate.
REFERENCES
- 1.Benito A, Ribó M, Vilanova M. On the track of antitumour ribonucleases. Mol. Biosyst. 2005;1:294–302. doi: 10.1039/b502847g. [DOI] [PubMed] [Google Scholar]
- 2.Arnold U, Ulbrich-Hofmann R. Natural and engineered ribonucleases as potential cancer therapeutics. Biotechnol. Lett. 2006;28:1615–1622. doi: 10.1007/s10529-006-9145-0. [DOI] [PubMed] [Google Scholar]
- 3.Arnold U. Aspects of the cytotoxic action of ribonucleases. Curr. Pharm. Biotechnol. 2008;9:1615–1622. doi: 10.2174/138920108784567263. [DOI] [PubMed] [Google Scholar]
- 4.Costanzi J, Sidransky D, Navon A, Goldsweig H. Ribonucleases as a novel pro-apoptotic anticancer strategy: review of the preclinical and clinical data for ranpirnase. Cancer Invest. 2005;23:643–650. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- 5.Pavlakis N, Vogelzang NJ. Ranpirnase—an antitumour ribonuclease: Its potential role in malignant mesothelioma. Expert Opin. Biol. Ther. 2006;6:391–399. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- 6.Lee JE, Raines RT. Ribonucleases as novel chemotherapeutics: The ranpirnase example. BioDrugs. 2008;22:53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raines RT. Ribonuclease A. Chem. Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- 8.Capasso S, Giordano F, Mattia CA, Mazzarella L, Zagari A. Refinement of the structure of bovine seminal ribonuclease. Biopolymers. 1983;22:327–332. doi: 10.1002/bip.360220142. [DOI] [PubMed] [Google Scholar]
- 9.Mosimann SC, Ardelt W, James MNG. Refined 1.7 Å X-ray crystallographic structure of P-30 protein, an amphibian ribonuclease with anti-tumor activity. J. Mol. Biol. 1994;236:1141–1153. doi: 10.1016/0022-2836(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 10.Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: Structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murthy BS, Sirdeshmukh R. Sensitivity of monomeric and dimeric forms of bovine seminal ribonuclease to human placental ribonuclease inhibitor. Biochem. J. 1992;281:343–348. doi: 10.1042/bj2810343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turcotte RF, Raines RT. Interaction of onconase with the human ribonuclease inhibitor protein. Biochem. Biophys. Res. Commun. 2008;377:512–514. doi: 10.1016/j.bbrc.2008.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutkoski TJ, Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr. Pharm. Biotech. 2008;9:185–199. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Alessio G, Parente A, Guida C, Leone E. Dimeric structure of seminal ribonuclease. FEBS Lett. 1972;27:285–288. doi: 10.1016/0014-5793(72)80642-2. [DOI] [PubMed] [Google Scholar]
- 15.Crestfield AM, Stein WH, Moore S. On the aggregation of bovine pancreatic ribonuclease. Arch. Biochem. Biophys. Supplement. 1962;1:217–222. [PubMed] [Google Scholar]
- 16.Gotte G, Laurents DV, Libonati M. Three-dimensional domain-swapped oligomers of ribonuclease A: Identification of a fifth tetramer, pentamers and hexamers, and detection of trace heptameric, octameric and nonameric species. Biochim. Biophys. Acta. 2006;1764:44–54. doi: 10.1016/j.bbapap.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Libonati M, Gotte G. Oligomerization of bovine ribonuclease A: Structural and functional features of its multimers. Biochem. J. 2004;380:311–327. doi: 10.1042/BJ20031922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libonati M. Biological actions of the oligomers of ribonuclease A. Cell. Mol. Life Sci. 2004;61:2431–2436. doi: 10.1007/s00018-004-4302-x. [DOI] [PubMed] [Google Scholar]
- 19.Leich F, Koditz J, Ulbrich-Hofman R, Arnold U. Tandemization endows bovine pancreatic ribonuclease with cytotoxic activity. J. Mol. Biol. 2006;358:1305–1313. doi: 10.1016/j.jmb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Hartman FC, Wold F. Cross-linking of bovine pancreatic ribonuclease A with dimethyl adipimidate. Biochemistry. 1967;6:2439–2448. doi: 10.1021/bi00860a021. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Wilson G, Moore S. Preparation of cross-linked dimers of pancreatic ribonuclease. Biochemistry. 1976;15:660–665. doi: 10.1021/bi00648a033. [DOI] [PubMed] [Google Scholar]
- 22.Bartholeyns J, Baudhuin P. Inhibition of tumor cell proliferation by dimerized ribonuclease. Proc. Natl. Acad. Sci. U.S.A. 1976;73:573–576. doi: 10.1073/pnas.73.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tarnowski GS, Kassel RL, Mountain IM, Blackburn P, Wilson G, Wang D. Comparison of antitumor activities of pancreatic ribonuclease and its cross-linked dimer. Cancer Res. 1976;36:4074–4078. [PubMed] [Google Scholar]
- 24.Bartholeyns J, Zenebergh A. In vitro and in vivo antitumor effect of dimerized ribonuclease A. Eur. J. Cancer. 1979;15:85–91. doi: 10.1016/0014-2964(79)90209-3. [DOI] [PubMed] [Google Scholar]
- 25.Bartholeyns J, Moore S. Pancreatic ribonuclease: Enzymic and physiological properties of a cross-linked dimer. Science. 1974;186:444–455. doi: 10.1126/science.186.4162.444. [DOI] [PubMed] [Google Scholar]
- 26.Gotte G, Testolin L, Costanzo C, Sorrentino S, Armato U, Libonati M. Cross-linked trimers of bovine ribonuclease A: Activity on double-stranded RNA and antitumor action. FEBS Lett. 1997;415:308–312. doi: 10.1016/s0014-5793(97)01147-2. [DOI] [PubMed] [Google Scholar]
- 27.Murdock AL, Grist KL, Hirs CHW. On the dinitrophenylation of bovine pancreatic ribonuclease A. Kinetics of the reaction in water and 8 M urea. Arch. Biochem. Biophys. 1966;114:375–390. [Google Scholar]
- 28.Messmore JM, Fuchs DN, Raines RT. Ribonuclease A: Revealing structure-function relationships with semisynthesis. J. Am. Chem. Soc. 1995;117:8057–8060. doi: 10.1021/ja00136a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bretscher LE, Abel RL, Raines RT. A ribonuclease A variant with low catalytic activity but high cytotoxicity. J. Biol. Chem. 2000;275:9893–9896. doi: 10.1074/jbc.275.14.9893. [DOI] [PubMed] [Google Scholar]
- 30.Green NS, Reisler E, Houk KN. Quantitative evaluation of the lengths of homobifunctional protein cross-linking reagents used as molecular rulers. Protein Sci. 2001;10:1293–1304. doi: 10.1110/ps.51201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J-S, Souček J, Matoušek J, Raines RT. Mechanism of ribonuclease cytotoxicity. J. Biol. Chem. 1995;270:31097–31102. doi: 10.1074/jbc.270.52.31097. [DOI] [PubMed] [Google Scholar]
- 32.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki M, Saxena SK, Boix E, Prill RJ, Vasandani VM, Ladner JE, Sung C, Youle RJ. Engineering receptor-mediated cytotoxicity into human ribonucleases by steric blockage of inhibitor interaction. Nat. Biotechnol. 1999;17:265–270. doi: 10.1038/7010. [DOI] [PubMed] [Google Scholar]
- 34.Farha OK, Julius RL, Hawthorne MF. Synthesis of a homotrifunctional conjugation reagent based on maledimide chemistry. Tetrahedron Lett. 2006;47:2619–2622. [Google Scholar]
- 35.Arai I, Sei Y, Maramatsu I. Preparation of 1,3,5-triaminobenzene by reduction of phloroglucinol trioxime. J. Org. Chem. 1981;46:4597–4599. [Google Scholar]
- 36.Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J. Mol. Biol. 1996;264:1028–1043. doi: 10.1006/jmbi.1996.0694. [DOI] [PubMed] [Google Scholar]
- 37.Leland PA, Schultz LW, Kim B-M, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc. Natl. Acad. Sci. USA. 1998;98:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J. Mol. Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Kothandaraman S, Hebert MC, Raines RT, Nibert ML. No role for pepstatin-A-sensitive acidic proteinases in reovirus infections of L or MDCK cells. Virology. 1998;251:264–272. doi: 10.1006/viro.1998.9434. [DOI] [PubMed] [Google Scholar]
- 40.Abel RL, Haigis MC, Park C, Raines RT. Fluorescence assay for the binding of ribonuclease A to the ribonuclease inhibitor protein. Anal. Biochem. 2002;306:100–107. doi: 10.1006/abio.2002.5678. [DOI] [PubMed] [Google Scholar]
- 41.Sica F, Di Fiore A, Zagari A, Mazzarella L. The unswapped chain of bovine seminal ribonuclease: Crystal structure of the free and liganded monomeric derivative. Proteins. 2003;52:263–271. doi: 10.1002/prot.10407. [DOI] [PubMed] [Google Scholar]
- 42.Lee JE, Raines RT. Cytotoxicity of bovine seminal ribonuclease: Monomer versus dimer. Biochemistry. 2005;44:15760–15767. doi: 10.1021/bi051668z. [DOI] [PubMed] [Google Scholar]
- 43.Gaur D, Swaminathan S, Batra JK. Interaction of human pancreatic ribonuclease with human ribonuclease inhibitor. Generation of inhibitor-resistant cytotoxic variants. J. Biol. Chem. 2001;276:24978–24984. doi: 10.1074/jbc.M102440200. [DOI] [PubMed] [Google Scholar]
- 44.Leland PA, Staniszewski KE, Kim B-M, Raines RT. Endowing human pancreatic ribonuclease with toxicity for cancer cells. J. Biol. Chem. 2001;276:43095–43102. doi: 10.1074/jbc.M106636200. [DOI] [PubMed] [Google Scholar]
- 45.Johnson RJ, McCoy JG, Bingman CA, Phillips GN, Jr., Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J. Mol. Biol. 2007;368:434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.delCardayré SB, Ribó M, Yokel EM, Quirk DJ, Rutter WJ, Raines RT. Engineering ribonuclease A: Production, purification, and characterization of wild-type enzyme and mutants at Gln11. Protein Eng. 1995;8:261–273. doi: 10.1093/protein/8.3.261. [DOI] [PubMed] [Google Scholar]
- 47.Kim J-S, Raines RT. Bovine seminal ribonuclease produced from a synthetic gene. J. Biol. Chem. 1993;268:17392–17396. [PubMed] [Google Scholar]
- 48.Seno M, Futami J, Kosaka M, Seno S, Yamada H. Nucleotide sequence encoding human pancreatic ribonuclease. Biochim. Biophys. Acta. 1994;1218:466–468. doi: 10.1016/0167-4781(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 49.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 50.Sela M, Anfinsen CB, Harrington WF. The correlation of ribonuclease activity with specific aspects of tertiary structure. Biochim. Biophys. Acta. 1957;26:502–512. doi: 10.1016/0006-3002(57)90096-3. [DOI] [PubMed] [Google Scholar]
- 51.D'Alessio G, Floridi A, De Prisco R, Pignero A, Leone E. Bull semen ribonucleases 1. Purification and physico-chemical properties of the major component. Eur. J. Biochem. 1972;26:153–161. doi: 10.1111/j.1432-1033.1972.tb01751.x. [DOI] [PubMed] [Google Scholar]
- 52.Lavis LD, Rutkoski TJ, Raines RT. Tuning the pKa of fluorescein to optimize binding assays. Anal. Chem. 2007;79:6775–6782. doi: 10.1021/ac070907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellman GL. A colorimetric method for determining low concentrations of mercaptans. Arch. Biochem. Biophys. 1958;74:443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- 54.Kim J-S, Raines RT. Dibromobimane as a fluorescent crosslinking reagent. Anal. Biochem. 1995;225:174–176. doi: 10.1006/abio.1995.1131. [DOI] [PubMed] [Google Scholar]
- 55.Ji TH. Bifunctional reagents. Methods Enzymol. 1983;91:580–609. doi: 10.1016/s0076-6879(83)91053-4. [DOI] [PubMed] [Google Scholar]
- 56.Kalia J, Raines RT. Catalysis of imido group hydrolysis in a maleimide conjugate. Bioorg. Med. Chem. Lett. 2007;17:6286–6289. doi: 10.1016/j.bmcl.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kelemen BR, Klink TA, Behlke MA, Eubanks SR, Leland PA, Raines RT. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999;27:3696–3701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JE, Raines RT. Contribution of active-site residues to the function of onconase, a ribonuclease with antitumoral activity. Biochemistry. 2003;42:11443–11450. doi: 10.1021/bi035147s. [DOI] [PubMed] [Google Scholar]
- 59.Hermans J, Jr., Scheraga HA. Structural studies of ribonuclease. V. Reversible change of configuration. J. Am. Chem. Soc. 1961;83:3283–3292. [Google Scholar]
- 60.Eberhardt ES, Wittmayer PK, Templer BM, Raines RT. Contribution of a tyrosine side chain to ribonuclease A catalysis and stability. Protein Sci. 1996;5:1697–1703. doi: 10.1002/pro.5560050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pace CN, Hebert EJ, Shaw KL, Schell D, Both V, Krajcikova D, Sevcik J, Wilson KS, Dauter Z, Hartley RW, Grimsley GR. Conformational stability and thermodynamics of folding of ribonucleases Sa, Sa2 and Sa3. J. Mol. Biol. 1998;279:271–286. doi: 10.1006/jmbi.1998.1760. [DOI] [PubMed] [Google Scholar]
- 62.Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klink TA, Raines RT. Conformational stability is a determinant of ribonuclease A cytotoxicity. J. Biol. Chem. 2000;275:17463–17467. doi: 10.1074/jbc.M001132200. [DOI] [PubMed] [Google Scholar]
- 64.Lee FS, Shapiro R, Vallee BL. Tight-binding inhibition of angiogenin and ribonuclease A by placental ribonuclease inhibitor. Biochemistry. 1989;28:225–230. doi: 10.1021/bi00427a031. [DOI] [PubMed] [Google Scholar]
- 65.Henderson PJF. A linear equation that describes the steady-state kinetics of enzymes and subcellular particles interacting with tightly bound inhibitors. Biochem. J. 1972;127:321–333. doi: 10.1042/bj1270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen MH, Johnson JR, Chen YF, Sridhara R, Pazdur R. FDA drug approval summary: Erlotinib (Tarceva®) tablets. Oncologist. 2005;10:461–466. doi: 10.1634/theoncologist.10-7-461. [DOI] [PubMed] [Google Scholar]
- 67.Hermanson GT. Bioconjugate Techniques. 2nd ed. Academic Press; San Diego, CA: 2008. [Google Scholar]
- 68.Kalia J, Raines RT. Advances in bioconjugation. Curr. Org. Chem. 2010;14:138–147. doi: 10.2174/138527210790069839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fasold H, Klappenberger J, Meyer C, Remold H. Bifunctional reagents for the crosslinking of proteins. Angew. Chem. Int. Ed. 1971;10:795–801. doi: 10.1002/anie.197107951. [DOI] [PubMed] [Google Scholar]
- 70.Chang FN, Flaks JG. The specific cross-linking of two proteins from the Escherichia coli 30 S ribosomal subunit. J. Mol. Biol. 1972;68:177–180. doi: 10.1016/0022-2836(72)90272-0. [DOI] [PubMed] [Google Scholar]
- 71.Peters K, Richards FM. Chemical cross-linking: Reagents and problems in studies of membrane structure. Annu. Rev. Biochem. 1977;46:523–551. doi: 10.1146/annurev.bi.46.070177.002515. [DOI] [PubMed] [Google Scholar]
- 72.Swaney JB. Use of cross-linking reagents to study lipoprotein structure. Methods Enzymol. 1986;128:613–626. doi: 10.1016/0076-6879(86)28095-7. [DOI] [PubMed] [Google Scholar]
- 73.Rappsilber J, Siniossoglou S, Hurt EC, Mann M. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal. Chem. 2000;72:267–275. doi: 10.1021/ac991081o. [DOI] [PubMed] [Google Scholar]
- 74.Kwaw I, Sun J, Kaback HR. Thiol cross-linking of cytoplasmic loops in the lactose permease of Escherichia coli. Biochemistry. 2000;39:3134–3140. doi: 10.1021/bi992509g. [DOI] [PubMed] [Google Scholar]
- 75.Tada H, Onizuka M, Muraki K, Masuzawa W, Futami J, Kosaka M, Seno M, Yamada H. Insertional-fusion of basic fibroblast growth factor endowed ribonuclease 1 with enhanced cytotoxicity by steric blockade of inhibitor interaction. FEBS Lett. 2004;568:39–43. doi: 10.1016/j.febslet.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 76.Futami J, Maeda T, Kitazoe M, Nukui E, Tada H, Seno M, Kosaka M, Yamada H. Preparation of potent cytotoxic ribonucleases by cationization: Enhanced cellular uptake and decreased interaction with ribonuclease inhibitor by chemical modification of carboxyl groups. Biochemistry. 2001;26:7518–7524. doi: 10.1021/bi010248g. [DOI] [PubMed] [Google Scholar]
- 77.Fuchs SM, Rutkoski TJ, Kung VM, Groeschl RT, Raines RT. Increasing the potency of a cytotoxin with an arginine graft. Protein Eng. Des. Sel. 2007;20:505–509. doi: 10.1093/protein/gzm051. [DOI] [PubMed] [Google Scholar]
- 78.Johnson RJ, Chao T-Y, Lavis LD, Raines RT. Cytotoxic ribonucleases: The dichotomy of coulombic forces. Biochemistry. 2007;46:10308–10316. doi: 10.1021/bi700857u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turcotte RF, Lavis LD, Raines RT. Onconase cytotoxicity relies on the distribution of its positive charge. FEBS J. 2009;276:3846–3857. doi: 10.1111/j.1742-4658.2009.07098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vasandani VM, Wu Y-N, Mikulski SM, Youle RJ, Sung C. Molecular determinants in the plasma clearance and tissue distribution of ribonucleases of the ribonuclease A superfamily. Cancer Res. 1996;56:4180–4186. [PubMed] [Google Scholar]
- 82.Lázníček M, Schiavon O, Caliceti P, Veronese FM. Pharmacokinetics and distribution of ribonuclease and its monomethoxypoly(ethylene glycol) derivatives in rats. Pharmacol. Res. 1993;28:153–161. doi: 10.1006/phrs.1993.1117. [DOI] [PubMed] [Google Scholar]
- 83.Maddox DA, Deen WM, Brenner BM. Glomerular filtration. In: Windhager EE, editor. Renal Physiology. Vol. 1. Oxford University Press; New York, NY: 1992. pp. 545–638. [Google Scholar]
- 84.Venturoli D, Rippe B. Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: Effects of molecular size, shape, charge, and deformability. Am. J. Physiol. Renal Physiol. 2005;288:F605–613. doi: 10.1152/ajprenal.00171.2004. [DOI] [PubMed] [Google Scholar]
- 85.Alpern RJ, Hebert SC, editors. Seldin and Geibisch's The Kidney: Physiology and Pathophysiology. 4th ed. Academic Press; New York, NY: 2007. [Google Scholar]
- 86.Maack T. Renal handling of low molecular weight proteins. Am. J. Med. 1975;58:57–64. doi: 10.1016/0002-9343(75)90533-1. [DOI] [PubMed] [Google Scholar]
- 87.Maack T, Johnson V, Kau ST, Figueiredo J, Sigulem D. Renal filtration, transport, and metabolism of low-molecular-weight proteins: A review. Kidney Int. 1979;16:251–270. doi: 10.1038/ki.1979.128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.