Abstract
Macrophages are important cells of the innate immune system, and their study is essential to gain greater understanding of the inflammatory nature of psoriasis. We used immunohistochemistry and double-label immunofluorescence to characterize CD163+ macrophages in psoriasis. Dermal macrophages were increased in psoriasis compared to normal skin and were identified by CD163, RFD7, CD68, LAMP2, Stabilin-1, and MARCO. CD163+ macrophages expressed C-lectins CD206/MMR and CD209/DC-SIGN, as well as co-stimulatory molecules CD86 and CD40. They did not express mature DC markers CD208/DC-LAMP, CD205/DEC205 or CD83. Microarray analysis of in vitro derived macrophages treated with IFNγ showed that many of the genes upregulated in macrophages were found in psoriasis, including STAT1, CXCL9, Mx1 and HLA-DR. CD163+ macrophages produced inflammatory molecules IL-23p19 and IL-12/23p40 as well as TNF and iNOS. These data demonstrate that CD163 is a superior marker of macrophages, and identifies a subpopulation of “classically activated” macrophages in psoriasis. We conclude that macrophages are likely to be contributing to the pathogenic inflammation in psoriasis, a prototypical Th1 and Th17 disease, by releasing key inflammatory products.
Keywords: Psoriasis, macrophages, myeloid dendritic cell, BDCA-1, Tip-DC, dermis, inflammation, skin
Introduction
Macrophages are important sentinels of the innate immune system. Their primary role is believed to be as phagocytic cells that participate in tissue homeostasis, and in the clearance of erythrocytes and removal of cellular debris generated during tissue remodeling (Mosser and Edwards, 2008). Macrophages have long been recognized as antigen presenting cells, capable of activating T cells during stimulation of the adaptive arm of the immune response.
The current classification of macrophages into classically activated (M1) and alternatively activated (M2) cells is parallel to the Th1/Th2 paradigm (Mantovani et al., 2004). Classically activated macrophages are activated by IFN-γ, alone or in concert with microbial products (e.g. LPS) or cytokines (e.g.TNF) and have high capacity to present antigen. Alternatively activated macrophages are induced by IL-4 and IL-13 and promote Type 2 responses. CD163, along with other macrophage markers such as Factor XIIIA (FXIIIA), CD206/macrophage mannose receptor (MMR), Macrophage Receptor with Collagenous structure (MARCO) and Stabilin-1 were classified as markers of alternatively activated macrophages (Djemadji-Oudjiel et al., 1996; Goerdt et al., 1999; Gratchev et al., 2005; Kzhyshkowska et al., 2006; Nickoloff et al., 2006; Torocsik et al., 2005).
In normal skin, we have identified CD163 as the most useful marker of macrophages (Zaba et al., 2007b). CD163 is a hemoglobin-haptoglobin complex-binding scavenger receptor that is expressed on most mature tissue macrophages (Fabriek et al., 2005). We have shown that macrophages in normal skin ingested tattoo pigment and were not able to stimulate T cell proliferation in an allogeneic mixed leukocyte reaction (allo-MLR) (Zaba et al., 2007b), consistent with their main role as phagocytic cells.
Psoriasis is a chronic inflammatory skin disease that results from the complex interplay between T cells, dendritic cells (DCs) and keratinocytes (Lowes et al., 2007). Recently, the pathogenesis of psoriasis has evolved from a purely classical type 1 (Th1) disease activated by IFNγ, to include a new T cell subset, T helper 17 (Th17) cells (Lowes et al., 2008). The function of macrophages in inflammatory skin diseases like psoriasis is not yet fully understood. CD163+ cells show a three-fold increase in psoriatic lesional skin and return to non-lesional skin levels after effective treatment with Etanercept (Zaba et al., 2007a). We have shown that in psoriasis, macrophages were not immunostimulatory and were unable to polarize T cells to produce IL-17 (Zaba et al., 2009).
We were interested in further characterizing macrophages in psoriasis. In this paper, we demonstrated that other macrophage markers were increased in psoriasis compared to normal skin, and co-expressed with CD163. The genomic profile of IFNγ-treated in vitro monocyte-derived macrophages was highly expressed in psoriasis transcriptome. We also showed that CD163+ macrophages expressed IFN-γ regulated genes such as signal transducer and activator of transcription-1 (STAT-1), CXCL9, Myxovirus resistance 1 (Mx1), and HLA-DR as well as other inflammatory mediators including IL-23p19, IL-12/23p40, TNF, and iNOS. Hence, we report that CD163+ macrophages, although previously classified as alternatively activated, also identify a subpopulation of classically activated macrophages in the presence of the IFNγ cytokine environment in psoriasis.
Results
Macrophages were more abundant in psoriasis compared to normal skin
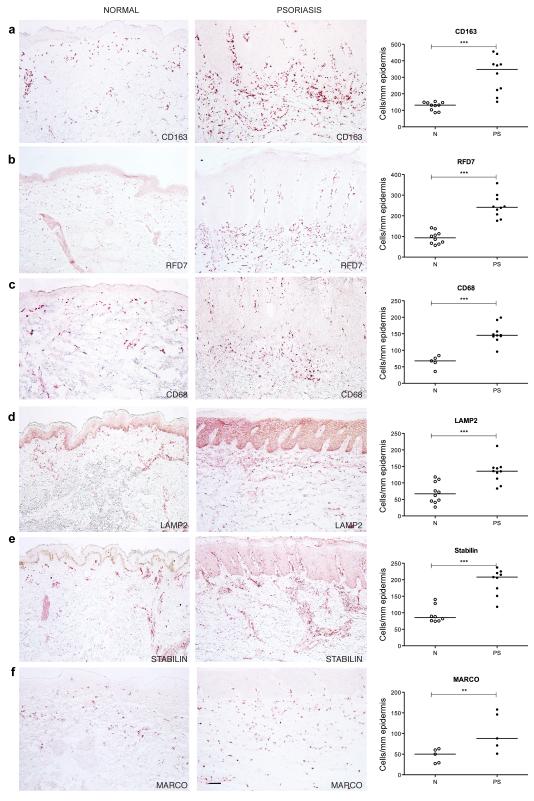
To evaluate the staining pattern and distribution of macrophages in normal skin and psoriasis lesional skin we performed immunohistochemistry using a panel of previously known macrophage markers: CD163 (Moestrup and Moller, 2004; Onofre et al., 2009), RFD7 (Pantelidis et al., 2001; Taams et al., 1999), CD68 (Ochoa et al., 2008; Yawalkar et al., 2009), LAMP2 (Eskelinen, 2006), Stabilin-1 (Kzhyshkowska et al., 2004; Politz et al., 2002) and MARCO (Arredouani et al., 2005; Sankala et al., 2002) (Figure 1). Table S1 summarizes the macrophage markers and their respective description/function. Positive cell counts per mm epidermis surface length (Figure 1), and per square area (Table S1) were all significantly increased in psoriasis compared to normal skin (p<0.05).
Figure 1. Macrophage markers were more abundant in psoriasis lesional skin compared to normal skin.
Representative immunohistochemistry staining and cell counts in normal and psoriasis skin of (a) CD163, (b) RFD7, (c) CD68, (d) LAMP2, (e) Stabilin and (f) MARCO, showing a significantly increased number of positive cells/per mm epidermis surface length in psoriasis compared to normal skin. Each dot represents a patient. Bar = 100 μm. **p<0.01; ***p<0.001.
CD163+ and RFD7+ macrophages were widely distributed over the papillary and reticular dermis of normal and psoriatic skin (Figure 1a, 1b), but CD163+ cells were more abundant. These numbers of CD163+ and RFD7+ macrophages were similar to the numbers of inflammatory DCs in psoriasis (Zaba et al., 2007a), and thus identify a second major myeloid leukocyte population in lesional skin.
Most of the CD68+ and LAMP2+ macrophages were located in the upper portion of both normal and psoriatic dermis (Figure 1c, 1d). There were some CD68+ cells near the dermo-epidermal junction (DEJ) particularly in psoriasis. The non-specific reactivity noted in the epidermis with LAMP2 antibody is common with antibodies conjugated to FITC. In both normal and psoriatic skin, Stabilin-1+ macrophages were observed in a perivascular distribution in the dermis (Figure 1e). MARCO+ cells were scattered throughout the dermis of normal and psoriatic skin (Figure 1f). While all these macrophage markers identified many dermal cells in psoriasis, they had slightly different staining patterns and distributions, suggesting some heterogeneity of surface antigens on macrophages.
CD163+ macrophages co-expressed additional macrophage markers in psoriasis
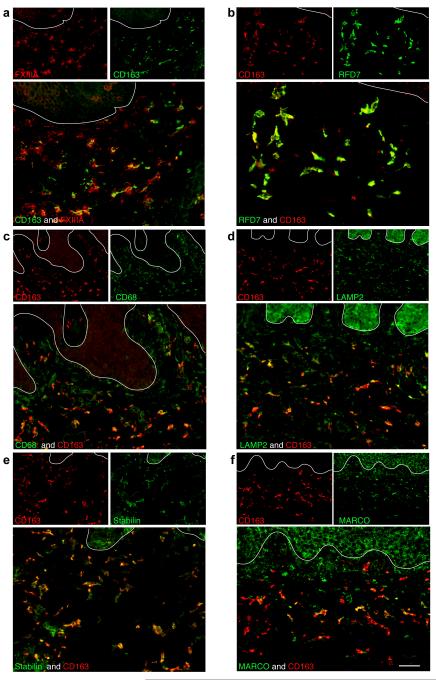
We further characterized the co-expression of these macrophage markers with CD163 in psoriatic skin using double-label immunofluorescence (Figure 2). We have previously reported that in normal skin FXIIIA+, CD68+ and RFD7+ cells co-localized with CD163 (Zaba et al., 2007b). In psoriatic skin, FXIIIA+ and RFD7+ macrophages almost completely co-expressed CD163 (Figure 2a, 2b). The majority of CD68+ cells co-expressed CD163 in the upper reticular dermis (Figure 2c) but the cells near the DEJ did not co-localize with CD163. We have previously demonstrated that these CD68+ cells near the DEJ are CD11c+ (Wang et al., 2006), and therefore this marker identifies both myeloid DCs and macrophages. Over half of LAMP2+ and Stabilin-1+ macrophages also co-expressed CD163 (Figures 2d, 2e). MARCO+ cells had the least co-expression with CD163 (Figure 2f).
Figure 2. CD163+ macrophages co-expressed other macrophage markers in psoriasis lesional skin.
CD163+ cells almost completely co-expressed (a) FXIIIA and (b) RFD7. (c) The majority of CD68+ cells in the reticular dermis were CD163+. (d) More than half of LAMP2+ cells and (e) Stabilin+ cells co-expressed CD163. (f) Some MARCO+ cells also coexpressed CD163. In all immunofluorescence figures, single-stained controls are above the merged image, white line denotes dermo-epidermal junction, dermal collagen fibers gave green autofluorescence, and antibodies conjugated with a fluorochrome often gave background epidermal fluorescence. Bar =100 μm.
To assess the potential for these markers to be exclusive for macrophages, we performed double-label immunofluorescence with CD11c (Figure S1). All these macrophage markers were co-expressed to some degree with CD11c. However, among all the markers studied, CD163 had the least co-expression with CD11c making it a superior marker of macrophages in psoriasis. We also showed that CD163+ macrophages and CD11c+ myeloid DCs co-expressed CD14, CD16 and LFA-1 (Figure S2).
CD163+ macrophages also expressed C-lectins and co-stimulatory molecules in psoriasis
We evaluated the expression of C-lectins and co-stimulatory molecules on CD163+ macrophages by double-label immunofluorescence (Figure S3). In normal skin, we have previously reported that CD206/MMR and CD209/DC-specific ICAM-3-grabbing non-integrin (DC-SIGN) were more abundant on macrophages than DCs (Zaba et al., 2007b). We now demonstrate that in psoriatic skin, almost all CD163+ macrophages were CD206+/MMR and approximately three quarters co-expressed CD-209/DC-SIGN+(Supplemental Figure 3a, 3b). It has been previously reported that macrophages constitutively express CD86 (Way et al., 2009) and CD40 (Suttles and Stout, 2009). Here, we demonstrated that in psoriasis less than half of CD163+ macrophages expressed CD86 and CD40 co-stimulatory molecules.
CD163+ macrophages did not express “classic” dendritic cell markers in psoriasis
In order to clearly distinguish macrophages from DCs, we performed double-label immunofluorescence with CD163 and the well-recognized DC markers CD208/DC-lysosomal-associated membrane glycoprotein (DC-LAMP), CD205/DEC-205, and CD83 (Figure S4). In psoriatic and normal skin, CD163+ macrophages did not co-express any of these DC maturation markers. These observations support that there are distinct markers for mature DCs and macrophages.
Genomic signature of in vitro “classically activated” (M1) macrophages was over-expressed in psoriasis
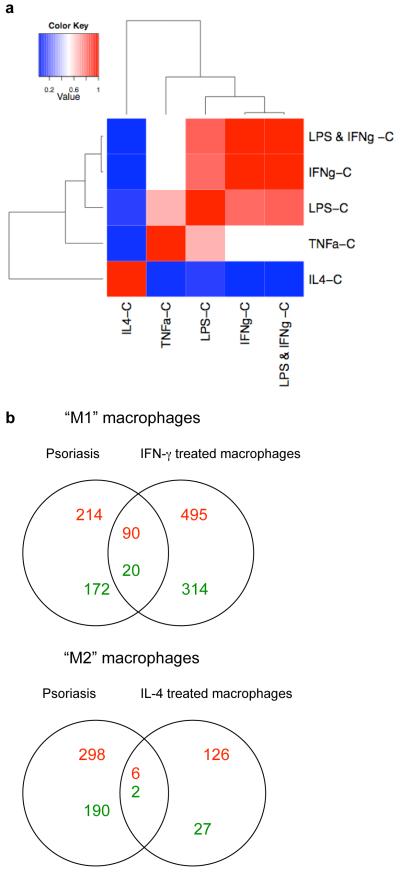
Previous studies have suggested that CD163 is a marker of alternatively activated macrophages in psoriasis (Djemadji-Oudjiel et al., 1996). However, we were interested to explore why alternatively activated cells would be present in the Th1 “classical” microenvironment of psoriasis. As we were unable to obtain sufficient quantities of macrophages from psoriatic lesional skin to study ex vivo, we turned to in vitro methods to further understand macrophage biology in chronic cutaneous inflammation. To determine the transcriptional profile associated with macrophage polarization to M1 or M2, we cultured macrophages derived from monocytes (n=7) for 7 days and treated them with IFNγ, IL-4, TNF, LPS, and LPS+IFNγ. Martinez et al used a similar experimental approach to generate LPS+IFNγ- and IL-4-treated macrophages (Martinez et al., 2006). However, we included additional conditions (IFNγ alone, LPS alone, and TNF) and also generated lists of differentially expressed genes (DEG) by comparing the effect of the cytokine on macrophages with control (Table S2). A heatmap of the estimated fold change of every condition versus control showed a tight relationship between the effect of LPS+IFNγ, IFNγ, LPS, and TNF treatment of macrophages, while IL-4 has a distinct effect (Figure 3a).
Figure 3. Genomic signature of in vitro “classically activated” (M1) macrophages was over-expressed in psoriasis.
(a) Heatmap of cytokine treated in-vitro derived macrophages, compared to control (C) for each condition. This heatmap showed that macrophages treated with LPS+IFNγ, IFNγ, LPS, and TNF were clustered together, while IL-4-treated macrophages were more distant. (b) Venn Diagrams of “M1” and “M2” macrophages showing the number of upregulated genes (in red) and downregulated genes (in green) and their overlap in psoriasis lesional skin compared to IFN-γ and IL-4 treated macrophages respectively.
We were particularly interested in the genomic profiles of “M1” macrophages induced with IFNγ, compared to “M2” macrophages treated with IL-4 (Figure 3b). We identified 919 DEGs between IFNγ-treated macrophages compared to control (FCH>2.0, FDR<0.01). There were 585 probes upregulated (in red) in the IFNγ-treated macrophages, and 334 down-regulated probes (in green). Results of Ingenuity analysis of up- and down-regulated M1 and M2 genes are provided in Table S3. As expected, Gene Ontology classified upregulated M1 genes with Inflammatory Response and Inflammatory Disease. M1 upregulated genes were significant in the Cannonical Pathways for Communication between Innate and Adaptive Immune Cells, Interferon Signaling, and Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis.
In psoriasis lesional skin, there were 304 upregulated probes and 192 downregulated probes compared to non-lesional skin, on the same Illumina Human HT-12 microarray chip (FCH>2.0, FDR<0.01). The overlap of these two DEG lists showed that 90/585 (15%) probes were upregulated in both IFNγ-treated macrophages and psoriasis, and 20/334 (6%) probes were downregulated in both IFNγ-treated macrophages and psoriasis. In contrast, there were 132 probes that were upregulated by IL-4 treated macrophages, and 29 probes down-regulated. Only 6/132 (4.5%) probes overlapped with the psoriasis transcriptome and 2 probes were in common in the downregulated set.
We generated cytokine-treated macrophage “pathways”, consisting of the genes expressed by the addition of a given cytokine to in vitro monocyte-derived macrophages. We then used Gene Set Enrichment Analysis (GSEA) to compare these macrophage pathways with the genomic profile of psoriasis lesional skin. We hypothesized that there should be greater expression of “M1” macrophage genes in psoriasis. The GSEA results indicated that the following gene sets were significantly enriched in psoriasis lesional tissue (compared with non-lesional): LPS-UP, IFNγ-UP, LPS+IFNγ-UP, TNFα-UP, IL-4-DOWN (FDR<0.013 for all pathways) (Table S4). The LPS and IFNγ “pathways” were the top 3, with a normalized enrichment score (NES) >2.3, showing that these pathways were the most enriched in the psoriasis gene set. The LPS+IFNγ-DOWN and IFNγ-DOWN gene sets were also enriched in non-lesional tissue (FDR=0.009 and 0.037 respectively).
Table 1 presents the fold change of 20 selected up-regulated genes in common between psoriasis and IFNγ-treated macrophages. CXCL9 (122.69), CXCL10 (81.22), Mx1 (18.97), CCL20 (8.85) and STAT1 (8.54) (FDR <10−6 for all genes) are well known psoriasis genes that were increased by IFNγ in macrophages. Table S5 lists the fold change for the few genes in common between psoriasis and IL-4-treated and TNF-treated macrophages.
Table 1.
Upregulated genes in IFN-γ-treated macrophages in common with Psoriasis
| Gene Symbol |
Description | Fold Change |
|---|---|---|
| CXCL9 | Chemokine (C-X-C motif) ligand 9 | 122.69 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | 81.22 |
| ISG 20 | Interferon stimulated exonuclease gene 20kDa | 58.32 |
| RSAD2 | Radical s-adenosyl methionine domain containing 2 | 25.15 |
| CCL5 | Chemokine (C-C motif) ligand 5 | 23.14 |
| MX1 | Myxovirus resistance 1, interferon-inducible protein p78 | 18.97 |
| CCL8 | Chemokine (C-C motif) ligand 8 | 18.21 |
| IL1b | Interleukin 1, beta | 9.65 |
| CCL20 | Chemokine (C-C motif) ligand 20 | 8.85 |
| STAT1 | Signal transducer and activator of transcription 1 | 8.54 |
| IL1F9 | Interleukin 1 family, member 9 | 6.44 |
| CCL4L2 | Chemokine (C-C motif) ligand 4-Like 2 | 6.34 |
| CCL4L1 | Chemokine (C-C motif) ligand 4-Like 1 | 4.89 |
| IL7R | Interleukin 7 receptor | 4.42 |
| TNIP3 | TNFAIP3 interacting protein 3 | 3.06 |
| IL8 | Interleukin 8 | 2.79 |
| CCL3L3 | Chemokine (C-C motif) ligand 3-Like 3 | 2.70 |
| IL1RN | Interleukin 1 receptor antagonist | 2.16 |
| IL19 | Interleukin 19 | 2.15 |
| S100A12 | S100 Calcium Binding protein A12 | 2.09 |
CD163+ macrophages expressed IFN-γ regulated genes
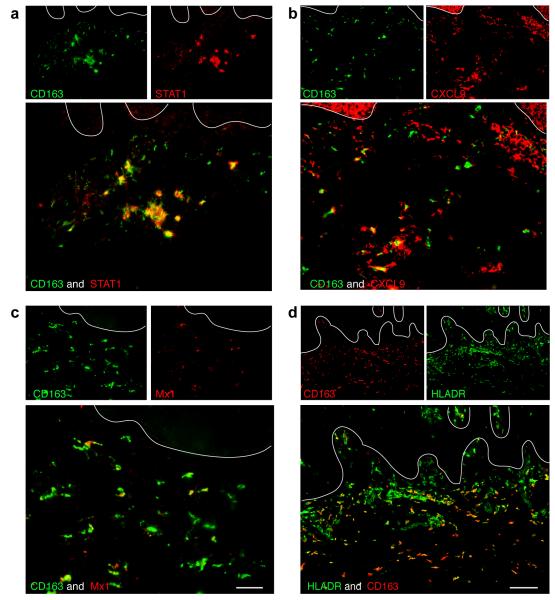
It is well appreciated that there is a dominant “Type 1” IFNγ signature in psoriasis (Kryczek et al., 2008; Lew et al., 2004) and our microarray results suggested that macrophages treated with IFNγ were well represented at the genomic level in psoriasis. To verify that macrophages in psoriasis lesions are indeed responsive to IFNγ, we performed double label immunofluorescence with CD163 and IFN-γ regulated genes identified by microarray, STAT-1, CXCL9, Mx1, and HLA-DR (Figure 4). We observed that the majority of macrophages expressed STAT-1 (Figure 4a), which is known as one of the most consistent transcription factor alterations in psoriasis (Lew et al., 2004). By immunohistochemistry, STAT-1 expression in normal skin was minimal, but was abundant and nuclear in psoriatic lesional skin (Figure S5a), indicating activation in psoriasis.
Figure 4. CD163+ macrophages expressed IFN-γ regulated genes.
Many CD163+ macrophages co-expressed (a) STAT1, (b) CXCL9, (c) Mx1 and, (d) HLA-DR. Bar =100 μm.
Approximately half of CD163+ macrophages expressed CXCL9 (Figure 4b). This chemokine is considered a key IFN-γ-regulated gene and is thought to bind to CXCR3-bearing activated T-cells, and may be involved in T-cell trafficking to psoriatic dermis and epidermis (Nograles et al., 2008; Rottman et al., 2001). There was low level CXCL9 expression in normal skin but was increased in psoriasis by immunohistochemistry (Figure S5b). Some CD163+ macrophages expressed Mx1 (Figure 4c), which is one of the genes induced by IFN-γ in keratinocytes (Haider et al., 2007). Almost three quarters of CD163+ macrophages expressed HLA-DR (Figure 4d), a molecule induced on many cell types by IFNγ (van den Oord et al., 1995).
CD163+ macrophages expressed products of classically activated macrophages
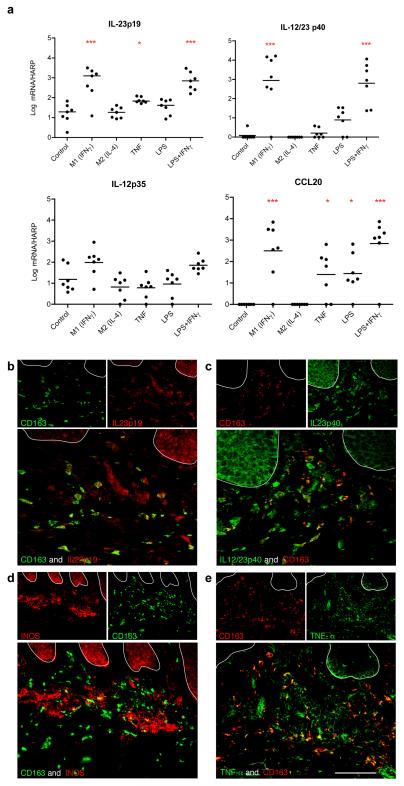
We were also interested in the expression of cytokines and chemokines important for psoriasis pathogenesis in macrophages, including IL-12 and IL-23 subunits, and CCL20 (Sanmiguel et al., 2009). There was increased gene expression of IL-23p19, IL-12/23p40 and CCL20 in M1 macrophages (+IFNγ) and LPS+IFNγ-treated macrophages. CCL20 was also increased in macrophages treated with TNF and LPS. IL-12p35 was filtered out from the gene lists due to low expression. We confirmed these findings by real time RT-PCR (Figure 5a). We demonstrated that IL-23p19 and IL-12/23p40 were significantly increased in IFNγ-treated and IFN-γ+LPS-treated macrophages compared to control (p<0.001). We attributed this increase to IFN-γ since macrophages treated with LPS alone did not show a significant increase compared to control. IL-23p19 was also increased significantly (p<0.05) in TNF-α treated macrophages compared to control. CCL20 was significantly increased in IFNγ-treated and IFN-γ+LPS-treated macrophages compared to control (p<0.001), and also in TNF-α and LPS treated macrophages compared to control (p<0.05).
Figure 5. Expression of cytokines in in vitro-derived macrophages and psoriasis by RT-PCR and immunofluorescence. (a).
RT-PCR analysis of mRNA expression of IL-23p19, IL-12/IL-23p40, IL-12p35 and CCL20 in cytokine treated (IFNγ, IL-4, TNF-α, LPS and LPS+IFN) in vitro macrophages (n=7) compared to control. mRNA expression normalized to human acidic ribosomal protein (HARP). * p<0.05, ***p<0.001. Some CD163+ macrophages co-expressed products of classically activated macrophages, (b) IL-23p19, (c) IL-12/IL-23p40, and inflammatory mediators (d) INOS and (e) TNF-α. Bar =100 μm.
We also confirmed these findings at the protein level showing that CD163+ cells produced IL23p19 and IL12/IL23p40 (Figure 5b, 5c). This has been recently shown by Yawalkar et al using an antibody that binds both subunits of IL-23 (Yawalkar et al., 2009). Having shown that macrophages can produce IL-23, we were also interested in whether they could produce other inflammatory products often ascribed to DCs. We have previously shown that myeloid DCs produce TNF and iNOS in psoriasis, and called these cells TNF-α and iNOS-producing DCs (Tip-DCs) (Lowes et al., 2005). We demonstrated that some CD163+ cells also had co-expression with iNOS (Figure 5d), and TNF (Figure 5e), and were thus able to produce products of classically activated macrophages (Mosser and Edwards, 2008).
Discussion
The role of macrophages in inflammation and tumorigenesis is a field of great interest to many investigators. Primary immunodeficiency of macrophages has been implicated as a cause of Crohn’s disease, which for many years has been considered a prototypic autoimmune disease (Casanova and Abel, 2009). Although much has been learned about DCs, a related myeloid population, studies on macrophages in human skin have been less common over recent years.
Macrophages and DCs share many properties: they are both cells of the innate arm of the immune system, of myeloid cell origin, are variably phagocytic, and capable of antigen presentation to T cells. The two key differences between these cells are that macrophages are non-migratory and live and die in the tissue in which they develop, and that mature DCs can present antigen to both naïve and memory T cells (Bryant and Ploegh, 2004). It has been surprisingly difficult to find markers to consistently distinguish these two cell populations in humans. There appear to be certain molecules that are restricted to mature DCs, such as CD205, CD208, and CD83, which we have confirmed in this study. Perhaps these molecules are important for antigen presentation to naïve T cells, a specialized DC function. However, immature DCs and macrophages both possess pattern recognition receptors such as C-lectins (Wu et al., 1996) and TLRs (Krutzik et al., 2005) for antigen binding, HLA-Class I and II molecules, and co-stimulatory molecules for antigen presentation. Differentiating macrophages and DCs is not simply academic, as understanding the properties and functions of these cells during inflammation may yield new cellular or cytokine therapeutic targets.
Our data suggest that the linear classification of macrophages based on surface phenotype may be over-simplified. It was originally proposed that tissue macrophages undergo local activation in response to various inflammatory and immune stimuli, classified as either “classically activated” or “alternatively activated”, leading to M1 and M2 macrophages, respectively (Gordon, 2003). The functional phenotype of macrophages may evolve in response to microenvironmental cues, as suggested by Stout et al (Stout et al., 2005). M1 and M2 macrophages can even be re-polarized by Th2 or Th1 cytokines, respectively (Gratchev et al., 2006). Recently, an alternative paradigm for macrophages was proposed to take into account the apparent plasticity of macrophages. In this circular model, three populations of macrophages were defined as either wound-healing, classically activated, or regulatory macrophages (Mosser and Edwards, 2008). This model suggests that macrophages might retain their original features and acquire new abilities in their response to environmental changes, for example during inflammation, cancer, or tissue remodeling. Thus there are macrophages in transit, which may have features of more than one subgroup.
We used in vitro macrophages as a surrogate to demonstrate that many of the genes upregulated by IFNγ in macrophages were present in psoriasis lesional skin. We also demonstrated that several of these known IFNγ regulated genes (STAT 1, CXCL9, Mx1, HLA-DR) were present in CD163+ cells, indicating that macrophages in psoriasis were responding to IFNγ in situ. An interpretation of our data is that steady state macrophages in the skin are all CD163+, perhaps in anticipation of their expected role as tissue phagocytes, and antigen presenting cells. As they develop in the Th1 inflammatory environment of psoriasis, characterized by abundant IFNγ, a subpopulation acquire new markers and properties, but retain their original surface markers. Therefore it may be more useful to consider the function of macrophages by the products they produce rather than surface markers.
Lately, the role of macrophages in psoriatic inflammation has become of more interest as the link between psoriasis skin disease and the metabolic syndrome has become appreciated. TNF may play a key role inducing systemic inflammation, and macrophages in the skin and adipose tissue may be crucial to this process (Gisondi et al., 2009; Nijsten and Wakkee, 2009). Macrophages have also recently been implicated in atherosclerosis, as cells that retain lipids in plaques, and are a promising target for treatment of atherosclerosis (Wilson et al., 2009). Characterization of the markers, capabilities, and functions of macrophages in psoriasis is essential to understanding the systemic manifestations associated with psoriasis.
In conclusion, our study provides new tools that can be used to study macrophages in inflammation, and contributes to the evolving paradigm of macrophage polarization. Furthermore, our results reinforce the important role of IFNγ driving macrophage activation and contributing to the maintenance phase of psoriasis pathogenesis.
Materials and Methods
Material and Methods, and statistical analysis are described in greater detail in Supplemental Materials and Methods (SMM).
Skin samples
Skin punch biopsies (6mm diameter) were obtained from normal volunteers and psoriasis patients under Rockefeller University IRB-approved protocols. Informed consent was obtained and the study was performed in adherence with the Principles of the Declaration of Helsinki. Biopsies were frozen in OCT (Sakura, Tokyo, Japan) and stored at −80 °C for immunohistochemistry and immunofluorescence.
Immunohistochemistry and Immunofluorescence
Standard procedures were used for immunohistochemistry and immunofluorescence as previously described (Zaba et al., 2009) and outlined in greater detail in SMM. Normal and psoriasis lesional skin sections (n=5-10) were stained with macrophage markers CD163, RFD7, LAMP2, CD68, Stabilin, and MARCO (Table S6). Immunofluorescence images were acquired using either Zeiss Axioplan 2 widefield fluorescence microscope, or upright confocal microscope. Methods for counting cells are described in Figure S6.
Macrophage cultures
Macrophages were produced from peripheral blood mononuclear cells (PBMCs; n=7) using M-CSF (50ng/ml) as described in SMM. Macrophages were polarized by adding 20 ng/ml IFN-γ (for M1 polarization), 25ng/ml IL-4 (for M2 polarization), 20ng/ml TNF-α, 1 ug/ml LPS, and 1 ug/ml LPS plus 20ng/ml IFN-γ.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
RNA was extracted from cultured macrophages (n=7), and paired non-lesional and lesional psoriasis samples (n=5 pairs) using the RNeasy Mini KIT (Qiagen, Valencia, CA, U.S.A.). RT-PCR was performed as previously described (Chamian et al., 2005) and outlined further in SMM. The results were normalized to HARP housekeeping gene.
Gene Array
RNA was amplified and labeled (Message Amp Premier RNA Amplification Kit, Ambion Inc., TX). A total of 750ng of biotinylated cRNA was hybridized to Illumina Human HT-12 Bead Chip (Illumina, Inc. San Diego, CA, USA). Results were analyzed as described in SMM. Briefly, to compare gene expression of in vitro cytokine treated macrophages versus control, and lesional versus non-lesional samples of psoriasis tissues, a moderated paired t-test available in limma package from R/Bioconductor was used. Genes were considered significant if they had a FCH > 2, and FDR<0.01. Gene Set Enrichment Analysis (GSEA) approach (Subramanian et al., 2005; Zaba et al., 2009) was used to correlate the response profile of the macrophage pathways in psoriasis lesional versus non-lesional skin. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE18686. Ingenuity analysis was performed using M1 and M2 macrophage DEGs.
Statistics
Cell counts were analyzed by Mann Whitney U test, significance was accepted as p<0.05. RT-PCR data was analyzed using a repeated measures ANOVA, and Dunnett’s Multiple Comparison Test was used to compare with control. The p values were designated as p < 0.05 (*), p< 0.01 (**) and p<0.001 (***).
Supplementary Material
Figure S1. CD11c coexpression with macrophage markers in psoriasis lesions. a) CD11c was coexpressed on a few CD163+ cells. There was greater co-expression of CD11c with (b) CD68, (c) RFD7, (d) LAMP2, (e) Stabilin, and (f) MARCO. Bar =100 μm
Figure S2. CD163+ macrophages also expressed CD14, CD16 and LFA-1 in psoriasis lesional skin. (a, b) CD163 showed greater coexpression of CD14 than CD11c+ DCs. CD16 (c, d) and LFA-1 (e, f) were present on both CD163+ macrophages and CD11c+ DCs. Bar =100 μm
Figure S3. CD163+ macrophages also expressed C-lectins and co-stimulatory molecules in psoriasis lesional skin. (a) Almost all CD163+ cells co-expressed CD206, and (b) the majority coexpressed CD209. Less than half of the CD163+ cells co-expressed the co-stimulatory molecules (c) CD86 and (d) CD40. Bar =100 μm
Figure S4. Macrophages in psoriasis lesions and normal skin did not express mature DC markers. CD 163+ macrophages did not c o-express well-recognized DC maturation markers (a) CD208/DC-LAMP, (b)CD205/DEC-205, and (c) CD83. Bar =100 μm
Figure S5. IFN-γ induced genes STAT-1 and CXCL9 immunohistochemistry in normal skin and psoriasis lesions. (a) STAT-1 expression was increased in psoriasis compared to normal skin. (b) CXCL9 showed low level expression in normal skin, and was increased in psoriasis. Bar =100 μm
Figure S6. Methods for counting cells. The diagrams illustrate two methods used for counting cells. In Method A, a representative image is captured, which is a field measuring 1000μm (1mm) × 1200μm (1.2mm). The cells are counted manually using computer-assisted image analysis (NIH Image 6.1;http://rsb.info.nih.gov/nih-image), and the number quantified per epidermis surface length. Examples for normal skin and psoriasis are shown. This method was used to obtain cell counts for Figure 1. In Method B, the same number of cells are counted, but cell number is calculated per area of dermis. Examples of the area of dermis are shown, but the image J program is used to calculate the actual area. This method was used to obtain cell counts for Table S1.
Table S1. Macrophage markers used in this study
Table S4. Gene Set Enrichment Analysis (GSEA) of cytokine-treated macrophage pathways in psoriasis compared to control
Table S5. Upregulated and downregulated genes in IL-4 and TNF-treated macrophages in common with Psoriasis
Table S6. Antibodies for Immunohistochemistry and Immunofluorescence
Acknowledgements
Research was supported by National Institutes of Health (NIH) grant UL1 RR024143 from the National Center for Research Resources (NCRR). MSF is partially supported by NIH grant UL1 RR024143 and Milstein Program in Medical Research, LCZ is supported by NIH MSTP grant GM07739; KEN is supported by the Clinical Scholars Program at The Rockefeller University; KCP is supported by the Dana Foundation (Human Immunology Consortium Grant); MAL is supported by K23 AR052404-01A1 and The Doris Duke Charitable Foundation. We thank Dr Leanne Johnson-Huang for her valuable comments on the manuscript. We appreciated assistance and advice from the Bio-imaging Resource Center (Dr A North) at Rockefeller University. We thank Patricia Gilleaudeau and Mary Whalen-Sullivan for excellent care of our patients.
Abbreviations
- LAMP2
Lysosomal Associated Membrane Protein 2
- MARCO
Macrophage Receptor with Collagenous structure
- MMR
Macrophage Mannose Receptor
- DC-SIGN
Dendritic Cell-Specific ICAM-3-Grabbing Non-integrin
- DC
Dendritic Cell
- BDCA
Blood Dendritic Cell Antigen
- DC-LAMP
Dendritic Cell-Lysosomal-Associated Membrane glycoprotein
- allo-MLR
allogeneic mixed leukocyte reaction
- Tip-DC
TNF-and-iNOS producing dendritic cell
- iNOS
inducible nitric oxide synthase
- GSEA
Gene Set Enrichment Analysis
- FDR
False Discovery Rate
- FCH
Fold change
- CS
Connectivity Score
- DEG
differentially expressed genes
- DEJ
dermo-epidermal junction
Footnotes
Conflict of Interest The authors do not have any financial interest related to this work.
References
- Arredouani MS, Palecanda A, Koziel H, Huang YC, Imrich A, Sulahian TH, et al. MARCO is the major binding receptor for unopsonized particles and bacteria on human alveolar macrophages. J Immunol. 2005;175:6058–6064. doi: 10.4049/jimmunol.175.9.6058. [DOI] [PubMed] [Google Scholar]
- Bryant P, Ploegh H. Class II MHC peptide loading by the professionals. Curr Opin Immunol. 2004;16:96–102. doi: 10.1016/j.coi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Casanova JL, Abel L. Revisiting Crohn’s disease as a primary immunodeficiency of macrophages. J Exp Med. 2009;206:1839–1843. doi: 10.1084/jem.20091683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamian F, Lowes MA, Lin SL, Lee E, Kikuchi T, Gilleaudeau P, et al. Alefacept reduces infiltrating T cells, activated dendritic cells, and inflammatory genes in psoriasis vulgaris. Proc Natl Acad Sci U S A. 2005;102:2075–2080. doi: 10.1073/pnas.0409569102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djemadji-Oudjiel N, Goerdt S, Kodelja V, Schmuth M, Orfanos CE. Immunohistochemical identification of type II alternatively activated dendritic macrophages (RM 3/1+3, MS-1+/−, 25F9-) in psoriatic dermis. Arch Dermatol Res. 1996;288:757–764. doi: 10.1007/BF02505293. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Fabriek BO, Dijkstra CD, van den Berg TK. The macrophage scavenger receptor CD163. Immunobiology. 2005;210:153–160. doi: 10.1016/j.imbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Gisondi P, Targher G, Zoppini G, Girolomoni G. Non-alcoholic fatty liver disease in patients with chronic plaque psoriasis. J Hepatol. 2009;51:758–764. doi: 10.1016/j.jhep.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Goerdt S, Politz O, Schledzewski K, Birk R, Gratchev A, Guillot P, et al. Alternative versus classical activation of macrophages. Pathobiology. 1999;67:222–226. doi: 10.1159/000028096. [DOI] [PubMed] [Google Scholar]
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- Gratchev A, Kzhyshkowska J, Kothe K, Muller-Molinet I, Kannookadan S, Utikal J, et al. Mphi1 and Mphi2 can be re-polarized by Th2 or Th1 cytokines, respectively, and respond to exogenous danger signals. Immunobiology. 2006;211:473–486. doi: 10.1016/j.imbio.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Gratchev A, Kzhyshkowska J, Utikal J, Goerdt S. Interleukin-4 and dexamethasone counterregulate extracellular matrix remodelling and phagocytosis in type-2 macrophages. Scand J Immunol. 2005;61:10–17. doi: 10.1111/j.0300-9475.2005.01524.x. [DOI] [PubMed] [Google Scholar]
- Haider AS, Lowes MA, Suarez-Farinas M, Zaba LC, Cardinale I, Blumenberg M, et al. Cellular Genomic Maps Help Dissect Pathology in Human Skin Disease. J Invest Dermatol. 2007;128:606–615. doi: 10.1038/sj.jid.5701067. [DOI] [PubMed] [Google Scholar]
- Krutzik SR, Tan B, Li H, Ochoa MT, Liu PT, Sharfstein SE, et al. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat Med. 2005;11:653–660. doi: 10.1038/nm1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Bruce AT, Gudjonsson JE, Johnston A, Aphale A, Vatan L, et al. Induction of IL-17+ T cell trafficking and development by IFN-gamma: mechanism and pathological relevance in psoriasis. J Immunol. 2008;181:4733–4741. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kzhyshkowska J, Gratchev A, Goerdt S. Stabilin-1, a homeostatic scavenger receptor with multiple functions. J Cell Mol Med. 2006;10:635–649. doi: 10.1111/j.1582-4934.2006.tb00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kzhyshkowska J, Gratchev A, Martens JH, Pervushina O, Mamidi S, Johansson S, et al. Stabilin-1 localizes to endosomes and the trans-Golgi network in human macrophages and interacts with GGA adaptors. J Leukoc Biol. 2004;76:1151–1161. doi: 10.1189/jlb.0504300. [DOI] [PubMed] [Google Scholar]
- Lew W, Lee E, Krueger JG. Psoriasis genomics: analysis of proinflammatory (type 1) gene expression in large plaque (Western) and small plaque (Asian) psoriasis vulgaris. Br J Dermatol. 2004;150:668–676. doi: 10.1111/j.0007-0963.2004.05891.x. [DOI] [PubMed] [Google Scholar]
- Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- Lowes MA, Chamian F, Abello MV, Fuentes-Duculan J, Lin SL, Nussbaum R, et al. Increase in TNF-alpha and inducible nitric oxide synthase-expressing dendritic cells in psoriasis and reduction with efalizumab (anti-CD11a) Proc Natl Acad Sci U S A. 2005;102:19057–19062. doi: 10.1073/pnas.0509736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowes MA, Kikuchi T, Fuentes-Duculan J, Cardinale I, Zaba LC, Haider AS, et al. Psoriasis Vulgaris Lesions Contain Discrete Populations of Th1 and Th17 T Cells. J Invest Dermatol. 2008;128:1207–1211. doi: 10.1038/sj.jid.5701213. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- Moestrup SK, Moller HJ. CD163: a regulated hemoglobin scavenger receptor with a role in the anti-inflammatory response. Ann Med. 2004;36:347–354. doi: 10.1080/07853890410033171. [DOI] [PubMed] [Google Scholar]
- Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff BJ, Bonish BK, Marble DJ, Schriedel KA, DiPietro LA, Gordon KB, et al. Lessons learned from psoriatic plaques concerning mechanisms of tissue repair, remodeling, and inflammation. J Investig Dermatol Symp Proc. 2006;11:16–29. doi: 10.1038/sj.jidsymp.5650010. [DOI] [PubMed] [Google Scholar]
- Nijsten T, Wakkee M. Complexity of the association between psoriasis and comorbidities. J Invest Dermatol. 2009;129:1601–1603. doi: 10.1038/jid.2009.55. [DOI] [PubMed] [Google Scholar]
- Nograles KE, Zaba LC, Guttman-Yassky E, Fuentes-Duculan J, Suarez-Farinas M, Cardinale I, et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br J Dermatol. 2008;159:1086–1091. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa MT, Loncaric A, Krutzik SR, Becker TC, Modlin RL. “Dermal Dendritic Cells” Comprise Two Distinct Populations: CD1(+) Dendritic Cells and CD209(+) Macrophages. J Invest Dermatol. 2008 doi: 10.1038/jid.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofre G, Kolackova M, Jankovicova K, Krejsek J. Scavenger receptor CD163 and its biological functions. Acta Medica (Hradec Kralove) 2009;52:57–61. [PubMed] [Google Scholar]
- Pantelidis P, McGrath DS, Southcott AM, Black CM, du Bois RM. Tumour necrosis factor-alpha production in fibrosing alveolitis is macrophage subset specific. Respir Res. 2001;2:365–372. doi: 10.1186/rr87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz O, Gratchev A, McCourt PA, Schledzewski K, Guillot P, Johansson S, et al. Stabilin-1 and -2 constitute a novel family of fasciclin-like hyaluronan receptor homologues. Biochem J. 2002;362:155–164. doi: 10.1042/0264-6021:3620155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottman JB, Smith TL, Ganley KG, Kikuchi T, Krueger JG. Potential role of the chemokine receptors CXCR3, CCR4, and the integrin alphaEbeta7 in the pathogenesis of psoriasis vulgaris. Lab Invest. 2001;81:335–347. doi: 10.1038/labinvest.3780242. [DOI] [PubMed] [Google Scholar]
- Sankala M, Brannstrom A, Schulthess T, Bergmann U, Morgunova E, Engel J, et al. Characterization of recombinant soluble macrophage scavenger receptor MARCO. J Biol Chem. 2002;277:33378–33385. doi: 10.1074/jbc.M204494200. [DOI] [PubMed] [Google Scholar]
- Sanmiguel JC, Olaru F, Li J, Mohr E, Jensen LE. Interleukin-1 regulates keratinocyte expression of T cell targeting chemokines through interleukin-1 receptor associated kinase-1 (IRAK1) dependent and independent pathways. Cell Signal. 2009;21:685–694. doi: 10.1016/j.cellsig.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–349. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttles J, Stout RD. Macrophage CD40 signaling: a pivotal regulator of disease protection and pathogenesis. Semin Immunol. 2009;21:257–264. doi: 10.1016/j.smim.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Taams LS, Poulter LW, Rustin MH, Akbar AN. Phenotypic analysis of IL-10-treated macrophages using the monoclonal antibodies RFD1 and RFD7. Pathobiology. 1999;67:249–252. doi: 10.1159/000028103. [DOI] [PubMed] [Google Scholar]
- Torocsik D, Bardos H, Nagy L, Adany R. Identification of factor XIII-A as a marker of alternative macrophage activation. Cell Mol Life Sci. 2005;62:2132–2139. doi: 10.1007/s00018-005-5242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Oord JJ, De Ley M, De Wolf-Peeters C. Distribution of interferon-gamma receptors in normal and psoriatic skin. Pathol Res Pract. 1995;191:530–534. doi: 10.1016/S0344-0338(11)80872-1. [DOI] [PubMed] [Google Scholar]
- Wang F, Lee E, Lowes MA, Haider AS, Fuentes-Duculan J, Abello MV, et al. Prominent production of IL-20 by CD68+/CD11c+ myeloid-derived cells in psoriasis: Gene regulation and cellular effects. J Invest Dermatol. 2006;126:1590–1599. doi: 10.1038/sj.jid.5700310. [DOI] [PubMed] [Google Scholar]
- Way KJ, Dinh H, Keene MR, White KE, Clanchy FI, Lusby P, et al. The generation and properties of human macrophage populations from hemopoietic stem cells. J Leukoc Biol. 2009;85:766–778. doi: 10.1189/jlb.1108689. [DOI] [PubMed] [Google Scholar]
- Wilson HM, Barker RN, Erwig LP. Macrophages: promising targets for the treatment of atherosclerosis. Curr Vasc Pharmacol. 2009;7:234–243. doi: 10.2174/157016109787455635. [DOI] [PubMed] [Google Scholar]
- Wu K, Yuan J, Lasky LA. Characterization of a novel member of the macrophage mannose receptor type C lectin family. J Biol Chem. 1996;271:21323–21330. doi: 10.1074/jbc.271.35.21323. [DOI] [PubMed] [Google Scholar]
- Yawalkar N, Tscharner GG, Hunger RE, Hassan AS. Increased expression of IL-12p70 and IL-23 by multiple dendritic cell and macrophage subsets in plaque psoriasis. J Dermatol Sci. 2009;54:99–105. doi: 10.1016/j.jdermsci.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Zaba LC, Cardinale I, Gilleaudeau P, Sullivan-Whalen M, Farinas MS, Fuentes-Duculan J, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007a;204:3183–3194. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, Abello MV, Novitskaya I, Pierson KC, et al. Psoriasis Is Characterized by Accumulation of Immunostimulatory and Th1/Th17 Cell-Polarizing Myeloid Dendritic Cells. J Invest Dermatol. 2009;129:79–88. doi: 10.1038/jid.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaba LC, Fuentes-Duculan J, Steinman RM, Krueger JG, Lowes MA. Normal human dermis contains distinct populations of CD11cBDCA-1 dendritic cells and CD163FXIIIA macrophages. J Clin Invest. 2007b;117:2517–2525. doi: 10.1172/JCI32282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. CD11c coexpression with macrophage markers in psoriasis lesions. a) CD11c was coexpressed on a few CD163+ cells. There was greater co-expression of CD11c with (b) CD68, (c) RFD7, (d) LAMP2, (e) Stabilin, and (f) MARCO. Bar =100 μm
Figure S2. CD163+ macrophages also expressed CD14, CD16 and LFA-1 in psoriasis lesional skin. (a, b) CD163 showed greater coexpression of CD14 than CD11c+ DCs. CD16 (c, d) and LFA-1 (e, f) were present on both CD163+ macrophages and CD11c+ DCs. Bar =100 μm
Figure S3. CD163+ macrophages also expressed C-lectins and co-stimulatory molecules in psoriasis lesional skin. (a) Almost all CD163+ cells co-expressed CD206, and (b) the majority coexpressed CD209. Less than half of the CD163+ cells co-expressed the co-stimulatory molecules (c) CD86 and (d) CD40. Bar =100 μm
Figure S4. Macrophages in psoriasis lesions and normal skin did not express mature DC markers. CD 163+ macrophages did not c o-express well-recognized DC maturation markers (a) CD208/DC-LAMP, (b)CD205/DEC-205, and (c) CD83. Bar =100 μm
Figure S5. IFN-γ induced genes STAT-1 and CXCL9 immunohistochemistry in normal skin and psoriasis lesions. (a) STAT-1 expression was increased in psoriasis compared to normal skin. (b) CXCL9 showed low level expression in normal skin, and was increased in psoriasis. Bar =100 μm
Figure S6. Methods for counting cells. The diagrams illustrate two methods used for counting cells. In Method A, a representative image is captured, which is a field measuring 1000μm (1mm) × 1200μm (1.2mm). The cells are counted manually using computer-assisted image analysis (NIH Image 6.1;http://rsb.info.nih.gov/nih-image), and the number quantified per epidermis surface length. Examples for normal skin and psoriasis are shown. This method was used to obtain cell counts for Figure 1. In Method B, the same number of cells are counted, but cell number is calculated per area of dermis. Examples of the area of dermis are shown, but the image J program is used to calculate the actual area. This method was used to obtain cell counts for Table S1.
Table S1. Macrophage markers used in this study
Table S4. Gene Set Enrichment Analysis (GSEA) of cytokine-treated macrophage pathways in psoriasis compared to control
Table S5. Upregulated and downregulated genes in IL-4 and TNF-treated macrophages in common with Psoriasis
Table S6. Antibodies for Immunohistochemistry and Immunofluorescence