Abstract
Extracellular and intracellular mediators of inflammation, such as Tumor Necrosis Factor alpha (TNFα) and NF-kappaB (NF-κB), play major roles in breast cancer pathogenesis, progression and relapse. SLUG, a mediator of the epithelial-mesenchymal transition process, is over-expressed in CD44+/CD24− tumor initiating breast cancer cells and in basal-like carcinoma, a subtype of aggressive breast cancer endowed with a stem cell-like gene expression profile. Cancer stem cells also over-express members of the pro-inflammatory NF-κB network, but their functional relationship with SLUG expression in breast cancer cells remains unclear. Here, we show that TNFα treatment of human breast cancer cells up-regulates SLUG with a dependency on canonical NF-κB/HIF1α signaling, which is strongly enhanced by p53 inactivation. Moreover, SLUG up-regulation engenders breast cancer cells with stem cell-like properties including enhanced expression of CD44 and Jagged-1 in conjunction with ERα down-regulation, growth as mammospheres and extracellular matrix invasiveness. Our results reveal a molecular mechanism whereby TNFα, a major pro-inflammatory cytokine, imparts breast cancer cells with stem cell-like features, which are connected to increased tumor aggressiveness.
Keywords: TNFalpha, SLUG, basal-like phenotype, breast cancer stem cells
Introduction
Over the past decade, it has become increasingly clear that inflammation plays a major role in cancer pathogenesis (Mantovani et al., 2008). Several lines of evidence indicate that in breast cancer patients, some serum biomarkers of chronic inflammation are associated with disease recurrence (Cole 2009; Pierce et al., 2009) and that normal mammary gland involution, a widely acknowledged pro-tumorigenic process, is an inflammatory phenomenon (O’Brien et al., 2009). Consistent with human evidence, mice models clearly indicate that inflammatory molecules released by resident immune cells or secreted systemically, are permissive for tumor development in cancer prone strains (O’Brien et al., 2009; Rao et al., 2006). Moreover, the knock down of extracellular and intracellular mediators of inflammation, such as Tumor Necrosis Factor alpha (TNFα) and NF-kappaB (NF-κB), are protective with respect to chemical induced mammary gland carcinogenesis (Warren et al., 2009; Cao et al., 2007). Further, the in vitro activation of the TNFα/NF-κB axis induces an invasive and malignant behaviour in breast cancer cells (Balkwill 2009).
The phenotype and gene expression profile of a subpopulation of CD44+/CD24− breast cancer cells, endowed with tumor initiating capability (referred to as breast cancer stem cells), has recently been characterized (Shipitsin et al., 2007; Al-Hajj et al., 2003; Mani et al., 2008). Such putative breast cancer stem cells over-express members of the pro-inflammatory NF-κB network, which predicts poor prognosis in breast cancer patients (Liu et al., 2007). In vitro, NF-κB was proven to sustain features of breast cancer stem cells, including the capacity to grow as multicellular spheroids, named mammospheres (MS), and to invade the extracellular matrix (Zhou et al., 2008a; Sansone et al., 2007a; Storci et al., 2008; Sheridan et al. 2006; Wu et al., 2009; Mani et al., 2008). In tissue specimens, NF-κB up-regulation was found in basal-like tumors, an aggressive subtype of breast cancer that presents a stem cell-like gene expression profile (Ben-Porath et al., 2008; Bertucci et al., 2009). Hallmarks of basal-like tumors include the lack of estrogen receptor alpha (ERα), the presence of the CD44 membrane marker and the expression of the Notch ligand Jagged-1 (Storci et al., 2008, Bertucci et al., 2009, Charafe-Jauffret et al., 2006; Honeth et al., 2008; Dickson et al., 2007; Reedijk et al., 2005). Notably this is also the gene expression profile of breast cancer stem cells and MS (Shipitsin et al., 2007; Storci et al., 2008; Mani et al., 2008; Horwitz et al., 2008). Up-regulation of the SLUG gene (also known as SNAI2) has been functionally linked to the onset of a basal-like gene expression profile in breast cancer cells and the genes of the SNAI family have been shown to activate a stem cell-like phenotype in human mammary gland derived epithelial cells (Storci et al., 2008; Mani et al., 2008). Accordingly, basal-like tumors and CD44+/CD24− breast cancer stem cells express SLUG and SNAI gene family members (Storci et al., 2008, Mani et al., 2008; Sarriò et al., 2008).
Recently, a SNAI family member SNAIL was shown to be up-regulated by the TNFα/NF-κB axis (Dong et al., 2007). Here, we have show that the expression of the breast cancer stem cell phenotype induced by TNFα is a consequence of the functional relationship between the NF-κB and SLUG regulatory pathways. In addition, we demonstrate that the NF-κB/SLUG circuit is up-regulated in cells with an inactive p53 protein, a very frequent event in basal-like breast carcinoma (Bertucci et al., 2009, Charafe-Jauffret et al., 2006).
Materials and Methods
Reagents and Cell Culture
Recombinant human TNFα was purchased from Sigma (St.Louis, MS, USA). Parthenolide, (an inhibitor of IκBα degradation), the IκBα phosphorylation inhibitor Bay 11–7082, and the IKKβ specific inhibitor sc-514 were purchased from Biomol (Plymouth Meeting, PA), Sigma and Calbiochem (San Diego, CA, USA), respectively. The efficiency of Parthenolide treatment in inhibiting NF-κB activity (Hehner et al., 1999) was assessed by a sensitive NF-κB-Luc reporter assay (Supplementary Figure 1A). MCF-7 cells were cultured in RPMI-1640 medium supplemented with 10% Fetal Bovine Serum, 100µg/ml streptomycin, 100IU/ml penicillin and 2mM L-Glutamine (Euroclone, Milan, Italy). All cells were grown at 37°C in a humidified (5% CO2) atmosphere.
Generation of mammospheres (MS) from MCF7 cells and normal breast tissues
MS were derived from single cell level re-suspension of MCF7 cells in complete mammary epithelial growth medium (MEGM), supplemented with 10 ng/ml Epidermal Growth Factor (EGF), 10 ng/ml basic Fibroblasts Growth Factor (bFGF), 10 µg/ml Insulin, 10−6 M hydrocortisone, Gentamycin and Amphotericine ad hoc aliquot (Cambrex, East Rutherford, NJ, USA) and plated at 1-5-10 ×103/cm2 in ultra-low attachment well (Corning, Lowell, MA, USA). TNFα, Parthenolide, Bay 11–7082 and sc-514 treatment were performed by exposing MCF7 cells for 24h to each molecule. Then, cells were resuspended at single cell level in MEGM and plated at 1-5-10 ×103/cm2 in ultra-low attachment well. MS were scored in triplicates after 7 days. Mammospheres from mammary gland tissues (N-MS) were obtained as previously described (Sansone et al., 2007a; Storci et al., 2008; Dontu et al., 2003). Briefly, tissues were placed in sterile Epicult medium (StemCell Technologies, Vancouver, Canada), minced with sterile scalpels, and incubated for 6–12 hours in the presence of 1,000 IU Collagenase/Hyaluronidase enzyme mix (StemCell Technologies). Cell suspension was filtered through a 40 µm nylon mesh (Becton Dickinson, San Josè, CA) re-suspended in complete MEGM and plated in 1cm2 ultra low attachment plates. Secondary MS were generated by of incubation primary MS with 1× Trypsin-EDTA solution (Cambrex) for 3 minutes, filtration throughout a 40 µm nylon mesh and single cell re-suspension in complete MEGM. Secondary MS were assessed at day 7. Written informed consent was obtained by patients whose tissues were used in the study.
Cell death assessment
Cell death was evaluated in triplicates by Trypan blue dye exclusion.
Gene silencing by transient siRNA and stable shRNA knock-down (KD)
SLUG specific siRNAs (Stealth™ select 3 RNAi set) and non-specific siRNA control oligonucleotides with a matched GC content were purchased from Invitrogen (Carlsbad, CA, USA). siRNAs were transfected into adherent MCF7 (105 cells in a 3 cm2 well) at a concentration of 1 µg/well, using Lipofectamine 2000 (Invitrogen). siRNA transfection in MS was performed by mixing 1µg of siRNA with in vitro JET-PEI reagent (Poly plus Transfection, Illkirch, France).
Stable SLUG knock down was achieved by retroviral transduction with the pCtoGMB moloney retroviral vector. pCtoGMB co-expresses an shOligo (embedded in a Mir30 expression cassette) and a GFP gene under the control of a tetracycline repressor responsive CMV promoter/enhancer (Cto), in addition to a Pgk promoter driven Blasticidin resistance gene. pCtoGMB was assembled by standard molecular cloning techniques from portions of several other plasmid vectors [including pShag.C2 (Paddison et al., 2004), pTMP (Dickins et al., 2005) and pcDNATO/Luc (Invitrogen)] (Marcu, KB, unpublished). Briefly, the PgkPuro-IRES-GFP cassette in pTMP was replaced by Pgk promoter-Blasticidin sequences to produce the pTMB retrovector. Separately, a 1.6 kB fragment containing Mir30-CAM sequences was released from pShag.c2 by SalI + AgeI double digestion and subcloned between the HindIII/XbaI sites of pcDNATO/Luc, generating the intermediate plasmid vector pcDNATO-Mir30CAM. Next, a 900 bp NruI/XhoI fragment specifying the CMVTO-5'Mir30 sequences in pcDNATO-Mir30CAM was subcloned between the unique XbaI/XhoI sites of pTMB generating the pCtoMB retrovector, in which a CMVTO-Mir30 expression cassette resides upstream of Pgk-Blasticidin sequences. Finally, a 745 bp fragment encoding a GFP ORF was inserted into a unique PmeI site between the CMVTO and Mir30 sequences of pCtoMB to yield pCtoGMB. Next, we generated a DNA fragment encoding a shRNA-mir fold sequence targeted to a specific 25 nt coding sequence (GGCTCATCTGCAGACCCATTCTGAT) in human SLUG mRNA by a PCR based subcloning strategy, as described by Hannon and colleagues (Paddison et al., 2004) and subcloned it within the miR30 expression cassette between unique XhoI/RI sites in pCtoGMB. Retroviral transductions were performed by spinoculation, as previously described (Dickins et al., 2005; Olivotto et al., 2008). Populations of stably transduced cells were selected in 1.5 µg/ml Blasticidin. pCtoMGB was similarly used to stably deliver human shIKKβ oligonucleotides (Olivotto et al., 2008). IKKβ KD efficiency was assessed by an NF-κB-Luc reporter assay in MCF-7 cells exposed to TNFα (Supplementary Figure 1B).
Plasmid and retroviral protein expression
Retroviral vectors expressing murine p65 and constitutively activated IKKβ (IKKβca) have been previously described (Penzo et al., 2009; Zhang et al., 2005). MCF7 cells stably transduced with a retroviral vector encoding a p53 dominant inactivating miniprotein (p53D) were previously described (Shaulian et al., 1992; Sansone et al., 2007b). A human SLUG encoding vector was kindly provided by Dr. Tony Ip (Massachusetts Medical School, Worcester, MA). Plasmid encoding HIF1α lacking the oxygen degradation domain (Huang et al., 1998) was obtained from Eric Huang (Department of Neurosurgery, University of Utah, Salt Lake City, Utah). Transient transfections of plasmid vectors were performed with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions.
Luciferase reporter gene assays
SLUG-Luc and Estrogen Response Element (ERE-Luc) plasmids were kindly provided by Dr. Togo Ikuta (Saitama Cancer Centre, Saitama, Japan) and Dr. Rakesh Kumar (Dept. of Molecular and Cellular Oncology, MD Anderson Cancer Center, Houston, Texas), respectively. Hypoxia response element (HRE-Luc) was obtained from Dr. Giovanni Melillo (Tumor hypoxia laboratory, National Cancer Institute, Frederick, MD, USA). TOPFLASH reporter vector was a gift of Dr. Rolf Kemler (Max Planck Institute, Heidelberg, Germany). Each of the above plasmids (1µg) were transfected with Lipofectamine 2000 (Invitrogen) in co-transfections with a thymidine kinase promoter driven Renilla luciferase (40ng) plasmid as a reference control (Promega, Madison, WI). Luciferase activity was assayed after 24 hours using the Dual-Luciferase® Reporter Assay System (Promega), according to the manufacturer’s instructions. Luciferase activity was normalized over Renilla activity and all reported experiments were performed in triplicates.
Immunofluorescence
Cultured cells were seeded onto glass cover-slips at 60% confluence, fixed with 2% paraformaldehyde and permeabilized with 0.2% Triton X-100. Cells were incubated with anti β-catenin (sc-7963) antibody (Santa Cruz Biotechnology, Santa Cruz, California) and secondary anti-mouse anti-rabbit antibody FITC conjugated (Dako Cytomation, Glostrup, DK).
Immunohistochemistry
Immunohistochemistry was performed in formalin-fixed, paraffin-embedded human breast carcinoma tissue (Supplementary Table 1) with the following rabbit polyclonal antibodies: anti-HIF1α (Upstate Technology, Billerica, MA, USA), anti-phosphoNF-κB(p65)-Ser276 (Cell Signaling Technology, Danvers, MA USA), and anti-SLUG (Santa Cruz Biotechnology) . The immunological reaction was developed using a 3,3’-tetrahydrochloride diaminobenzidine (DAB)/H2O2-PBS solution and counterstained with haematoxylin. Written informed consent was obtained by patients whose tissues were used in the study.
Boyden chamber invasion assays
Cell invasion assays were performed in triplicate as previously described (Storci et al., 2008).
RNA extraction, Reverse Transcription and cDNA amplification
Total RNA was extracted from cells using TRIzol® Reagent according to the manufacturer’s protocol (Invitrogen). Primers and PCR conditions are reported in Supplementary Table 2.
Statistical Analysis
Data are expressed as mean + standard deviation (S.D.). Sample means were compared using Student’s t test or Anova followed by post-hoc (Bonferroni or Dunnet’s) comparisons. Calculations were executed with the SPSS statistical package (SPSS INC, Chicago, IL).
RESULTS
TNFα up-regulates CD44 and Jagged-1 and down-regulates ERα expression via SLUG
Ductal breast carcinoma MCF7 cells were transfected with a human SLUG expression vector which up-regulated CD44 and Jagged-1 gene expression, in conjunction with the down-regulation of ERα mRNA expression and transcriptional activity (Figure 1A). TNFα, a potent NF-κB activating pro-inflammatory stimulus, up-regulated endogenous SLUG expression and simultaneously induced the up-regulation of NF-κB binding site and SLUG promoter driven luciferase reporters in MCF7 cells (Shaulian et al., 1992; Figure 1B). TNFα also elicited the up-regulation of CD44 and Jagged-1 in conjunction with the down-regulation of ERα mRNA and activity; these latter effects were inhibited by the transient transfection of a SLUG specific siRNA (Figure 1C). These data indicate that SLUG expression, under TNFα exposure, induces a basal-like gene profile in breast cancer cells.
FIGURE 1. TNFα up-regulates CD44 and Jagged-1 and down-regulates ERα expression via SLUG.
(A): RT-PCR analysis of CD44, Jagged-1 and ERα mRNA level, and ERE-Luc in MCF7 cells transiently transfected with empty (pcDNA3.1) and human SLUG encoding (pSLUG) vector (1µg each, 24h); (B): NF-κB-Luc, SLUG promoter driven luciferase activity assay (SLUG-Luc) and RT-PCR analysis of SLUG mRNA in MCF7 cells exposed to TNFα (10ng/ml, 24h); (C): CD44, Jagged-1 and ERα mRNA level and ERE-Luc assay in MCF7 cells transiently transfected with SCR or SLUG specific siRNA (1µg, 48h) exposed or less to TNFα (10ng/ml, 24h). Data are presented as mean +/− S.D. of three replicates, p values of unpaired t tests: *<0.05, #<0.01 and § <0.005. β-Actin was used as reference control (RT-PCR analysis in panel A and B are normalized on the same β-Actin).
TNFα/NF-κB signalling promotes MS formation and invasive capacity of breast cancer cells via SLUG up-regulation
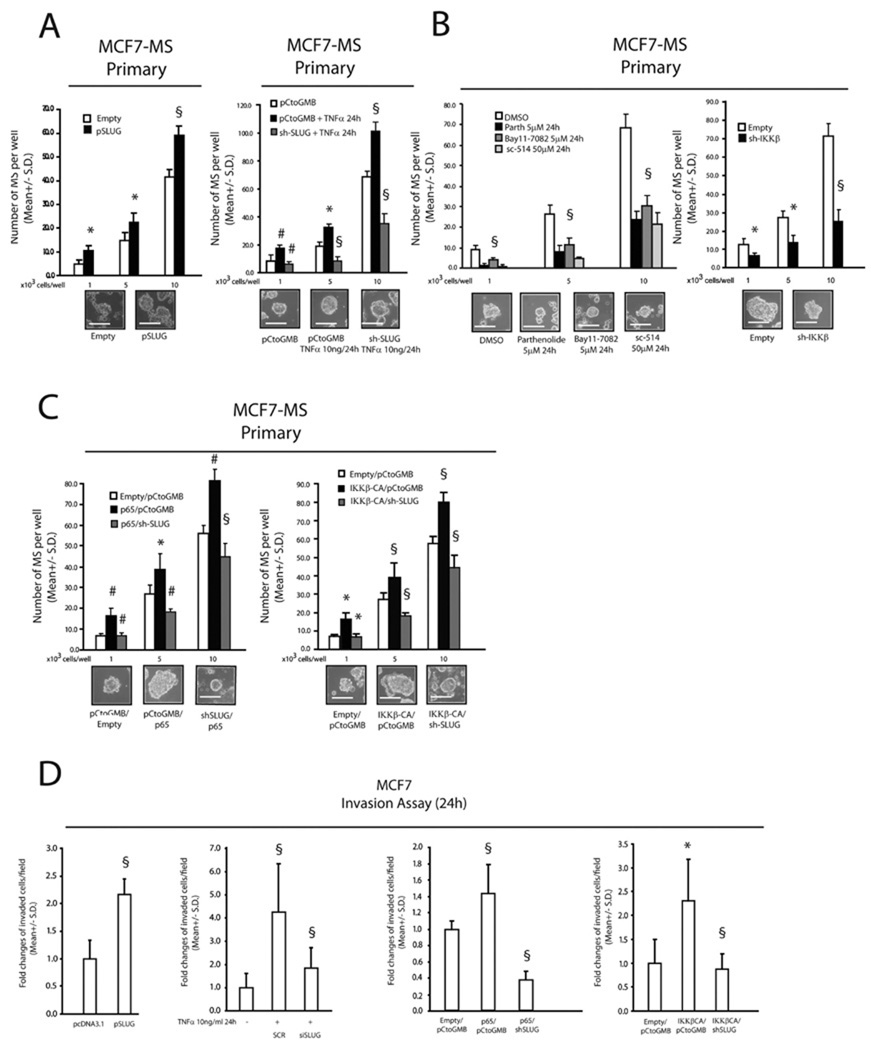
The SNAI-induced breast cancer stem cell gene expression profile has recently been associated with MS forming capacity (Mani et al., 2008). In line with these findings, we observed that MCF7 cells acquired enhanced MS forming ability in response to either a SLUG expression vector or TNFα exposure, and the latter response was blunted in SLUG shRNA knock-down cells (SLUG KD) (Figure 2A). Recent findings indicate that NF-κB activity promotes MS forming ability and invasion (Zhou et al., 2008; Wu et al., 2009). We observed a dramatic decrease in MS forming capacity in MCF7 cells exposed to the IαBκ degradation inhibitors Parthenolide and Bay11–708, the IKKβ specific inhibitor sc-514, or stably transduced with an IKKβ specific shRNA expressing retroviral vector (Figure 2B). Moreover, MS forming capacity was increased in MCF7 cells stably transduced with the p65 NF-κB subunit or the constitutively active IKKβ mutant (IKKβca), and these enhancing effects on MS formation were inhibited in SLUG-KD cells (Figure 2C). In agreement with these results, SLUG-KD MCF7 cells impaired their invasive capacity, even after the exposure to TNFα or after stable transduction with p65/IKKβca vectors (Figure 2D). Finally, the transfection of SLUG siRNA into mammospheres, from normal human mammary glands reduced secondary MS formation, blunted invasive capacity, reduced CD44 and Jagged-1 mRNA levels and increased ERα expression (Supplementary Figure 2).
FIGURE 2. TNFα/NF-κB signalling promotes MS formation and invasive capacity of breast cancer cells via SLUG up-regulation.
(A): MS forming capacity of empty/pSLUG transiently transfected (1µg, 24h) MCF7 cells and pCtoGMB/ shSLUG stably transduced MCF7 cells exposed or less to TNFα (10 ng/ml, 24h); (B): MS forming capacity of MCF7 cells exposed to the IκBα degradation inhibitors Parthenolide or Bay 11-7082 (5µM, 24h each), the specific IKKβ inhibitor sc-514 (5µM, 24h each), or stably transduced with an IKKβ specific shRNA/empty expressing retroviral vector; (C): MS forming capacity of pCtoGMB/shSLUG MCF7 cells transduced with empty or p65/IKKβ-CA encoding vector, representative MS pictures are also reported. The scale bar inset corresponds to 100µm; (D): Invasion assay in pSLUG transfected (1µg), TNFα exposed (10ng/ml, 24h), p65/IKKβ-CA transduced pCtoGMB/shSLUG MCF7 cells. Data are presented as mean +/− S.D. of three replicates, p values of unpaired t tests: *<0.05, #<0.01 and § <0.005. The scale bar represents 100 µm.
HIF1α promotes CD44 and Jagged-1 expression, MS formation and breast cancer cellular invasiveness via SLUG
Recently, we found that SLUG is up-regulated by the hypoxia mimetic Desferroxamine, a potent activator of Hypoxia Inducible Factor 1 alpha (HIF1α) which is also transcriptionally induced by canonical NF-κB signalling and TNFα exposure (Storci et al., 2008; Gorlach et al., 2008; Rius et al., 2008). Therefore, we investigated if HIF1α might reside within the NF-κB/SLUG MS regulatory circuit. Indeed, enforced expression of a stable, HIF1α mutant protein, lacking the oxygen degradation domain (HIF1αODD), up-regulated SLUG-Luc activity and endogenous SLUG, CD44 and Jagged-1 mRNA expression (Figure 3A). HIF1αODD transfection in MCF7 cells also enhanced their MS formation and invasive capability (Figure 3B). However, these phenotypic changes were not induced when HIF1αODD was transfected into SLUG-KD cells (Figure 3C), suggesting that HIF1α acts up-stream of SLUG.
FIGURE 3. HIF1α is necessary for the SLUG-dependent stem cell-like gene signature, MS formation and invasive capacity of breast cancer cells.
(A): SLUG-Luc activity assay and RT-PCR analysis of SLUG, CD44 and Jagged-1 mRNA level in MCF7 cells transiently transfected with pCDNA3.1 and HIF1α encoding vector (HIF1αODD, 1µg, 24h); (B): MS forming and invasive capacity assay of MCF7 cells transiently transfected with HIF1αODD (1µg); (C): MS forming and invasive capacity assay of pCtoGMB/shSLUG MCF7 cells transiently transfected with HIF1αODD (1µg). Representative MS pictures are also reported. Data are presented as mean +/− S.D. of three replicates, p values of unpaired t tests *<0.05; #<0.01; §<0.005. The scale bar represents 100µm. β-Actin was used as reference control.
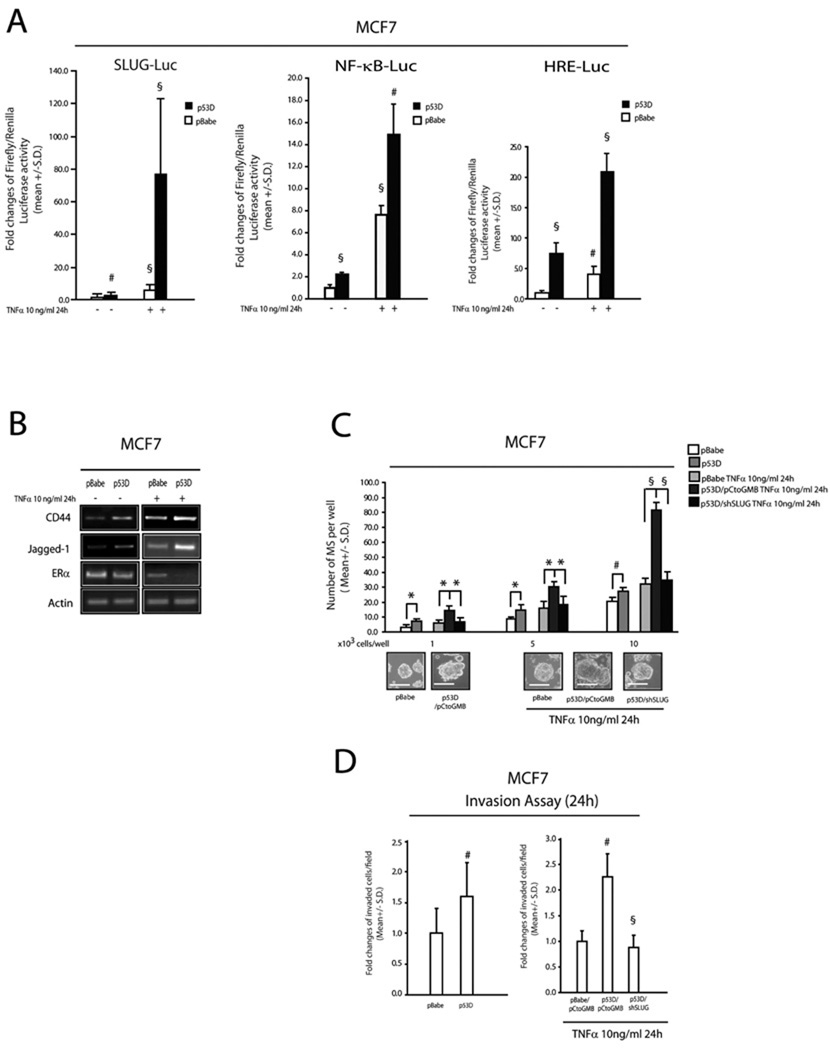
p53 compromised cells have an up-regulated NF-κB/HIF1α axis in conjunction with increased SLUG expression, which are further enhanced by TNFα exposure
p53 gene inactivating mutations are a salient feature of basal-like tumors (Bertheau et al., 2007) and are also known to up-regulate NF-κB and HIF1α activity (Weisz et al., 2007; Hammond et al., 2006). Interestingly, MCF7 cells stably transduced with a retroviral vector encoding a p53 inactivating miniprotein (hereafter referred to as p53D) had higher SLUG-Luc, NF-κB-Luc and HRE-Luc activities compared to a matched pBabe-empty retroviral vector control cell population (Figure 4A). Upon exposure to TNFα, both control and p53D cells showed increased SLUG-Luc, NF-κB-Luc and HRE-Luc activities, which were all more pronounced in p53D cells (Figure 4A). In addition, Jagged-1 and CD44 mRNA expression was up-regulated and ERα mRNA was down-regulated in p53D cells, particularly in response to TNFα (Figure 4B). Enhanced MS forming capacity was also observed in p53D cells, compared to their matched control; and this effect was amplified by TNFα exposure and blunted in SLUG KD cells (Figure 4C). The latter phenotypic changes occurred in conjunction with a comparable modulation of invasive capacity (Figure 4D). Taken together, these results show that p53 deficiency facilitates the up-regulation of the NF-κB/HIF1α/SLUG axis in a pro-inflammatory environment.
FIGURE 4. p53 compromised cells show an over-activation of the NF-κB/HIF1α axis and SLUG expression in response to TNFα.
(A): SLUG-Luc, NF-κB-Luc and HRE-Luc activity assays in pBabe/p53 dominant negative (p53D) transduced MCF7 cells exposed or less to TNFα 10 ng/ml, 24h; (B): RT-PCR analysis of CD44, Jagged-1, ERα in pBabe/p53D and pBabe/p53D exposed or less to TNFα 10ng/ml, 24h; (C): MS forming capacity of pBabe/p53D and pCtoGMB/shSLUG transduced MCF7 cells exposed or less to TNFα (10 ng/ml, 24h); representative MS pictures are also reported (D): Invasion assay of pBabe/p53D and pCtoGMB/shSLUG transduced MCF7 cells exposed or less to TNFα 0 (10 ng/ml, 24h). Data are presented as mean +/− S.D. of three replicates, p values of unpaired t tests: *<0.05; #<0.01 and §<0.005. Reference scale bar is 10µm. β-Actin was used as reference control.
Discussion
TNFα and NF-κB are required in different models of mammary gland carcinogenesis, reinforcing the concept that inflammation is involved in the initiation and promotion of breast cancer (Pratt et al., 2009; Lerebours et al., 2008; Van Laere et al., 2007). Here we have shown that breast cancer cells respond to the pro-inflammatory cytokine TNFα by inducing stem cell features through a mechanism involving the up-regulation of SLUG expression. In particular, TNFα-exposed cells require SLUG to acquire a basal-like gene expression profile (Jagged-1, CD44 over-expression, ERα down-regulation), that has been associated with tumor initiating cells and with an aggressive subtype of breast cancers that present a gene expression profile bearing similarities to embryonic stem cells (Shipitsin et al., 2007; Mani et al., 2008; Storci et al., 2008; Sansone et al., 2007a; Honeth et al., 2008; Dickson et al., 2007; Reedijk et al., 2005). Notably, Jagged-1 has been previously reported as a CD44+/CD24− MS growth promoting factor (Sansone et al., 2007) and was also found to be up-regulated by SLUG over-expression (Moreno Bueno et al., 2006).
On the functional side, TNFα treatment of breast cancer cells increases their MS forming ability, a proven assay that has been extensively employed to expand breast cancer and normal mammary gland stem cells in vitro (Dontu et al., 2003; Storci et al., 2008, Sansone et al., 2007a; Mani et al., 2008; Ponti et al., 2007; Cariati et al., 2008) and also engenders breast cancer cells with enhanced invasiveness in association with a CD44+/CD24− stem cell-like phenotype (Sheridan et al., 2006). In addition, SLUG is part of the proteomic profile of MCF7 cells that have been cultured in presence of TNFα and became resistant to TNFα-induced cell death (Zhou et al., 2007b). In this regard, we found that long term (1 week) TNFα exposure of adherent MCF7 cells triggers their spontaneous MS formation. The latter phenotypic change occurs in conjunction with the induction of a basal-like gene expression profile, which lasts three weeks post TNFα withdrawal, and subsequently reverts to control levels after an additional week (Supplementary Figure 3). Thus, we speculate that a SLUG dependent aggressive stem cell-like phenotype may arise as a consequence of the acquired capability of cancer cells to survive in an inflammatory environment.
Jagged-1 and CD44 are putative β-Catenin targets (Schwartz et al., 2003; Estrach et al., 2006) and basal-like carcinomas disclose a cytoplasmic localization of β-Catenin (Sarriò et al., 2008; McCarthy et al., 2007; Hayes et al., 2008). In this regard, we observed that TNFα exposure, as well as SLUG over-expression, induced the partial cytoplasmic and nuclear localization of β-Catenin, which was accompanied by an increased β-Catenin-Luc reporter gene activity reduced by siSLUG trasfection (Supplementary Figure 4). Therefore, we posit that β-Catenin plays a functional role in the induction of the basal/stem cell-like phenotype.
A NF-κB gene expression signature predicts poor prognosis in breast cancer patients (Liu et al., 2007). Intriguingly, SLUG expressing basal-like tumors and CD44+/CD24− breast tumor initiating cells over-express NF-κB (Shipitsin et al., 2007; Bertucci et al., 2009; Charafe-Jauffret et al., 2006). We have shown that HIF1α, a central regulator of the hypoxia response, is a crucial mediator of TNFα/NF-κB-dependent SLUG up-regulation and stem cell induction, thereby connecting these two pathways in the genesis of aggressive breast cancer cells. Our observations are in agreement with and extend other observations suggesting that NF-κB and HIF1α each play a role in regulating SLUG gene transcription (Dong et al., 2007; Ikuta et al., 2006; Laffin et al., 2008). Our data reinforce the notion that, after exposure to inflammatory mediators, HIF1α activity is up-regulated in the absence of hypoxia (Gorlach et al., 2006; Rius et al., 2008). The association between HIF1α and the stem cell-like phenotype is also consistent with hypoxic environments playing a major role in normal stem cell maintenance and promoting a de-differentiation program (Gustafsson et al., 2005; Simon et al., 2008; Eliasson et al., 2010). Moreover, HIF1α is over expressed in basal-like tumors and in CD44+/CD24−breast cancer stem cells along with NF-κB and SLUG (Shipitsin et al., 2007; Storci et al., 2008; Bertucci et al., 2009). Recently, a breast cancer stem cell-like phenotype has been documented in lymph-vascular tumor emboli arising from inflammatory breast carcinomas (Xiao et al., 2008). Of considerable interest, we also find that SLUG, p65-NF-κB and HIF1α are over-expressed in lymph-vascular tumor emboli in ductal breast carcinoma samples (Supplementary Figure 5). Indeed, lymphatic metastatic cells migrate via lymphatic fluids from the tumor mass into the axillary node, which is a compartment most devoid of oxygen supply (Hangai-Hoger et al., 2007). If we consider that basal-like carcinomas present large necrotic/hypoxic areas (Fulford et al., 2006; Livasy et al., 2006) and that SLUG and basal-like gene expression are up-regulated by the hypoxia mimetic Desferroxamine (Storci et al., 2008), it is conceivable that the expression of a stem cell-like gene profile in breast tissues could result from an hypoxic environment with inflammation hijacking the hypoxia-regulated mechanisms that promote the stem cell phenotype.
Most (80–90%) of basal-like tumors carry p53 mutations (Bertheau et al., 2007); our results show that the loss of p53 function up-regulates SLUG expression by unleashing NF-κB/HIF1α activity (Weisz et al., 2007; Hammond et al., 2006). The inability of p53 compromised breast cancer cells to restrain the expression of NF-κB/HIF1α/SLUG axis is particularly relevant in an inflammatory environment. Indeed, we observed that TNFα-exposed p53-deficient MCF7 cells exhibit enhanced survival in comparison to their p53 wild type, parental MCF7 counterparts cells and also display morphologic changes reminiscent of a mesenchymal phenotype (Supplementary Figure 6). These data suggest that p53-inactivating mutations confer an advantage to attain an aggressive stem cell-like phenotype, particularly in a permissive inflammatory environment.
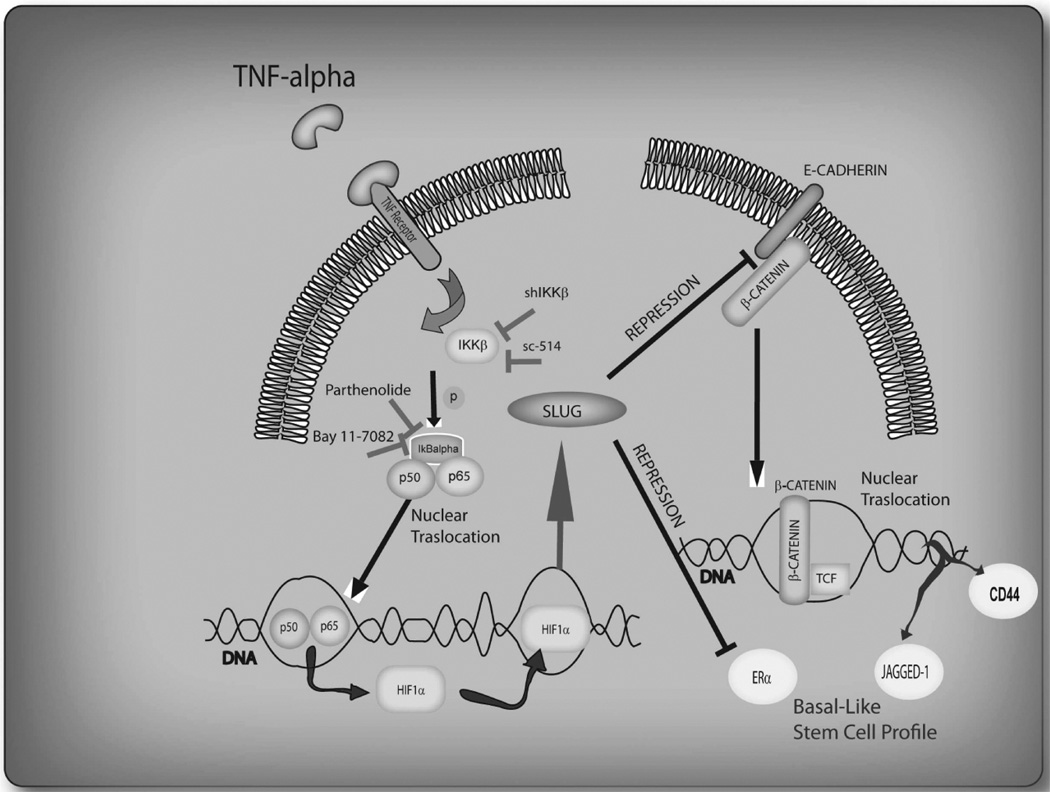
In conclusion, our results provide compelling evidence that the aggressive stem cell-like phenotype of breast cancer cells is induced by inflammatory cytokines that activate SLUG via NF-κB/HIF1α, which is further enhanced by the loss of p53 function (Figure 5).
FIGURE 5.
Inflammatory environment activation of NF-kappaB/HIF1α/SLUG/β-Catenin axis drives the up-regulation of the basal/stem cell-like gene expression profile in breast cancer cell.
p53 loss of function up-regulates the outlined interplay.
Supplementary Material
Acknowledgments
This work has been supported by grant PRIN 2008KTRN38 “Clinical, diagnostic and therapeutic implications of studies on breast cancer stem cells”, RFO funds ex 60%, Cornelia Pallotti and Roberto Pallotti Foundation to M. Bonafè, FIRB project RBNE03KZRJ to P. Chieco and also in part by the MAIN FPVI EU network of excellence (KBM) and USA NIH grant GM066882 (KBM). We also thank Fondazione Cassa di Risparmio in Bologna and Fondazione Banca del Monte e Ravenna for supporting the Center for Applied Biomedical Research.
Footnotes
Conflit of interest: All authors declare no conflict of interest.
References
- Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. Review. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertheau P, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4(3):e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, et al. How different are luminal A and basal breast cancers? Int J Cancer. 2009;124:1338–1348. doi: 10.1002/ijc.24055. [DOI] [PubMed] [Google Scholar]
- Cao Y, et al. IkappaB kinase alpha kinase activity is required for self-renewal of ErbB2/Her2-transformed mammary tumor-initiating cells. Proc Natl Acad Sci U S A. 2007;104:15852–15857. doi: 10.1073/pnas.0706728104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariati M, et al. Alpha-6 integrin is necessary for the tumourigenicity of a stem cell-like subpopulation within the MCF7 breast cancer cell line. Int J Cancer. 2008;122:298–304. doi: 10.1002/ijc.23103. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, et al. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25:2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- Cole SW. Chronic inflammation and breast cancer recurrence. J Clin Oncol. 2009;27:3418–3419. doi: 10.1200/JCO.2009.21.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- Dickson BC, et al. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- Dong R, et al. Role of nuclear factor kappa B and reactive oxygen species in the tumor necrosis factor-alpha-induced epithelial-mesenchymal transition of MCF7 cells. Braz J Med Biol Res. 2007;40:1071–1078. doi: 10.1590/s0100-879x2007000800007. [DOI] [PubMed] [Google Scholar]
- Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson P, Jönsson JI. The hematopoietic stem cell niche: Low in oxygen but a nice place to be. J Cell Physiol. 2010;222:17–22. doi: 10.1002/jcp.21908. [DOI] [PubMed] [Google Scholar]
- Estrach S, et al. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133:4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- Fulford LG, et al. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology. 2006;49:22–34. doi: 10.1111/j.1365-2559.2006.02453.x. [DOI] [PubMed] [Google Scholar]
- Görlach A, Bonello S. The cross-talk between NF-kappaB and HIF-1alpha: further evidence for a significant liaison. Biochem J. 2008;412:17–19. doi: 10.1042/BJ20080920. [DOI] [PubMed] [Google Scholar]
- Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Hammond EM, Giaccia AJ. Hypoxia-inducible factor-1 and p53: friends, acquaintances, or strangers? Clin Cancer Res. 2006;12:5007–5009. doi: 10.1158/1078-0432.CCR-06-0613. Review. [DOI] [PubMed] [Google Scholar]
- Hangai-Hoger N, et al. Terminal lymphatics: the potential "lethal corner" in the distribution of tissue pO2. Lymphat Res Biol. 2007;5:159–168. doi: 10.1089/lrb.2007.5303. [DOI] [PubMed] [Google Scholar]
- Hayes MJ, et al. Genetic changes of Wnt pathway genes are common events in metaplastic carcinomas of the breast. Clin Cancer Res. 2008;14:4038–4044. doi: 10.1158/1078-0432.CCR-07-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehner SP, et al. The antiinflammatory sesquiterpene lactone parthenolide inhibits NF-kappa B by targeting the I kappa B kinase complex. J Immunol. 1999;163:5617–5623. [PubMed] [Google Scholar]
- Honeth G, et al. CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;17:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz KB, et al. Rare steroid receptor-negative basal-like tumorigenic cells in luminal subtype human breast cancer xenografts. Proc Natl Acad Sci U S A. 2008;105:5774–5779. doi: 10.1073/pnas.0706216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE, et al. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T, Kawajiri K. Zinc finger transcription factor Slug is a novel target gene of aryl hydrocarbon receptor. Exp Cell Res. 2006;312:3585–3594. doi: 10.1016/j.yexcr.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Laffin B, et al. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28:1936–1946. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerebours F, et al. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer. 2008;8:41. doi: 10.1186/1471-2407-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- Livasy CA, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, et al. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. Review. [DOI] [PubMed] [Google Scholar]
- McCarthy A, et al. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211:389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
- Moreno-Bueno G, et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res. 2006;66:9543–9556. doi: 10.1158/0008-5472.CAN-06-0479. [DOI] [PubMed] [Google Scholar]
- O'Brien J, Schedin P. Macrophages in breast cancer: do involution macrophages account for the poor prognosis of pregnancy-associated breast cancer? J Mammary Gland Biol Neoplasia. 2009;14:145–157. doi: 10.1007/s10911-009-9118-8. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivotto E, et al. Differential requirements for IKKalpha and IKKbeta in the differentiation of primary human osteoarthritic chondrocytes. Arthritis Rheum. 2008;58:227–239. doi: 10.1002/art.23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat Meth. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- Penzo M, et al. Sustained NF-kappaB activation produces a short-term cell proliferation block in conjunction with repressing effectors of cell cycle progression controlled by E2F or FoxM1. J Cell Physiol. 2009;218:215–227. doi: 10.1002/jcp.21596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, et al. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J Clin Oncol. 2009;27:3437–3444. doi: 10.1200/JCO.2008.18.9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponti D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Pratt MA, et al. The canonical NF-kappaB pathway is required for formation of luminal mammary neoplasias and is activated in the mammary progenitor population. Oncogene. 2009;28:2710–2722. doi: 10.1038/onc.2009.131. [DOI] [PubMed] [Google Scholar]
- Rao VP, et al. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res. 2006;66:7395–7400. doi: 10.1158/0008-5472.CAN-06-0558. [DOI] [PubMed] [Google Scholar]
- Reedijk M, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- Rius J, et al. NF-kappaB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1alpha. Nature. 2008;453:807–811. doi: 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007a;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansone P, et al. The p53 codon 72 proline allele is endowed with enhanced cell-death inducing potential in cancer cells exposed to hypoxia. Br J Cancer 2007. 2007b;96:1302–1308. doi: 10.1038/sj.bjc.6603723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrió D, et al. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- Schwartz DR, et al. Novel candidate targets of beta-Catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63:2913–2922. [PubMed] [Google Scholar]
- Shaulian E, et al. Identification of a minimal transforming domain of p53: negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, et al. CD44/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Simon MC, Keith B. The role of oxygen availability in embryonic development and stem cell function. Nat Rev Mol Cell Biol. 2008;9:285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storci G, et al. The basal-like breast carcinoma phenotype is regulated by SLUG gene expression. J Pathol. 2008;214:25–37. doi: 10.1002/path.2254. [DOI] [PubMed] [Google Scholar]
- Van Laere SJ, et al. NF-kappaB activation in inflammatory breast cancer is associated with oestrogen receptor downregulation, secondary to EGFR and/or ErbB2 overexpression and MAPK hyperactivation. Br J Cancer. 2007;97:659–669. doi: 10.1038/sj.bjc.6603906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren MA, et al. Tumor necrosis factor deficiency inhibits mammary tumorigenesis and a tumor necrosis factor neutralizing antibody decreases mammary tumor growth in neu/erbB2 transgenic mice. Mol Cancer Ther. 2009;8:2655–2663. doi: 10.1158/1535-7163.MCT-09-0358. [DOI] [PubMed] [Google Scholar]
- Weisz L, et al. Mutant p53 enhances nuclear factor kappaB activation by tumor necrosis factor alpha in cancer cells. Cancer Res. 2007;67:2396–2401. doi: 10.1158/0008-5472.CAN-06-2425. [DOI] [PubMed] [Google Scholar]
- Wu Y, et al. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15:416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, et al. The lymphovascular embolus of inflammatory breast cancer expresses a stem cell-like phenotype. Am J Pathol. 2008;173:561–574. doi: 10.2353/ajpath.2008.071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. Inhibition of MAPK and NF-kappa B pathways is necessary for rapid apoptosis in macrophages infected with Yersinia. J Immunol. 2005;174:7939–7949. doi: 10.4049/jimmunol.174.12.7939. [DOI] [PubMed] [Google Scholar]
- Zhou J, et al. NF-kappaB pathway inhibitors preferentially inhibit breast cancer stem-like cells. Breast Cancer Res Treat. 2008;111:419–427. doi: 10.1007/s10549-007-9798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, et al. (2). Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10:R105. doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.