Abstract
CRISPR/Cas, bacterial and archaeal systems of interference with foreign genetic elements such as viruses or plasmids, consist of DNA loci called CRISPR cassettes (a set of variable spacers regularly separated by palindromic repeats) and associated cas genes. When a CRISPR spacer sequence exactly matches a sequence in a viral genome, the cell can become resistant to the virus. The CRISPR/Cas systems function through small RNAs originating from longer CRISPR cassette transcripts. While laboratory strains of Escherichia coli contain a functional CRISPR/Cas system (as judged by appearance of phage resistance at conditions of artificial co-overexpression of Cas genes and a CRISPR cassette engineered to target a λ phage), no natural phage resistance due to CRISPR system function was observed in this best-studied organism and no E. coli CRISPR spacer matches sequences of well-studied E. coli phages. To better understand the apparently “silent” E. coli CRISPR/Cas system, we systematically characterized processed transcripts from CRISPR cassettes. Using an engineered strain with genomically located spacer matching phage λ we show that endogenous levels of CRISPR cassette and cas genes expression allow only weak protection against infection with the phage. However, derepression of the CRISPR/Cas system by disruption of the hns gene leads to high level of protection.
Introduction
CRISPR (clustered regularly interspaced short palindromic repeats) cassettes are present in virtually every archeaon studied and in ~40% of known bacteria (Mojica et al., 2000; Jansen et al., 2002b, Horwath and Barrangou, 2010). A CRISPR cassette consists of almost identical direct repeats of 23–47 bp interspersed with spacers (Jansen et al., 2002a). In every cassette, the length of spacers is similar, while their sequence varies. In some organisms, spacers are enriched with sequences matching sequences of bacteriophages infecting this or closely related organisms (Bolotin et al., 2005; Mojica et al., 2005; Pourcel et al., 2005). CRISPR cassettes are often associated with cas genes (Jansen et al., 2002b; Haft et al., 2005). While there is considerable variation in cas genes, the products of several core cas genes are related to proteins involved in eukaryotic RNAi pathways (Makarova et al., 2006).
On theoretical grounds, it has been proposed that CRISPR/Cas is an adaptive immunity system, which excludes viruses and other mobile genetic elements that contain sequences precisely matching those of CRISPR cassette spacers (Bolotin et al., 2005; Mojica et al, 2005; Pourcel et al., 2005; Makarova et al., 2006). This notion has been confirmed experimentally, and it has been shown that in Streptococcus thermophilus, a match between a single CRISPR spacer and invading phage sequence (called the protospacer) can be sufficient to provide immunity to infection (Barrangou et al., 2007). A perfect spacer-protospacer match is not sufficient for CRISPR mediated immunity, at least is some bacteria. Conserved protospacer-adjacent sequences (PAMs) have been identified bioinformatically and it has been shown that at least in the case of S. thermophlius mutations in PAM prevent CRISPR-mediated immunity even in the presence of matching spacer-protospacer pairs (Deveau et al., 2008).
In at least one eubacterial system, it has been shown that CRISPR/Cas targets DNA rather than RNA of mobile genetic elements, suggesting that the original analogy with RNAi may be misguided (Marraffini and Sontheimer, 2008). On the other hand, in archaea, CRISPR RNAs target invading RNA (Hale et al., 2009). Irrespective of the precise mechanism of CRISPR/Cas function, there is a consensus that transcription of CRISPR cassette followed by processing with the help of Cas proteins is necessary for the defensive action of CRISPR systems (Tang et al., 2002; Lillestøl et al., 2006). Further, it is clear that new CRISPR cassette spacers accumulate mainly on one side of the CRISPR cassette (Pourcel et al., 2005; Horvath et al., 2008; Semenova et al., 2009; Díez-Villaseñor et al., 2010). At least in some experimental systems, newly acquired spacers can provide resistance to phages whose genomes contain protospacers that match exactly the sequence of a spacer (Barrangou et al., 2007; Deveau et al., 2008). Some CRISPR cassettes are very extensive (Semenova et al., 2009). It remains to be determined whether “older” spacers remain functional in phage defense or are vestiges of host-phage interactions from distant past on their way to elimination from the genome. In this work, we studied the expression of CRISPR cassette and cas genes in E. coli, the best-studied microbe. Previous work from Van der Oost laboratory demonstrated that the set of cas genes from Escherichia coli K12 is functional, since when co-overexpressed from a plasmid, the cas genes can provide resistance to phage λ, but only when the cell harbors a plasmid from which an artificial CRISPR cassette containing spacers matching the λ phage genome is transcribed by T7 RNA polymerase (Brouns et al., 2008). No phage protection was observed in the absence of induction of plasmid-borne cas genes co-overexpression, indicating that endogenous levels of chromosomal cas genes expression are not sufficient for phage resistance.
While in many bacteria CRISPR spacers contain a large proportion of sequences matching fully or partially genomic sequences of phages that prey on this particular bacterium, very few sequences originating from known E. coli phages-and no sequences from well-characterized E. coli phages that have been extensively studied in laboratories over the last fifty or more years—are present in E. coli CRISPR cassettes (Díez-Villaseñor et al., 2010). This result may mean that i) the E. coli CRISPR system is not functional, at least in phage defense, possibly due to the lack of CRISPR transcription, since experiments with artificial CRISPR cassette mentioned above indicate that transcription of CRISPR cassette is required for phage resistance to manifest itself and/or ii) a very large proportion (in fact, most) of E. coli phages are not yet known (Semenova et al., 2009).
The levels of processed CRISPR transcripts in E. coli indeed appear to be very low. In fact, processed chromosomal CRISPR transcripts can only be reliably detected when upstream genes casA or casB are deleted (Brouns et al., 2008). Despite this apparently stimulatory effect of casA and casB absence on processed CRISPR transcripts abundance, overexpression of E. coli casA and casB is required for phage resistance. To get a better understanding of CRISPR/Cas function in E. coli, in this work we measured the abundance of processed and unprocessed CRISPR transcripts and determined the reasons for the dramatically increased abundance of processed CRISPR transcripts caused by cas gene disruptions observed by Brouns et al., (2008). We also directly tested the effect of a genomically located functional CRISPR spacer matching a protospacer in phage λ on the ability of the phage to infect an E. coli cell. While E. coli is known to contain two kinds of CRISPR cassettes (cse and csy-subtypes), characterized by different repeat sequences and cas genes sets (see Díez-Villaseñor et al., 2010 for a recent comprehensive survey), we here concentrate on the cse-subtype, which was shown to be functional in phage defense when overexpressed (Brouns et al., 2008).
RESULTS
Detection of processed CRISPR I transcripts
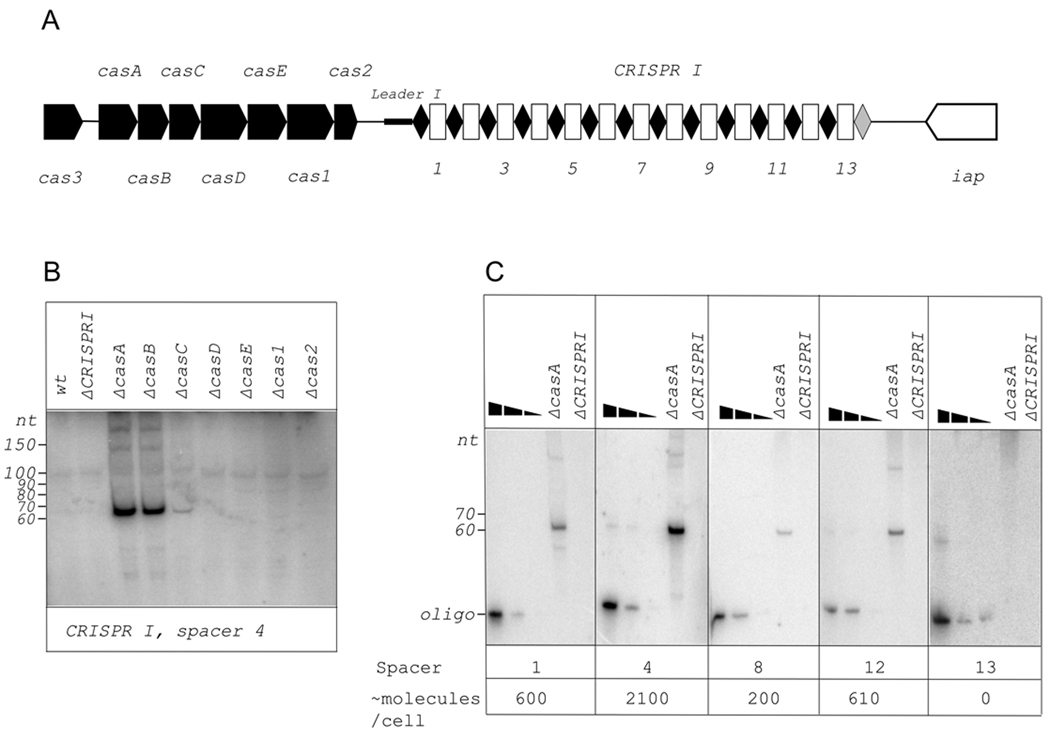
Previously, a processed CRISPR transcript corresponding to one spacer, spacer 4 of the 13-spacer CRISPR I cassette present in the E. coli K12 genome associated with a set of the so-called E. coli type cas genes (Fig. 1A), was detected using Northern blotting (Brouns et al., 2008). The amount of processed transcript in wild-type cells appeared to be very low. The amount of processed transcript was dramatically increased in isogenic cells lacking functional casA, casB, and, to a lesser extent, casC genes (ibid.). We used the same set of E. coli cas genes deletion strains (obtained from the Keio collection, Baba et al., 2006) to determine the abundance of processed transcripts corresponding to each CRISPR I cassette spacer. We confirmed the result of Brouns et al., (2008) with spacer 4 probe (Fig. 1B). We next wanted to determine if there exists a decreasing gradient in abundance of processed CRISPR transcripts related to spacer distance from the leader end. This expectation is a reasonable one since the CRISPR precursor transcript is i) not translated and ii) contains multiple palindromic repeats that may function as transcription terminators. Total RNA was prepared from wild-type and cas mutant cell cultures and used in Northern blotting experiments with oligonucleotide probes specific for each strand of the spacer sequence. An engineered strain lacking the entire CRISPR I cassette (ΔCRISPR1) was used as a negative control. Overall, results identical to those obtained with spacer 4-specific probe (strong hybridization signal from a ~65 base-long RNA in casA and casB mutants; less strong signal in casC strain; no signal in wild-type or other cas mutants) were observed with each probe complementary to CRISPR I RNA transcribed in the direction from cas genes to iap genes (rightward in Fig. 1A). The only exception was spacer 13 probe, with which no hybridization signal was obtained even in casA or casB mutant (data not shown, see also Fig. 1C).
Figure 1. Detection of processed transcripts of E. coli CRISPR I cassette.
A. The E. coli CRISPR I locus is schematically shown. Genes are indicated by arrows and identified. In CRISPR I cassette, repeats are indicated by black rhombuses, spacers – by white rectangles. Numbers below the scheme indicate spacer numbers. The last repeat of the cassette is colored grey, to indicate that its sequence differs from other repeats. The leader sequence is located between the CRISPR I cassette and the cas2 gene and is indicated by a thicker black line.
B. A Northern blot showing changes in abundance of spacer 4 transcript in E. coli strains with indicated cas genes disruptions. The probe was designed to reveal RNA produced by rightward (see Fig. 1A) transcription.
C. Northern blots with total RNA purified from E. coli cells with disrupted casA gene were performed with probes specific for spacers indicated (direction of transcription the same as in Fig. 1B). In each panel, the first three lanes contained increasing amounts of DNA oligonucleotides complementary to probes used for blotting. Approximate calculated numbers of processed CRISPR transcript per cell are shown below.
Most probes complementary to RNA produced by transcription from iap to cas direction (leftward in Fig. 1A) failed to hybridize. However, in several cases, hybridizing RNAs whose apparent sizes ranged from 75 to 150 were revealed (spacer 1, 3, 4, 5, and 7 specific probes, data not shown). However, the intensity of hybridization signal did not depend on the presence or absence of cas genes and was also observed in the ΔCRISPR1 strain (data not shown). We hypothesize that these cas-independent signals originate from non-CRISPR small cellular RNAs that hybridize with some of our probes due to fortuitous complementarity. Therefore, transcription of E. coli CRISPR I cassette appears to proceed unidirectionally from the cas genes side of the cassette.
To estimate the amount of short RNAs hybridizing to various spacers, we compared the intensities of hybridization signals with signals from known amounts of DNA oligos fully complementary to radioactive probes used in Northern blotting. Only RNA from casA mutants was used in this analysis, since transcript level in wild-type cells was too low. Several representative results are shown in Fig. 1C. The overall conclusion is that there is no significant decrease in processed CRISPR I transcripts abundance, at least in casA mutant, as the distance from a CRISPR I promoter located on the cas side of CRISPR cassette (see below) increases from spacer 1 to spacer 12. Based on intensities of hybridization signals, we estimate that ca. 0.3 – 1 ng of processed CRISPR I RNA was present per electrophoretic lane in Fig. 1C. This translates to ~200–2100 copies of individual processed CRISPR I transcripts per cell with disrupted casA. The abundance of processed CRISPR transcripts in wild-type cells is at least 100-fold lower.
Detection of processed CRISPR II and III transcripts
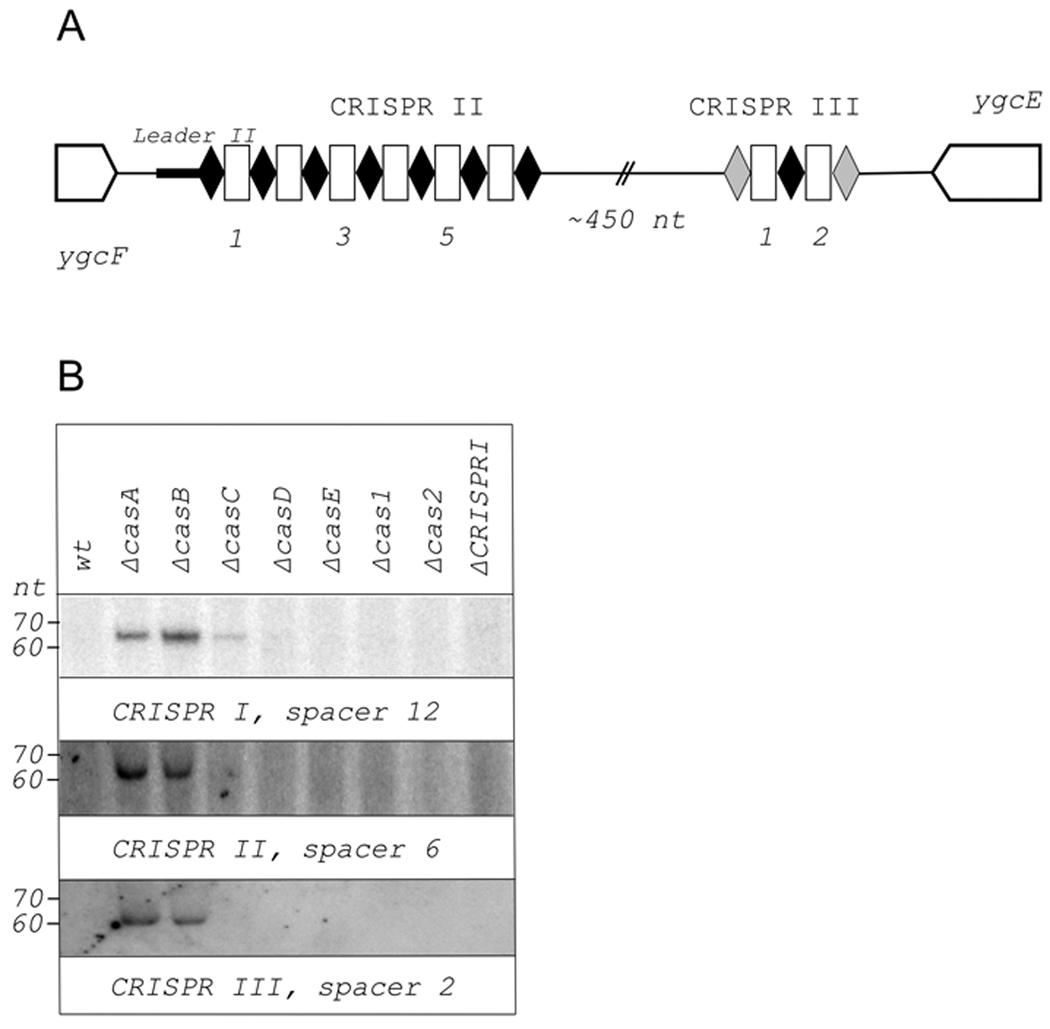
The CRISPR cassette associated with the set of cas genes, which we call CRISPR I, is not the only CRISPR cassette of this type present in E. coli K12. A locus located downstream of the ygcF gene contains an additional cassette with seven repeats identical to CRISPR I cassette repeats and six unique spacers between them (Díez-Villaseñor et al., 2010). This cassette is referred to as CRISPR II (Fig. 2A). Ca. 500 bp downstream of CRISPR II cassette (assuming it is transcribed in the same direction as CRISPR I cassette, above), there is an additional CRISPR repeat. On both sides, this repeat is separated by spacers from degenerated but still recognizable (four and seven mismatches) repeat copies (Díez-Villaseñor et al., 2010). This two-spacer cassette is referred to as CRISPR III (Fig. 2A). The orientation of CRISPR III is the same as that of CRISPR II. The intervening sequence between the two cassettes contains no recognizable ORFs. Neither cassette is associated with recognizable cas genes. The results of Northern blot analysis with CRISPR II and CRISPR III-specific spacer probes are presented in Fig. 2B. Representative results with a CRISPR I specific probe (spacer 12) are also shown for comparison. As can be seen, processed transcripts are generated both from CRISPR II and CRISPR III cassettes. Importantly, the abundance of these transcripts depends on mutations in cas genes in the same way as the abundance of CRISPR I cassette processed transcripts. Since CRISPR II and CRISPR III are not associated with cas genes, it can be safely concluded that the increased abundance of processed CRISPR transcripts observed in some cas mutants is not due to in cis effects of cas gene mutations.
Figure 2. Detection of processed transcripts of E. coli CRISPR II and CRISPR III cassettes.
A. The E. coli CRISPR II/CRISPR III locus is schematically shown. Genes are indicated by arrows and identified. Repeats are indicated by black rhombuses, spacers – by white rectangles. Numbers below the scheme indicate spacer numbers. The first and last repeats of CRISPR III cassette are colored in grey, to indicate that their sequence differs from other repeats. The leader sequence is located between the CRISPR II cassette and the ygcF gene and is indicated by a thicker black line.
B. Northern blots showing changes in abundance of indicated CRISPR I, CRISPR II, and CRISPR III spacer transcripts in E. coli strains with cas genes disruptions. The probes were designed to reveal RNA produced by rightward (see Figs. 1A and 2A) transcription.
As was the case with CRISPR I cassette, CRISPR II and III RNAs originating from transcripts in only one direction, namely from ygcF to ygcE and therefore matching the direction of CRISPR I transcription from cas genes to iap, responded to casA, casB, or casC disruption. In several cases, short RNAs were detected with oligo probes designed to reveal transcripts originating from the opposite strand of CRISPR II cassette (data not shown). However, the abundance of these RNAs did not depend on cas genes and they were deemed non-specific, i. e., arising from loci other than CRISPR.
Identification of processed CRISPR leader transcripts and CRISPR promoters
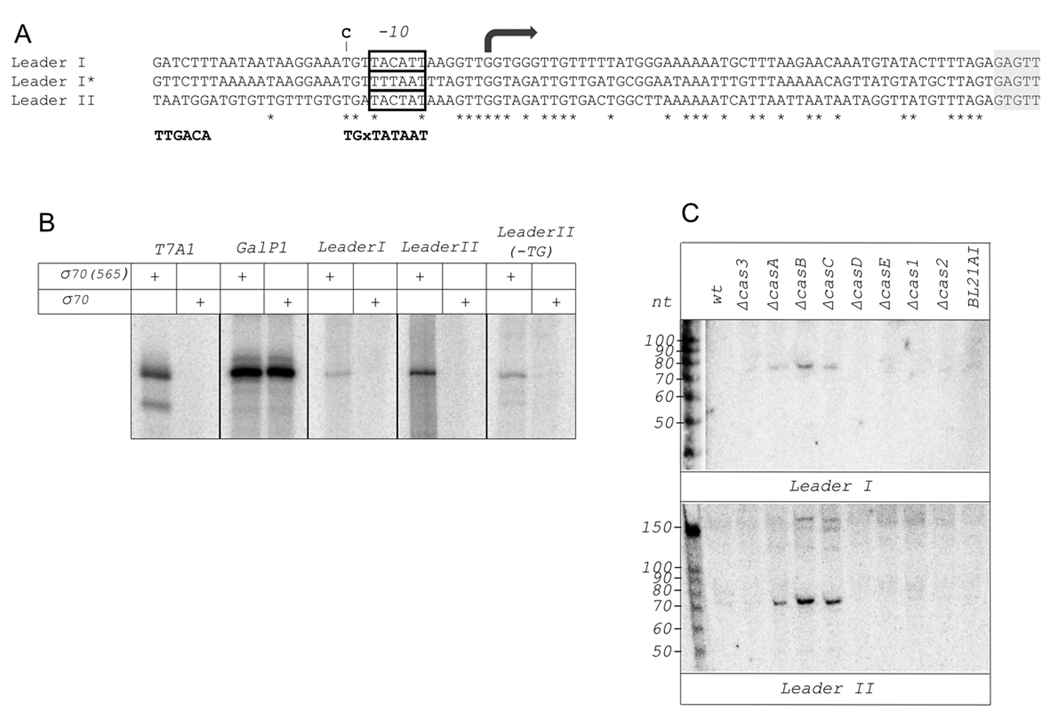
In many bacteria, AT rich, so-called leader sequences are present upstream (with respect to the direction of transcription) of CRISPR cassettes (Jansen et al., 2002b). Comparison of E. coli K12 CRISPR I and CRISPR II leader sequences (leader I and leader II, respectively) reveals extensive regions of sequence identities up to position −69 with respect to the beginning of the first repeat in each cassette (Fig. 3A). A variant leader I found in some E. coli isolates (KP, A. Kaznadzej, KS, unpublished observations) is also included in the alignment of Fig. 3A. As can be seen, all three sequences share identical regions that may control CRISPR cassette transcription and/or processing. The sequence upstream of CRISPR III cassette has no similarities to CRISPR I/CRISPR II leader sequences (data not shown) and is therefore not included in the alignment. The most upstream region of sequence conservation in leader sequences is similar to the extended −10 promoter element consensus sequence (TGxTATAAT). This region was recently shown by Pul et al., (2010) to function as a σ70 RNA polymerase promoter in both leader I and leader II. Using 5’-RACE we identified 5’ ends of leader transcripts that matched the start sites of CRISPR leader promoters revealed by Pul et al., 2010 (shown by an arrow in Fig. 3A).
Figure 3. Detection of processed CRISPR leader transcripts and CRISPR promoters.
A. An alignment of CRISPR I and CRISPR II leader sequences is presented. The sequence of a variant CRISPR I leader present in some E. coli strains (Leader I*) is also included in the alignment. Identities are indicated by asterisks below the alignment. The alignment area at far right with grey background corresponds to the beginning of the first CRISPR repeat. A rightward arrow above the alignment highlights a nucleotide corresponding to leader I and leader II transcripts 5’ ends determined by 5’-RACE. An upstream conserved region with similarity to −10 promoter consensus element is boxed. The consensus sequences of extended −10 promoter element is shown below the alignment. The consensus sequence of the −35 promoter element (TTGACA) is also shown at appropriate distance from the transcription start site. A T to C substitution in the extended −10 motif of the CRISPR II leader promoter obtained in this work is shown above the alignment.
B. The results of in vitro transcription with σ70 RNAP holoenzyme, or holoenzyme reconstituted with σ70(1–565), a mutant lacking region 4.2, from DNA fragments containing indicated promoters is shown. T7A1 and galP1 are included as examples of strong −10/−35 and extended −10 class promoters, respectively.
C. Northern blots showing changes in abundance of leader I and leader II transcripts in E. coli strains with cas genes disruptions. The probes were designed to reveal RNA produced by rightward (see Figs. 1A and 2A) transcription.
Neither CRISPR promoter contains sequences similar to the −35 promoter consensus element sequence. DNA fragments containing both leader sequences were combined with σ70 RNA polymerase and a steady-state in vitro transcription assay was performed. In both cases, transcripts of expected lengths were detected (data not shown, see also Fig. 3B). Primer extension experiments conducted with in vitro transcribed RNA proved that the start point corresponded to 5’ ends of leader RNA determined in vivo (data not shown). In vitro transcription experiments with RNA polymerase holoenzyme reconstituted with σ70(1–565), a σ70 mutant that lacks conserved region 4.2 required for recognition of the −35 promoter element and indispensable for promoter complex formation on −10/−35 class promoters (Minakhin and Severinov, 2003), demonstrated that neither CRISPR promoter was recognized by the mutant RNA polymerase (Fig. 3B). A control extended −10 promoter, galP1, behaved in the expected way and was active even with the σ70(1–565) holoenzyme. Nevertheless, analysis of a leader II fragment containing a substitution in the extended −10 promoter TG motif revealed that the substitution strongly decreased promoter strength. Thus, our data show that both CRISPR I and II cassettes are transcribed from weak extended −10 class σ70 promoters located in leader regions. Both promoters appear to be weak, with CRISPR II promoter being the stronger of the two.
Northern blotting experiments with oligonucleotide probes specific for leader I and leader II were performed. In both cases, a short RNA of ca. 80 bases, i.e., longer than internal processed CRISPR transcripts, was detected in casA, casB, and casC mutants (Fig. 3C). CRISPR leader promoters are located at an appropriate distance from the first CRISPR repeat in both cassettes to account for ~80 base long transcripts revealed by leader-specific probes in Northern experiments. Thus, transcripts revealed by CRISPR I and CRISPR II leader-specific probes must correspond to 5’ end proximal fragments of CRISPR transcripts.
No signal corresponding to RNA originating from the region in-between CRISPR II and III cassette could be detected by Northern blotting (data not shown). However, reverse-transcription PCR analysis with primer pairs annealing to one of the CRISPR II spacers and to a CRISPR III spacer revealed an amplicon whose size suggested that CRISPR III cassette may be transcribed together with CRISPR II (data not shown). Our inability to detect the combined transcript by Northern blot analysis is likely explained by different sensitivities of the two methods, RT-PCR being a more sensitive one.
Increased expression of casE is the reason for increased processed CRISPR transcript abundance in casA, casB, and casC mutants
We next investigated the reasons for a peculiar increase in the abundance of processed CRISPR transcripts in some cas mutants. At this juncture it is important to note that cas mutant strains used by us and earlier by Brouns et al., came from the Keio collection of strains (Baba et al., 2006) and contained a kanamycin resistance gene in place of the deleted gene. The kanamycin resistance gene is fitted with its own promoter and a downstream transcription terminator. In all cas mutants, the direction of transcription of the kanamycin resistance gene matches the direction of cas genes transcription. Starting with the original set of Keio deletions, we created a set of “clean” cas gene deletions with the kanamycin resistance gene removed. Total RNA was prepared from these strains and the amount of processed CRISPR transcripts was determined. A representative result is shown in Fig. 4A. As can be seen, there was no increase in the abundance of processed CRISPR transcripts in any of the clean deletions strains. The result thus indicates that the observed increases in processed CRISPR transcripts abundance in original casA, casB, and casC gene disruptions strains were not due to deletions of cas genes but were instead caused by the presence of the kanamycin resistance cassette, which likely caused increased expression of downstream cas genes (the effects of read-through transcription into the CRISPR I cassette can be excluded based on the fact that transcripts from unlinked CRISPR II and CRISPR III cassettes respond to replacement of casA, casB, or casC with kanamycin resistance cassette the same way as CRISPR I, above). We performed semi-quantitative reverse transcription PCR analysis of cas genes transcripts levels in several cas mutants with and without the kanamycin resistance cassette. The results are shown in Fig. 4B. A control RT-PCR analysis with cas3 specific primers showed that transcript levels of this gene, the most upstream gene in the cas cluster, were not affected by casA, casB, or casC deletions, whether with or without the kanamycin resistance cassette. The level of casC transcript was strongly increased in casA and casB mutants harboring the kanamycin resistance cassette. No such effect was observed in “clean” casA and casB deletions. Weaker, but still significant stimulation of expression of more downstream genes (casD, casE, cas2, and cas1) due to the presence of the kanamycin resistance cassette instead of casA or casB was also observed. Introduction of the kanamycin resistance gene instead of casC had a similar effect on downstream (casD, casE, cas2, and cas1) genes expression. The result thus establishes that substantial read-through transcription from the kanamycin resistance cassette in previously studied cas mutants occurs.
Figure 4. Effect of cas genes disruption, deletions and overproduction of processed CRISPR transcripts abundance.
A. A Northern blot showing abundance of spacer 4 transcript in E. coli strains with indicated cas genes “clean” deletions lacking the kanamycin-resistance gene present in original strains used in experiments shown in Figs. 1–3. RNA from one such strain, labeled ΔcasA_KanR, was loaded on the second lane. The probe was designed to reveal RNA produced by rightward (see Fig. 1A) transcription.
B. Changes in abundance of cas gene transcripts in cas disruption strains from the Keio collection were revealed by semi-quantitative RT-PCR as described in Experimental Procedures. In each vertical raw, the amounts of transcripts corresponding to genes indicated in the left are shown. The gyrB transcript was chosen as a control, whose level was not expected to depend on cas genes. Lane 10 shows results obtained with oligonucleotide pairs used for RT-PCR in lanes 1–9 using genomic DNA as a template (the reverse transcription step was omitted in this case)
C. A Northern blot showing abundance of spacer 4 transcripts in E. coli strains overexpressing indicated cas genes from ASKA library plasmids.
The results presented above are consistent with an idea that increased expression of cas genes located downstream of casB and, possibly, casC may be responsible for the increased abundance of processed CRISPR transcripts. To check this notion directly, we used cas overexpression plasmids from the ASKA library (Kitagawa et al., 2006). Plasmids from this collection contain individual E. coli genes under control of strong T5-lac promoter. We introduced individual ASKA cas expression plasmids into wild-type E. coli cells and determined the levels or processed CRISPR transcripts in these cells. Control experiments demonstrated that all Cas proteins were induced to approximately the same level (data not shown). The results, presented in Fig. 4C, showed that expression of casE was sufficient to lead to a significant increase in the abundance of processed CRISPR I transcripts. The effect was also observed for processed transcripts from CRISPR II and CRISPR III cassettes (data not shown). Overproduction of other Cas proteins had no effect on processed CRISPR transcripts abundance.
CasE directs specific processing of primary CRISPR transcripts
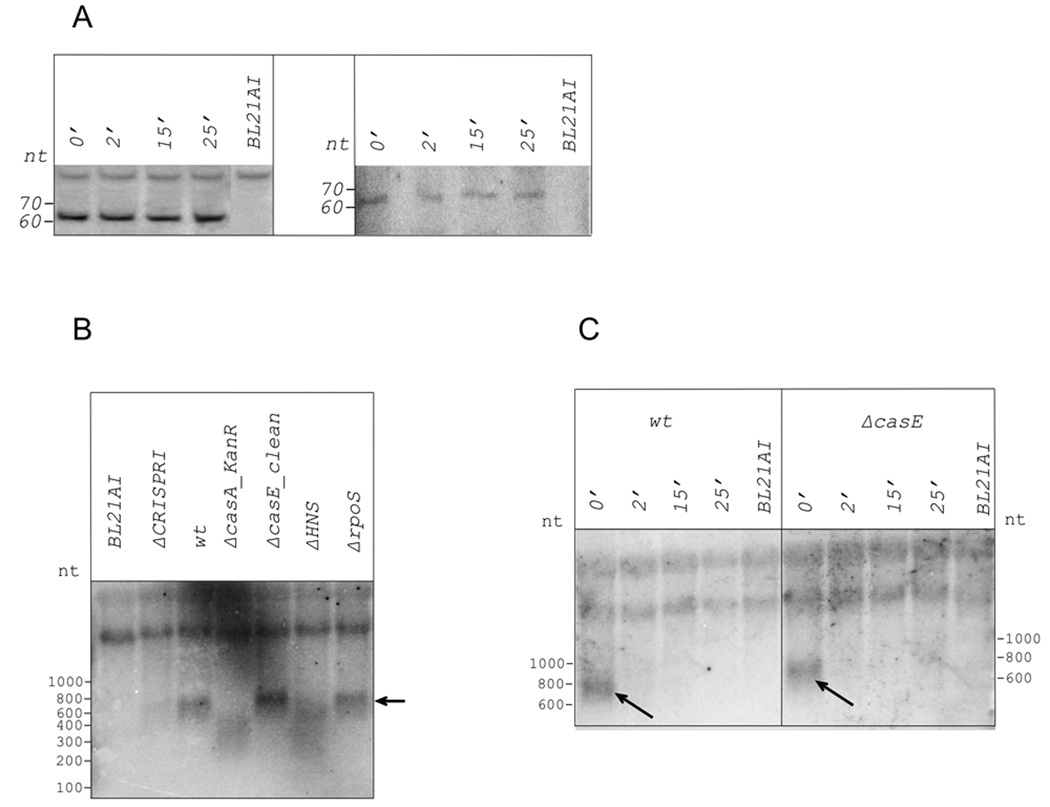
The observed increase in processed CRISPR transcripts abundance in the presence of increased amounts of CasE could be due to i) increased stability of processed CRISPR transcripts; ii) increased processing rate of unprocessed CRISPR transcripts; iii) increased abundance of unprocessed CRISPR transcripts due to increased activity of CRISPR promoters. Combinations of these possibilities are also possible. Transcription inhibition by rifampicin (Fig. 5A, left panel) indicated that processed CRISPR I transcripts are stable in cells with disrupted casA (and overexpressing casE) gene. However, similar analysis conducted with wild-type cells revealed that processed CRISPR transcripts were also stable (Fig. 5A, right panel; much longer exposures were required in this case to reveal CRISPR transcripts whose abundance was much lower). Thus, increased stability of processed CRISPR transcripts is an unlikely reason for CasE-dependent increase in processed transcripts abundance.
Figure 5. Determination of CRISPR transcripts half-lives.
A. Rifampicin was added to wild-type and ΔcasA_KanR E. coli cultures. Cells continued to grow for indicated times, after which total RNA was prepared and amount of processed spacer 4 CRISPR I transcript analyzed by Northern blotting. BL21AI is a control, these cells do not have CRISPR cassettes.
B. A Northern blot showing detection of full-sized CRISPR transcripts in RNA prepared from indicated cells. A repeat-specific probe was used for hybridization. Full-sized CRISPR transcript is indicated by an arrow.
C. Cultures of cells indicated were treated as in A and were analyzed as in B.
In the absence of pronounced differences in processed CRISPR transcripts stability, increased transcription and/or processing rate of primary CRISPR transcripts remain as the only explanations of observed dramatic difference in processed CRISPR transcript abundance caused by overproduction of CasE. The steady-state levels of primary CRISPR transcripts were determined by Northern blotting at conditions designed to reveal large (i.e., unprocessed) transcripts. Since probing with spacer-specific oligos did not reveal any full-sized transcript, probing with repeat-specific oligo was performed. The results revealed the presence of a ~800 nt band (Fig. 5B, lane 3). The intensity of this band strongly decreased in RNA prepared from ΔCRISPR I strain (Fig. 5B, lane 2). No band at all was detected in RNA prepared from BL21AI E. coli cells that lack both cassettes (Jeong et al. 2009, Fig. 5B, lane 1). We therefore propose that the residual full-sized CRISPR RNA detected in RNA from ΔCRISPR I cells is due to CRISPR II transcript. While a combined CRISPR II-CRISPR III transcript detectable by RT-PCR should be larger (~1100 nt) than the CRISPR I (~800 nt) transcript, we hypothesize that endonuclease cleavage of the combined CRISPR II–III transcript in the insert separating the two cassettes may be responsible for ~800 nt transcript that we observe. The absence of the primary CRISPR II-CRISPR III transcript detected by RT-PCR in Northern blotting experiments could be caused by differential sensitivities of the two methods (RT-PCR being a more sensitive method) or could be a trivial consequence of poor transfer of the longer transcript during blotting. The results seem to suggest that steady-state levels of the partially processed CRISPR II transcript are lower than those of CRISPR I transcript, despite the apparently higher strength of the leader II promoter. This may be an indication that the CRISPR II transcript is processed and/or degraded more rapidly than the CRISPR I transcript.
The band corresponding to unprocessed CRISPR transcript was absent in cells containing a kanamycin resistance cassette instead of the casA gene or in cells carrying an ASKA plasmid expressing casE (Fig. 5B, lane 4). Instead, a smear corresponding to smaller-sized transcripts was observed. In the “clean” casE deletion, the amount of unprocessed CRISPR I transcript was the same as in or slightly higher than in wild-type cells (Fig. 5B, lane 5). Thus, increased processed CRISPR transcript abundance caused by overproduction of CasE correlates with increased processing rate of full-sized transcript. Analysis of RNA purified from cells lacking rpoS, a gene encoding RNAP σS subunit, revealed no change in unprocessed CRISPR transcript levels (Fig. 5B, lane 7). Thus, σS does not appear to be involved in CRISPR cassette transcription.
Pul et al., (2010) reported that cas gene promoters are strongly inhibited by the histone-like protein H-NS. A much weaker effect with CRISPR leader promoters was also observed. Hommais et al., (2001) also reported that the abundance of RNA coding for individual Cas proteins is increased in the mutant strain lacking the hns gene. Cells harboring a deletion of the hns gene did not contain full-sized CRISPR transcript (Fig. 5B, lane 6) and at least in this respect were indistinguishable from cells overproducing CasE. Thus, deletion of hns decreases the abundance of full-sized CRISPR transcript (and increases abundance of processed transcripts, see below) most likely by increasing the intracellular concentration of CasE.
A rifampicin challenge experiment (Fig. 5C) revealed that full-sized CRISPR transcripts are short-lived (disappear after a 2-min incubation with rifampicin) even in the absence of CasE. Together, these results indicate that the strongly increased abundance of processed CRISPR transcripts in cells overexpressing the casE gene is caused by accumulation of stable processed CRISPR transcripts from unstable unprocessed transcript that is normally degraded by an as yet unidentified nuclease. Increased amounts of CasE lead to processing of the unstable primary CRISPR transcript (as opposed to degradation that does not lead to accumulation of specific short CRISPR transcripts) resulting in accumulation of stable processed CRISPR RNAs.
Endogenous levels of CRISPR I processed transcript and cas genes are sufficient to provide partial phage resistance
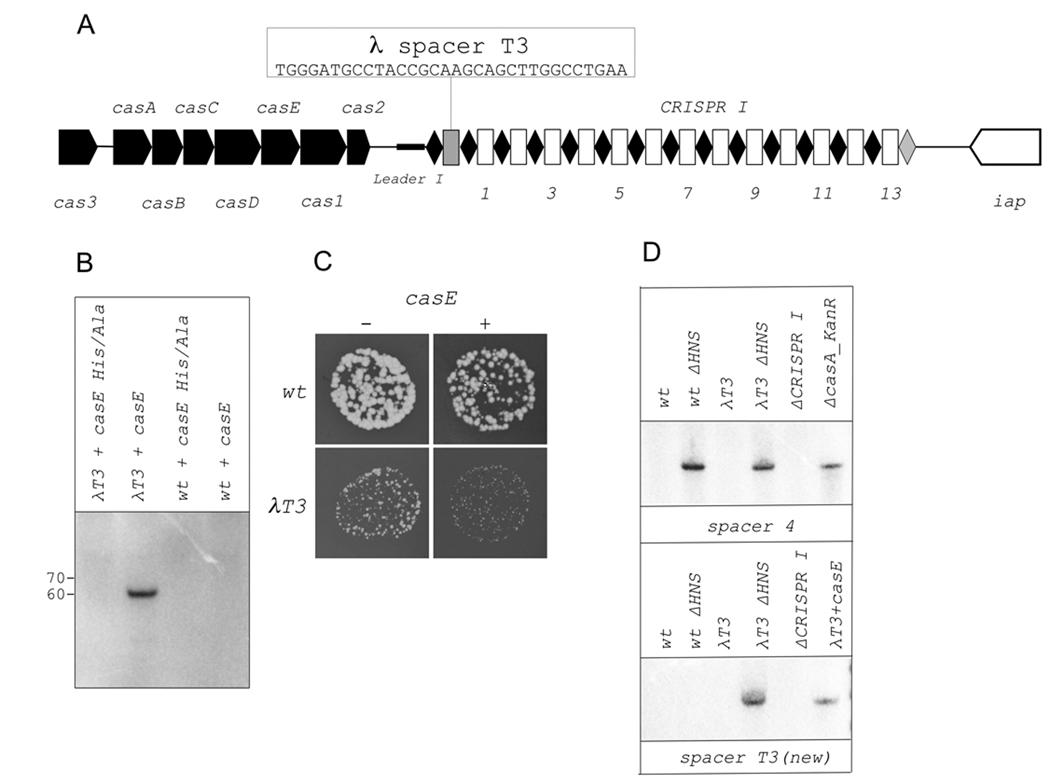
Previously, Brouns et al. (2008) showed that a CRISPR plasmid carrying four spacers matching phage λ genome provides high-level resistance to λ infection in a plaque assay. The artificial plasmid-borne cassette was transcribed by T7 RNAP from a promoter located upstream of the CRISPR I leader. For resistance to be manifested, the entire complement of cas genes products had to be overexpressed from a compatible plasmid. Despite numerous attempts, we were unable to isolate E. coli mutants resistant to λ or several other phages due to the expansion of CRISPR cassettes (data not shown). Attempts by other laboratories were similarly unsuccessful (Díez-Villaseñor et al., 2010). Low levels of processed CRISPR transcript abundance could be responsible for the lack of known cases of natural E. coli phage resistance due to CRISPR function. To directly test if endogenous levels of CRISPR/Cas expression are sufficient for phage resistance, we engineered an E. coli strain λT3 containing a new spacer in CRISPR I cassette. The new spacer is identical to one of the spacers (T3) used by Brouns et al., (2008) and was located upstream of the first spacer of the CRISPR I cassette (Fig. 6A). Northern blot analysis confirmed that the new spacer was processed in the expected way in the presence of CasE overproducing plasmid (Fig. 6B, lane 2) but not when CasEH20A, a CasE mutant with a substitution in the catalytic site that abolished preCRISPR-transcript processing in vitro (Brouns et al., 2008) was overproduced (Fig. 6B, lane 1). The engineered E. coli strain, as well as control wild-type E. coli strain, were infected with λ and plaque formation was monitored. The results, shown in Fig. 6C, show that while the number of phage plaques on λT3 and wild-type strains was the same, the size of plaques formed on engineered cell lawns was reproducibly smaller than on wild-type cells. The effect was stronger when CasE was overproduced (Fig. 6C). Thus, a CRISPR I spacer matching a λ protospacer negatively affects λ development at endogenous levels of CRISPR I cassette and cas genes expression.
Figure 6. Function of genomic E. coli CRISPR in phage protection.
A. The structure of engineered CRISPR cassette in λT3 (BW39651) strain is shown (see Fig. 1A for details).
B. Northern blot analysis of processed transcripts using spacer T3-specific probe in RNA prepared from wild-type or λT3 strains carrying indicated casE expression plasmids.
C. The size or plaques formed by bacteriophage λ on indicated cell lawns. Identical dilutions of phage λ were deposited on lawns of indicated cells. Results of overnight growth at 30 °C are presented. The colors of plate photographs were inverted to make phage plaques better visible. The plaques appear white, while cell lawns appear black.
D. Northern blot analysis of processed transcripts using spacer 4 (top) and spacer T3 (bottom) specific probes in RNA prepared from wild-type or λT3 strains with or without the hns gene. ΔCRISPRI and casA_KanR are controls (see Fig. 1 and Fig. 4 legends).
E. Plaque formation by bacteriophage λ on indicated cell lawns. See panel C legend for details.
We next created a Δhns derivative of λT3 strain. In the new strain, the abundance of CRISPR I spacer 4 processed transcripts was increased to the same level as in cells overproducing CasE (Fig. 6D, top) or in wild-type cells carrying an hns deletion, a result consistent with data of Pul et al., (2010) and our results shown in Fig. 5B. In λT3 strain lacking hns, the abundance of T3 spacer processed transcripts was increased in the absence of cas overexpression (Fig. 6D, bottom). In phage infection assay, no λ plaques were observed on lawns of λT3 strain lacking hns, while normal size plaque were formed on hns mutant with wild-type CRISPR I cassette (Fig. 6E). We therefore conclude that a matching genomic CRISPR spacer can provide effective protection from phage infection, but only in the absence of hns. Despite the apparent functionality of the genomic spacer in phage protection (in the context of the hns deletion) we were unable to recover λ-resistant mutants from our Δhns strain using a protocol that worked well for S. thermophilus selections (data not shown). The reason(s) for this failure remain unknown but the result may indicate that an important aspect of the adaptation part of the CRISPR pathway (which involves the acquisition of new spacers) is somehow compromised in E. coli.
DISCUSSION
In this work, we systematically studied the abundance of processed cse-subtype CRISPR cassette transcripts in E. coli. Our results indicate that there is no significant decrease in abundance of processed CRISPR transcripts as the distance from the leader located between the cas genes cluster and the CRISPR cassette is increased. Thus, whatever the biological function of CRISPR locus of E. coli is, “older” spacers should be as functional as new ones, at least in E. coli. It is possible however that a gradient in abundance of processed CRISPR transcripts exists in organisms with long cassettes.
Since CRISPR transcripts are not translated, it is likely that special antitermination mechanisms exist to ensure efficient transcription of CRISPR RNA. In the case of rDNA transcription, a poorly characterized antitermination system that relies on an upstream rut element ensures efficient production of full-sized transcripts (Arnvig et al., 2008). No sequences matching the rut sequence are present in CRISPR transcripts. However, conserved elements present in CRISPR I and CRISPR II leader sequences between the transcription start point and beginning of the first repeat may act in cis to ensure efficient transcription of the CRISPR cassette. Further experimentation in this direction seems to be warranted.
The apparent absence of processed CRISPR I spacer 13 transcript could be due to the fact that spacer 13 is followed by the most degenerate and probably the “oldest” repeat of CRISPR I cassette. The sequence of this repeat differs from other CRISPR repeat sequences. It is possible that the distinct sequence of the oldest repeat that is shared by all E. coli strains carrying CRISPR I cassette i) ensures transcription termination to prevent production of antisense transcripts of the iap gene located immediately downstream and ii) destabilizes the processed spacer 13 transcript that must arise upon the upstream processing event that generates processed spacer 12 transcript. Again, further experimentation will be needed to understand the events at this side of the cassette.
We identified promoters responsible for E. coli CRISPR transcription. Both CRISPR I and CRISPR II promoters are quite weak. Absence of sequence conservation upstream of extended −10 elements of CRISPR I and CRISPR II promoters argues against the presence of additional DNA binding factors that ensure co-regulation of transcription of the two cassettes. Both CRISPR promoters contain a TG motif characteristic of promoters of the extended −10 class. The TG motif clearly contributes to CRISPR promoter strength, as fortuitous down mutation in the TG motif of CRISPR II promoter demonstrates. Yet, neither CRISPR promoter can function in the absence of σ70 conserved region 4.2 that is responsible for the recognition of the −35 promoter consensus element. Neither promoter contains sequences similar to the −35 consensus element, indicating that σ70 conserved region 4.2 makes favorable but non-specific interactions with CRISPR promoter DNA. While our work was in progress, Pul et al., (2010) reported the mapping of E. coli CRISPR promoters that matches our results. These authors went on to show that the activity of CRISPR promoters is negatively affected by H-NS, a histone-like architectural protein that appears to be employed by the cell to keep CRISPR transcription low.
The previous observation of Brouns et al., (2008) on the very strong increase in the abundance of processed CRISPR transcripts upon disruption of casA, casB, and, to a lesser extent, casE, was difficult to rationalize. Here, we show this effect to be an artifact caused by read though transcription from the kanamycin resistance cassette that was used to disrupt cas genes, which leads to the increased synthesis of CasE. The result shows that caution should be exercised when interpreting phenotypes of Keio collection strains harboring disruptions of genes, which are part of operons. CasE is a nuclease that is responsible for CRISPR RNA processing (Brouns et al., 2008). Once processed, CRISPR transcripts are very stable, whether increased amounts of CasE are present or not. Disruption of casC with a kanamycin resistance cassette has a similar stimulatory effect on casE transcript abundance as disruption of casA or casB. Yet, disruption of casC leads to much smaller increase in processed CRISPR transcript abundance. This effect suggests that CasC needs to be present for efficient CasE-dependent processing of the full-sized CRISPR transcript to occur. The strong increase of processed CRISPR transcripts abundance mentioned by Pul et al., (2010) and confirmed by us is likely a consequence of increased casE expression, since transcription of cas operon is negatively regulated by H-NS (Pul et al., 2010).
Since overexpression of CasE alone is sufficient for complete processing of the CRISPR transcript (and, conversely, virtually no processing occurs at basal levels of casE expression), it follows that other cas gene products are not limiting the processing rate at their physiological concentrations. A very strong increase in abundance of processed CRISPR transcripts upon CasE overexpression occurs despite very low steady-state levels of full-sized CRISPR transcripts observed in wild-type cells. We provide experimental data that explain this dramatic accumulation by differential stabilities of processed and unprocessed CRISPR transcripts. The short half-life of unprocessed CRISPR transcripts is determined by an as yet unidentified RNase, which may be an important negative regulator of CRISPR system function.
The principal result of our work is the demonstration that E. coli CRISPR system can function in phage defense. Using an engineered strain with expanded CRISPR cassette containing a λ-specific spacer, we show that development of phage λ is inhibited in the context of wild-type cells and prevented in the context of an hns mutant. The result provides an answer to a perplexing observation that CRISPR system was never recovered as a functional phage-resistance mechanism despite years of research by some of the giants of early molecular biology (Young, 2008). It now appears that E. coli CRISPR system, while clearly active based on the enormous variability of the locus (Díez-Villaseñor et al., 2010 and our unpublished observations), is kept inactive at laboratory conditions by (at least) H-NS and an unidentified RNase that degrades full-sized CRISPR transcripts. Determination of physiological conditions that activate the E. coli CRISPR system remain the subject of ongoing experiments in our laboratory.
Proto-spacer adjacent motifs (PAMs) within target sequences constitute a cornerstone component of at least some of CRISPR/Cas immune systems (Deveau et al., 2008), allowing a self vs. non-self discrimination of target DNA molecules. Mutations in PAM have been shown to prevent CRISPR mediated immunity even at conditions of a perfect match between the spacer and protospacer sequences (Deveau et al., 2008). The absence of PAM within the CRISPR locus (and the presence of repeats) explains why the CRISPR locus itself is not recognized by the cognate small CRISPR RNAs that are generated inside the cell (Marraffini and Sontheimer, 2010). Analysis of matches between known E. coli spacers and E. coli phage and, most notably, plasmid sequences allowed Mojica et al. (2009) to identify a likely PAM consensus AWG. On the other hand, Brouns et al. (2008) had shown, that at least at conditions of artificial CRISPR cassette and cas genes co-overexpression, engineered CRISPR spacers provide efficient protection from phage λ infection even in the absence of PAM-like sequences adjacent to protospacer. In particular, the protospacer matching their T3 spacer that was also used in our work has a TGG sequence instead of PAM, with only one position (a G immediately adjacent to the protospacer) matching the proposed PAM consensus. While the relaxed requirement for PAM in the Brouns et al. experimental system could have been caused by co-overexpression of CRISPR/Cas components, in our case efficient interference with phage λ infection is achieved even with genomically located T3 spacer. The result thus may suggest that the stringency of requirements for PAM vary in bacteria (less stringent in E. coli, more stringent in S. thermophilus) or, alternatively, that elements within the spacer itself can override the requirement for PAM. These questions are currently being investigated in our laboratory.
EXPERIMENTAL PROCEDURES
Bacteria strains, media, and primers
Escherichia coli strains used in this work are described in Table 1. The BW39651 strain containing bacteriophage λ specific spacer T3 in CRISPR I cassette was made by using a simple two-step mutagenesis procedure that is based on use of the Red recombinase (Datsenko and Wanner, 2006). In the first step, a PCR product encoding a toxin gene under tight control by rhaBp (Datsenko and Wanner, 2006) was recombined onto the chromosome. In the second step, the toxin gene cassette was replaced with dsDNA fragment by selecting rhamnose-resistant transformants. Details of this two-step protocol will be described elsewhere (Datsenko and Wanner, manuscript in preparation). Cells were grown in LB medium (1% bactotryptone, 1% NaCl, 0.5% yeast extract, with or without 1.5% bactoagar) supplemented with appropriate antibiotics.
Table 1.
E. coli strains used in this work.
| HB101 | F-, hsdS20(rB-, mB-), xyl5, l-, recA13, galK2, ara14, supE44, lacY1, rpsL20(strr), leuB6, mtl-1, thi-1 |
(AllianceBio) |
| BW25113 |
rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1 |
(6) |
| BW38985 | like BW25113, except ΔrpoS::kan | (2) |
| BW39121 | like BW25113, except Δhns::kan | (2) |
| BW39182 | like BW25113, except ΔygbF::kan | (2) |
| BW39183 | like BW25113, except ΔygbT::kan | (2) |
| BW39184 | like BW25113, except ΔygcH::kan | (2) |
| BW39185 | like BW25113, except ΔygcI::kan | (2) |
| BW39186 | like BW25113, except ΔygcJ::kan | (2) |
| BW39187 | like BW25113, except ΔygcK::kan | (2) |
| BW39188 | like BW25113, except ΔygcL::kan | (2) |
| BW39189 | like BW25113, except ΔygcB::kan | (2) |
| BW39292 | like BW25113, except ΔCRISPR-1::kan | This study |
| BW39371 | like BW25113, except ΔygbF | BW39182 KanS with pCP20a |
| BW39372 | like BW25113, except ΔygbT | BW39183 KanS with pCP20 |
| BW39373 | like BW25113, except ΔygcH | BW39184 KanS with pCP20 |
| BW39374 | like BW25113, except ΔygcI | BW39185 KanS with pCP20 |
| BW39375 | like BW25113, except ΔygcJ | BW39186 KanS with pCP20 |
| BW39376 | like BW25113, except ΔygcK | BW39187 KanS with pCP20 |
| BW39377 | like BW25113, except ΔygcL | BW39188 KanS with pCP20 |
| BW39378 | like BW25113, except ΔygcB | BW39189 KanS with pCP20 |
| BW39651 | like BW25113, but with T3 spacer | This study |
| BW39671 | like BW39651, except Δhns::kan | This study |
Elimination of antibiotic resistance gene was performed as described elsewhere (6)
Escherichia coli HB101 (AllianceBio) and McConkey agar base plates containing 1% galactose were used as host strains in experiments to study pLeadI and pLeadII promoter activities in vivo.
Primers used in this work can be found in Table S1.
5’ RACE
5 mg DNase I-treated total RNA from BW28357 E. coli strain (wt) was combined with 5 U Tobacco Acid Pyrophosphatase (TAP) (Epicentre) in a 25 µl reaction volume according to manufacturer’s instructions with the addition of 20 U RNaseOUT (Invitrogen). RNA was phenol/chloroform extracted, ethanol precipitated, and dried. The pellet was resuspended in a 50 µl ligation buffer containing 150 ng of RNA oligonucleotide and 120 U T4 RNA Ligase I (New England Biolabs) according to manufacturer’s instructions. Following phenol/chloroform extraction and ethanol precipitation with 50 pmoles of cDNA primer, the RNA/primer pellet was resuspended in 10 µl water, heated at 70 °C for 10 min, and immediately transferred on ice for 5 min. The cDNA synthesis was carried out with 100 U of Super Script III from the First-Strand Synthesis System for RT-PCR (Invitrogen) according to manufacturer’s instructions for 50 min at 50 °C. cDNA was then purified by the Qiaquick PCR cleanup method (Qiagen), and PCR-amplified with Taq DNA polymerase (New England Biolabs) for 40 cycles (95 °C 35 sec, 49 °C 45 sec, 72 °C 45 sec). PCR reactions were visualized on 2% agarose gels; bands were excised from the gel and cloned into pT7Blue cloning vector from the pT7Blue Perfectly Blunt Cloning Kit (Novagen). Inserts were then sequenced using standard T7-promoter oligonucleotide. The 5’ ends of transcripts were determined by the position of the sequence junction between the RNA oligonucleotide and genomic sequence.
Northern blotting
Detection of processed CRISPR transcripts
In all experiments but the RNA stability determination in wild-type strain (Fig. 5A), total RNA from 1.5 ml of mid-log phase E. coli cell cultures was isolated using the TRIzol reagent (Invitrogen). Northern blots were carried out by running 5 µg of RNA on 12 % polyacrylamide gels with 8 M urea in 1× TBE buffer. RNA was then transferred to a Hybond XL membrane (GE Healthcare) by semi-dry blotting using a Trans-blot SD (Bio-Rad). Membrane was dried and then UV cross-linked. ExpHyb hybridization solution (Clontech) was used for hybridization according to manufacturer’s instructions. Membrane was then washed 3 times with 2×SSC containing 0.1% SDS for 30 min and once with 0.1×SSC containing 0.1 % SDS for 30 min. Blots were visualized with a Storm Phosphorimager (GE Healthcare). RNA sizes were estimated by comparison with 32P-labeled Decade RNA marker (Ambion).
In the case of BW28357 (wild-type) E. coli, total RNA was extracted from 10 ml of culture with TRIzol reagent and small RNA fraction was isolated from the total RNA using Mirvana miRNA isolation kit (Ambion).
Detection of unprocessed CRISPR transcripts
For detection of longer CRISPR transcripts, a standard Northern blotting protocol (Sambrook et al., 1989) was employed. Total RNA from 1.5 ml of exponentially growing cell culture was extracted with RNeasy Mini kit (Qiagen) and up to 10 µg was loaded onto each lane.
In vitro transcription and primer extension
Genomic DNA of E. coli BW28357 was used as a template for PCR amplification of DNA fragments encompassing leader sequences of both cassettes (Leader I and Leader II fragments). Purified PCR fragments were next used as templates for in vitro transcription reactions. For in vitro transcription, promoter complexes were allowed to form for 10 min at 37 °C in 10 µl reactions containing 40 mM Tris-HCl (pH 8.0), 40 mM KCl, 10 mM MgCl2, 1mM DDT, 5% glycerol, 100 nM E. coli RNA polymerase σ70 holoenzyme (Epicentre) and 10 nM DNA fragments containing promoters of CRISPR cassettes I and II. To initiate transcription reaction, a mixture of 200 µM ATP, GTP, CTP and UTP was added to promoter complexes. After incubation at 37 °C for 15 min, reaction products were extracted with phenol/chloroform and ethanol precipitated. The RNA pellet was dissolved in water. For a single extension reaction up to 0.5 µg of in vitro transcribed RNA was reverse-transcribed with 100 U of SuperScript III enzyme of First-Strand Synthesis Kit for RT-PCR (Invitrogen) according to the manufacturer’s protocol in the presence of 1 pmoles of [32P] 5’-end-labelled primers. The reaction products were dissolved in a loading buffer containing 7 M urea-formamide and resolved on 10% sequencing gels. Sequencing reactions performed with the same end-labelled primers and LeaderI and LeaderII PCR-products as templates using fmol DNA Cycle Sequencing System (Promega) according to manufacturer’s protocol were run alongside primer extension reactions. The reaction products were revealed using Storm PhosphorImager.
Semi quantitative RT-PCR
Total RNA was isolated with RNeasy Mini kit (Qiagen). Up to 5 µg of total RNA was used in reverse transcription reaction with 150 ng random hexamer primer either in the presence or in the absence of 100 U of Super Script III enzyme. The resulting cDNA was used as a template for amplification with pairs of primers specific for a housekeeping gene (gyrB) or cas genes. Aliquots of each PCR reactions were withdrawn in the middle of the exponential phase of the amplification reaction and run on agarose gels in the presence of ethidium bromide.
Phage sensitivity test
Cell sensitivity to λ phage infection was determined using the spot test method. Plates with LB agar were overlaid with 3 ml LB soft agar containing 0.3 ml of cells grown overnight in TBM medium (10 g tryptone, 4 g NaCl per liter, supplemented with 1 ml of 20% maltose). After solidification for 5 min, 5 µl of the phage lysate diluted to ~105 pfu/ml were dropped on soft agar plate. Plates were allowed to dry and incubated overnight at 30°C.
Supplementary Material
Acknowledgements
This work was supported by NIH R01 grant GM59295 and a Russian Academy of Sciences Molecular and Cellular Biology grant to KS. We are indebted to Ryland Young for comments and advice.
References
- 1.Arnvig KB, Zeng S, Quan S, Papageorge A, Zhang N, Villapakkam AC, Squires CL. Evolutionary comparison of ribosomal operon antitermination function. J Bacteriol. 2008;190:7251–7257. doi: 10.1128/JB.00760-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 5.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2006;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJ. Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010;156:1351–1361. doi: 10.1099/mic.0.036046-0. [DOI] [PubMed] [Google Scholar]
- 9.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1(6):e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, Danchin A, Bertin P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 12.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 14.Jansen R, van Embden JD, Gaastra W, Schouls LM. Identification of a novel family of sequence repeats among prokaryotes. OMICS. 2002a;6:23–33. doi: 10.1089/15362310252780816. [DOI] [PubMed] [Google Scholar]
- 15.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002b;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 16.Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Ségurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3) J Mol Biol. 2009;394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2006;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 18.Lillestøl R, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minakhin L, Severinov K. On the role of the Escherichia coli RNA polymerase σ70 region 4.2 and a subunit C-terminal domains in promoter complex formation on the extended −10 galP1 promoter. J Biol Chem. 2003;278:29710–29718. doi: 10.1074/jbc.M304906200. [DOI] [PubMed] [Google Scholar]
- 23.Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 24.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 25.Mojica FJ, Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 26.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 27.Pul U, Wurm R, Arslan Z, Geißen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–1512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. CSHL Press; 1989. Electrophoresis of RNA through gels containing formaldehyde; pp. 7.43–7.45. [Google Scholar]
- 29.Semenova E, Nagornykh M, Pyatnitskiy M, Artamonova I, Severinov K. Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol Letts. 2009;296:110–116. doi: 10.1111/j.1574-6968.2009.01626.x. [DOI] [PubMed] [Google Scholar]
- 30.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young RF., 3rd Molecular biology. Secret weapon. Science. 2008;32:922–923. doi: 10.1126/science.1162910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.