Summary
Transport of Plasmodium falciparum Erythrocyte Membrane Protein 1 (PfEMP1) variants to the red blood cell (RBC) surface enables malarial parasite evasion of host immunity by modifying the antigenic and adhesive properties of infected RBCs. In this study, we applied the Bxb1 integrase system to integrate transgenes encoding truncated PfEMP1-GFP fusions into cytoadherent A4 parasites and characterize their surface transport requirements. Our studies revealed that the semi-conserved head structure of PfEMP1 proteins, in combination with the predicted transmembrane region and cytoplasmic tail, encodes sufficient information for RBC surface display. In contrast, miniPfEMP1 proteins with truncated head structures were exported to the RBC cytoplasm but were not detected at the RBC surface by flow cytometry or immuno-electron microscopy. We demonstrated the absence of a mechanistic barrier to having native and miniPfEMP1 proteins displayed simultaneously at the RBC surface. However, surface exposed miniPfEMP1 proteins did not convey cytoadherence properties to their host cells, implicating potential steric considerations in host-receptor interactions or the need for multiple domains to mediate cell binding. This study establishes a new system to investigate PfEMP1 transport and demonstrates that the PfEMP1 semi-conserved head structure is under selection for protein transport, in addition to its known roles in adhesion.
Introduction
Clinical manifestations of malaria result from the asexual blood stage forms of Plasmodium parasites that undergo cycles of invasion, replication, and egress of red blood cells (RBCs) (Miller et al., 2002). RBCs are terminally differentiated cells that lack secretory organelles, and P. falciparum extensively modifies these host cells to enable growth inside a parasitophorous vacuole, acquire intracellular and extracellular nutrients, and allow infected RBCs to cytoadhere to host endothelium. This latter modification causes microvascular sequestration of infected RBCs, which thereby avoid clearance in the spleen. These requirements make it necessary for the parasite to elaborate a sophisticated extracellular trafficking machinery capable of transporting parasite proteins across the parasitophorous vacuole membrane (PVM) and to different destinations within the host cell, including to the RBC surface.
The mechanisms that facilitate trafficking are still incompletely understood, however exported proteins usually have a hydrophobic N-terminal sequence (NTS) that directs them to the parasite endoplasmic reticulum (ER). A default trafficking pathway then transports them to the parasitophorous vacuole (Wickham et al., 2001; Lopez-Estrano et al., 2003). Transport across the PVM is enabled by a five amino acid sequence called the P. falciparum Export Element (PEXEL) (Marti et al., 2004) or the Vacuolar Transport Signal (VTS) (Hiller et al., 2004). Cleavage and acetylation of this sequence occurs in the ER (Chang et al., 2008; Boddey et al., 2009). Protein export appears to involve unfolding of proteins in the parasitophorous vacuole (Gehde et al., 2009) and their subsequent movement through a translocon machinery embedded in the PVM (de Koning-Ward et al., 2009). Several models have been put forth to explain protein transport within the infected RBC (Templeton and Deitsch, 2005; Lingelbach and Przyborski, 2006). These include transport through vesicles (Trelka et al., 2000), complex membrane networks (Elmendorf et al., 1992; Wickert et al., 2003), or non-lipid enclosed protein aggregates (Knuepfer et al., 2005). Many exported proteins associate with lipid enclosed structures known as Maurer's clefts, which appear to be protein sorting points within the RBC (Wickham et al., 2001; Kriek et al., 2003). Among the least understood aspects are the requirements for the final steps of protein transport to the infected RBC membrane.
A key molecule at the infected RBC surface is P. falciparum erythrocyte membrane protein 1 (PfEMP1), a large clonally variant adhesion protein (Baruch et al., 1995; Smith et al., 1995; Su et al., 1995). This protein is anchored at parasite-induced, ‘knob-like’ protrusions at the RBC membrane where it can interact with host adhesins including CD36, intercellular adhesion molecule-1 (ICAM-1) or chondroitin sulfate A (CSA), enabling infected RBCs to sequester within the microvasculature (Kraemer and Smith, 2006). The extracellular portion of PfEMP1 consists of multiple adhesion domains, classified as Duffy binding-like (DBL) domains or cysteine-rich interdomain regions (CIDR), which are highly variable between family members. The resulting, distinct adhesion properties can lead to widely different clinical outcomes, including cerebral or placental malaria (Miller et al., 2002). Although variable in sequence and domain composition, nearly all PfEMP1 proteins have a semi-conserved protein “head structure” consisting of an ∼60 amino acid N-terminal segment and a DBL and CIDR tandem domain (Gardner et al., 2002; Kraemer et al., 2007). This region of the protein appears to be under strong selection for host receptor binding (Robinson et al., 2003) and may have a role in protein transport (Hiller et al., 2004; Marti et al., 2004). In contrast to the extracellular region, the PfEMP1 cytoplasmic tail is relatively conserved, and interacts with knob associated histidine rich protein (KAHRP) and potentially with the RBC cytoskeleton proteins actin and spectrin at the knobs (Kilejian et al., 1991; Waller et al., 1999; Oh et al., 2000; Waller et al., 2002).
The sequence requirements for PfEMP1 surface display are incompletely understood. Unlike most exported proteins, PfEMP1 lacks an N-terminal hydrophobic signal sequence that directs proteins to the ER for further export. The PfEMP1 transmembrane (TM) domain might potentially fulfill this trafficking role (Knuepfer et al., 2005). In addition, two different PEXEL-like motifs have been proposed to reside in the PfEMP1 N-terminus (Hiller et al., 2004; Marti et al., 2004). Recently, trafficking to the RBC surface was achieved by expressing a chimeric protein consisting of a PfEMP1 transmembrane and C-terminal sequence fused to an upstream 120 amino acid sequence from KAHRP (Knuepfer et al., 2005). The KAHRP N-terminal sequence presumably provided a N-terminal hydrophobic signal sequence that is not present in the native PfEMP1 protein and a PEXEL element for protein export. Nevertheless, the native sequence requirements for PfEMP1 display at the infected RBC surface have remained uncharacterized.
PfEMP1s are encoded by the heterogeneous family of var genes, present at ∼60 copies per P. falciparum genome. These operate under an allelic exclusion mechanism that prevents expression of more than a single var gene per parasite and enables parasites to switch expression between different var genes (Scherf et al., 1998; Frank and Deitsch, 2006). It is not clear whether mechanistic barriers preclude having two PfEMP1 proteins simultaneously displayed at the RBC surface, a situation that could potentially occur in multiply infected RBCs. Although highly variable, PfEMP1 proteins are major targets of acquired host immunity, and represent prime candidates for a pregnancy-associated malaria vaccine (Salanti et al., 2004) and other potential therapeutic interventions. A system that would enable the study of PfEMP1 transport and exposure at the RBC surface would therefore be of clear benefit. However, genetic engineering approaches to analyze var genes face a range of problems. Most var genes average 8-10 kb in length, making it technically challenging to episomally express the full-length sequences from transgene expression plasmids. Allelic exchange strategies have been used to disrupt specific var genes by single or double recombination (Andrews et al., 2003; Horrocks et al., 2004; Viebig et al., 2005), but the large size of these genes and their switching properties have hindered structure-function studies and phenotypic analysis.
In this study we have established a transgenic system, based on Bxb1 integrase-mediated recombination (Nkrumah et al., 2006), which enables truncated PfEMP1-GFP fusions or miniPfEMP1-GFP proteins to be expressed from attP containing mini-var plasmids integrated into the attB docking site of a genetically engineered cytoadherent parasite line. Using this system, we define sequence requirements for PfEMP1 surface display and demonstrate that two PfEMP1 proteins can be simultaneously expressed at the infected RBC surface.
Results
Insertion of the attB site into the Type 3 var gene by homologous recombination
To develop a system to express recombinant, full-length PfEMP1, we first engineered an attB site into the P. falciparum A4 parasite line, a highly cytoadherent subclone of IT4/25/5 (Roberts et al., 1992). As the recipient var gene, we chose the type 3 IT4var3 sequence because it is the smallest known PfEMP1 protein (Gardner et al., 2002; Kraemer et al., 2007). We designed the plasmid pDC-attB-var3(HA) (Supplementary Table S1) to enable homologous recombination and single site crossover into its first exon (Fig. 1A). This plasmid harbored an attB site just upstream of the IT4var3 start codon, which would serve as the docking site for incoming attP containing plasmids and permit site-specific integration. Our initial intention was to introduce an attB site and a hemagglutinin (HA) epitope into the native IT4var3 gene, and use a constitutive ef-1α promoter to drive gene expression. This would allow us to test whether a small atypical PfEMP1 protein encoded sufficient information for RBC surface transport. This strategy would also enable expression of var genes with selected PfEMP1 adhesion domains, independent of var gene activation mechanisms, via a second round of integrase-mediated attB × attP recombination.
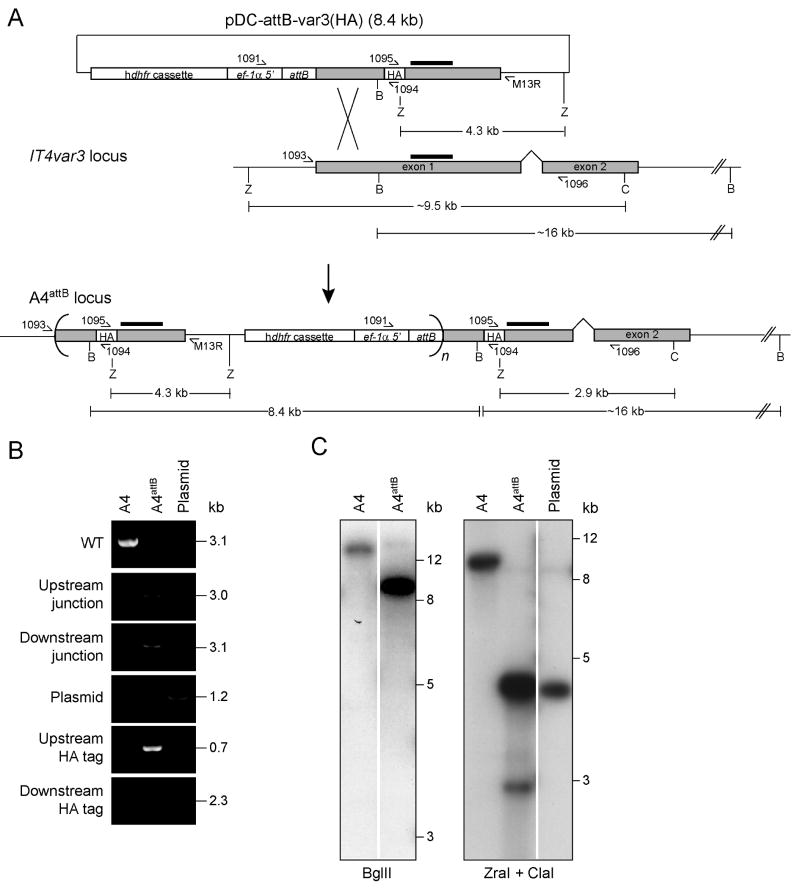
Fig. 1. Generation and confirmation of the A4attB parasite line.
A. Schematic diagram of the pDC-attB-var3(HA) integration plasmid, the IT4var3 locus, and the resulting A4attB locus after homologous recombination and single-site crossover (figure not drawn to scale). This shows the positions of PCR primers, enzyme digestion sites (C, ClaI; B, BglII; Z, ZraI); fragment sizes, and the IT4var3 probe hybridization sites (black bars). Concatameric plasmid integration is indicated by curved brackets, where n ≥ 1. Gray shading indicates the two-exon structure of the IT4var3 coding sequence. The human dhfr selectable marker cassette, elongation factor-1α promoter (ef-1α 5′), the integrated attB site, and the HA epitope tag are also indicated.
B. PCR analysis of the IT4var3 locus after homologous recombination and single-site crossover. The following primers were used: 1093 and 1096 to detect the 3.1 kb wild-type (WT) IT4var3 locus; 1093 and M13R to detect the 3.0 kb recombinant upstream junction; 1091 and 1096 to detect the 3.1 kb recombinant downstream junction; 1091 and M13R to detect plasmid (also faintly present in the A4attB lane); 1093 and 1094 to detect the 0.7 kb recombinant upstream HA epitope tag; and 1095 and 1096 to detect the 2.3 kb recombinant downstream HA tag. “Plasmid” refers to pDC-attB-var3(HA).
C. Southern blot analysis of the A4attB parasite line using an exon 1 IT4var3 probe. BglII digestion of genomic DNA from the A4 parental line liberated nearly the entire IT4var3 gene as a ∼16 kb fragment. Digestion of the A4attB parasite with BglII liberated full-length (8.4 kb) pDC-attB-var3(HA) plasmids in addition to the ∼16kb fragment. The presence of the HA epitope in the up- and downstream loci was confirmed by digestion with ZraI plus ClaI, which liberated 4.3 kb (upstream) and 2.9 kb (downstream) fragments in A4attB. Images were generated from the same exposure but intervening lanes were cropped for spacing.
We electroporated the pDC-attB-var3(HA) plasmid into A4 parasites and applied the antimalarial antifolate WR99210 to select for a recombinant parasite line expressing the plasmid-borne human dihydrofolate reductase (dhfr) selectable marker (Fidock et al., 1998). Following a period of episomal plasmid replication, we observed integration into the IT4var3 exon 1 sequence in these parasites by PCR screens that specifically detected the upstream and downstream junctions of homologous recombination and single-site crossover (Fig. 1B). This line was named A4attB. Evidence that the plasmid had integrated was provided by the detection of a PCR product that amplified over the crossover site of the transfection plasmid, and the lack of a product for the wild-type (WT) locus (Fig. 1B). By PCR, we also detected the presence of an HA tag in the IT4var3 exon1, both upstream and downstream of the integration site (Fig. 1B). This likely reflects further rearrangements that must have occurred after the initial single-site crossover event, in which the original wild-type exon 1 was either looped out during in situ DNA recombination, or corrected via DNA repair mechanisms to incorporate the HA epitope. Southern blot analysis of genomic or plasmid DNA digested with BglII and hybridized with an IT4var3 exon 1 probe showed an ∼16 kb band in A4 parasites, and an additional band of 8.4 kb in A4attB parasites, as predicted following plasmid integration into this exon (Fig. 1C). Hybridization, with the exon 1 probe, of DNA digested with ZraI plus ClaI yielded a 9.5 kb band for the A4 parental line and a 4.3 kb band for the digested plasmid. The A4attB line yielded a 2.9 kb band, corresponding to the 3′ integration site, and a more intense 4.3 kb band indicating that the plasmid had integrated as a concatemeric array (indicated in Fig. 1A by curved brackets). The resulting locus thus consisted of multiple truncated IT4var3 genes, each with a preceding attB site, and one full-length IT4var3 gene under the control of the P. berghei ef-1a promoter and harboring an attB site located between the promoter and the IT4var3 start codon. As expected, the IT4var3 transcript could be detected by reverse transcriptase PCR following plasmid integration, independently of the mutually exclusive control of var gene expression (data not shown). However, Western blot analysis using monoclonal antibodies directed against the HA epitope failed to detect recombinant protein (data not shown), possibly because of interference of the attB site with the initiation of translation. Therefore, to study PfEMP1 function, we adapted our original strategy and generated attP-containing constructs carrying a second promoter (selected from the highly expressed hrp3 gene, see below) to drive the expression of mini-var genes after plasmid integration into the attB sites.
The A4attB locus is highly amenable to integrase-mediated recombination
To assess attB × attP recombination in the A4attB parasite line, we co-transfected the integrase-expressing plasmid pINT with pLN-GFP, a GFP-expressing attP plasmid shown previously to integrate into the Dd2attB parasite line (Nkrumah et al., 2006). Parasites were selected with 100 μg/mL of neomycin (NEO) (to select for pINT) and varying concentrations of blasticidin (BSD) (for pLN-GFP), ranging from 0 to 2.0 μg/mL. On day 34 post-transfection, we confirmed integration by PCR (Supplementary Fig. S1A, B). All lines were positive for integration and negative for unrecombined attP. Parasites selected on NEO alone displayed no growth delay when later selected on BSD (data not shown) and no significant difference in the numbers of GFP-expressing cells were detected by live cell fluorescence (Supplementary Fig. S1C), indicating that successful attB × attP integration was not dependent on drug selection of the attP plasmid.
Generation of integrated mini-var transgenes
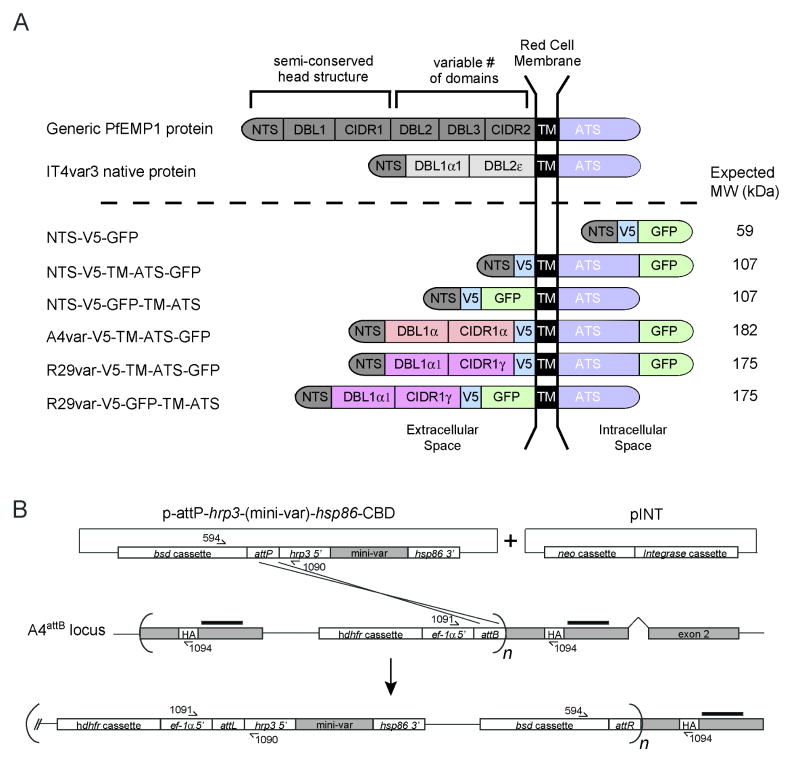
Previous work has suggested that PfEMP1 proteins might have two different PEXEL-like motifs, located either in the NTS segment 19 to 25 amino acids after the protein N-terminus (Marti et al., 2005), or about two thirds into the DBL1 domain (Hiller et al., 2004). To investigate specific PfEMP1 requirements for protein trafficking, we designed mini-var constructs that were expressed during the ring stages by an hrp3 promoter, and contained both a V5 epitope and a GFP protein tag to facilitate detection. A listing of these plasmid constructs is provided in Figure 2 and Table S1. These included one mini-var construct (p-attP-hrp3-NTS-V5-GFP) based on the first 197 amino acids (referred to herein as NTS) of IT4VAR3 PfEMP1 containing the N-terminal sequence with the first predicted PEXEL-like motif and the first half of the DBL1 domain, but not the second predicted PEXEL-like motif. The corresponding protein product is designated NTS-V5-GFP in Fig. 2A. We also generated two constructs harboring this NTS sequence fused to the IT4VAR3 TM domain and cytoplasmic tail or acidic terminal sequence (ATS), with GFP fused downstream or upstream of the TM domain (to attempt to locate GFP in the intracellular or extracellular space respectively; protein products designated NTS-V5-TM-ATS-GFP and NTS-V5-GFP-TM-ATS; Fig. 2A). Finally, we generated larger miniPfEMP1 proteins incorporating the semi-conserved head structures of either a Group B PfEMP1 variant (A4var, amino acids 1 to 846 containing the DBL1α and CIDR1γ domains) (Smith et al., 1998; Smith et al., 2000a) or a Group A PfEMP1 variant (R29var, amino acids 1 to 787 containing the DBL1α1 and CIDR1α1 domains) (Rowe et al., 1997). These larger transgene products possessed both predicted PEXEL-like motifs (Fig. 2A). These constructs all encoded the IT4VAR3 TM domain and cytoplasmic tail (amino acids 849 to 1324), with the GFP tag placed either just prior to the TM or following the ATS domain (products designated A4var-V5-TM-ATS-GFP, R29var-V5-TM-ATS-GFP and R29var-V5-GFP-TM-ATS; Fig. 2A).
Fig. 2. Integration of mini-var transgenes into the IT4var3 locus.
A. Schematic of a native PfEMP1 and miniPfEMP1 proteins depicting their predicted organization and topology. The top row shows a generic PfEMP1 with its protein head structure (composed of NTS, DBL1 and CIDR1 head domains, a variable number of internal domains, a TM and an ATS). The Type 3 var encodes the smallest PfEMP1 protein and lacks a CIDR1 domain (Gardner et al., 2002; Kraemer et al., 2007). The red cell membrane is indicated by two black bars, with predicted extracellular sequence to the left. The miniPfEMP1 recombinant molecule NTS-V5-GFP is depicted as residing within the intracellular space (note that the parasite vacuolar and plasma membranes have been omitted for clarity). NTS, N-terminal sequence; DBL, Duffy binding like; CIDR, cysteine-rich interdomain region; TM, transmembrane, ATS, acidic terminal sequence (cytoplasmic tail); V5, V5 epitope.
B. Schematic diagram depicting Bxb1 integrase-mediated recombination of a mini-var transgene into the attB-tagged IT4var3 locus. Coding regions are indicated by their gray shading. This schematic also indicates the probe hybridization site (black bars), the human dhfr cassette, the blasticidin resistance marker (bsd cassette), ef-1α 5′ UTR, hrp3 5′ UTR, hsp86 3′ UTR, the attP site, the attB element, the neomycin resistance cassette (neo cassette), the Bxb1 integrase cassette (Integrase cassette), and the HA tag. This schematic also shows the positions of primers used to verify integration.
Plasmid constructs were transfected in combination with pINT into the A4attB parasite line as described above, and recombined parasite lines were obtained 14 to 23 days post-transfection following selection with 100 μg/mL NEO (to select for the pINT plasmid) ± 1.0 μg/mL BSD (to select for the attP plasmid). A schematic of the attB × attP recombination event, leading to expression of the mini-var genes under control of the strong hrp3 promoter (which is not subject to allelic exclusion), is presented in Fig. 2B. For ease of interpretation, the resulting recombinant lines were assigned the same name as the protein products represented schematically in Fig. 2A, and were consistent with the naming of the corresponding transfection constructs listed in Table S1.
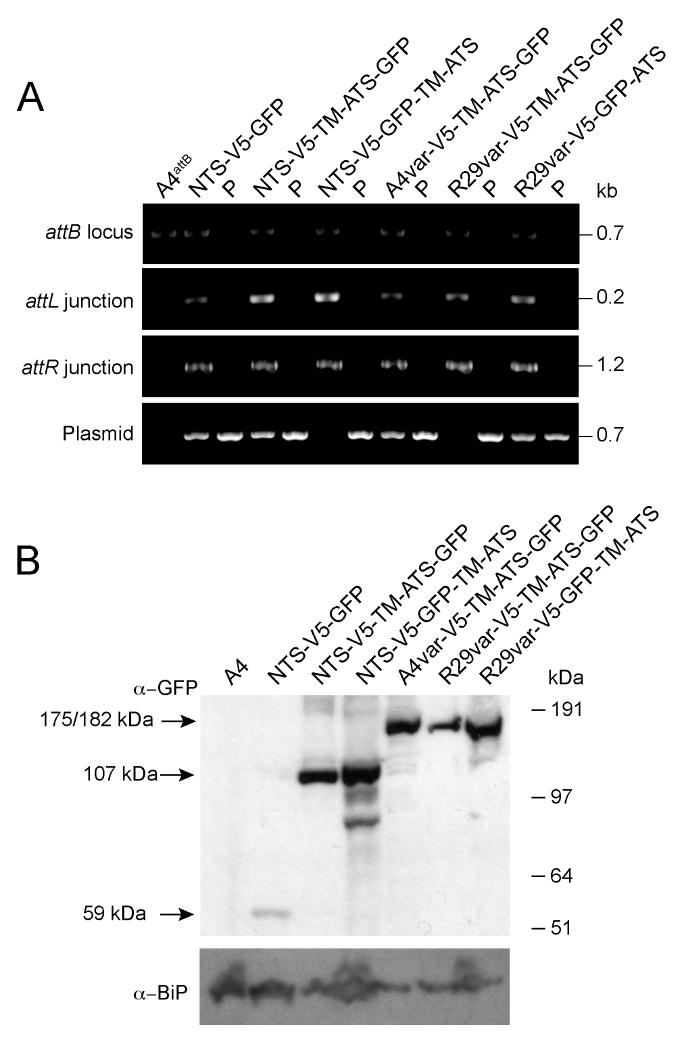
Integration was confirmed by PCR amplification over the attL and attR junctions, which represent the chimeric products of attB × attP integrase-mediated recombination (Fig. 3A). Unrecombined attP and attB sites could also be detected in several of the parasite lines. This indicated that integration events did not occur in all of the multiple attB sites present and that the attP plasmid had likely concatemerized prior to integration, because these PCR products were also detected in the absence of BSD selection on the attP plasmid (arguing against the presence of episomal plasmid).
Fig. 3. Confirmation of mini-var transgene integration into the IT4var3 locus and miniPfEMP1 expression.
A. PCR analysis of transfected lines was performed using the following primers: 1091 and 1094 to detect unrecombined attB sites; 1091 and 1090 to amplify the 5′ attL junction; 594 and 1094 to amplify the 3′ attR junction; and 594 and 1090 to detect unrecombined plasmid or plasmid that integrated as a concatemer. PCR product sizes are indicated. “P” indicates the attP plasmid used in the generation of each line.
B. Western blot of protein extracts from transgenic parasite lines expressing miniPfEMP1s. These extracts were probed with antibodies to GFP (top panel), or BiP (used as a loading control and shown on the bottom panel). The expected miniPfEMP1 product sizes are listed to the left of panel.
To investigate protein expression, trophozoite extracts were prepared from parasite cultures harvested by gelatin flotation. MiniPfEMP1 proteins were detected by Western blot using anti-GFP antibodies. In each case, the anti-GFP antibodies detected a dominant band of the correct predicted size (Fig. 2A and 3B). Antibodies specific to the ER-Golgi marker BiP (Kumar et al., 1991) were used as a control to ensure protein was loaded in all lanes, including those negative for GFP staining (Fig. 3B).
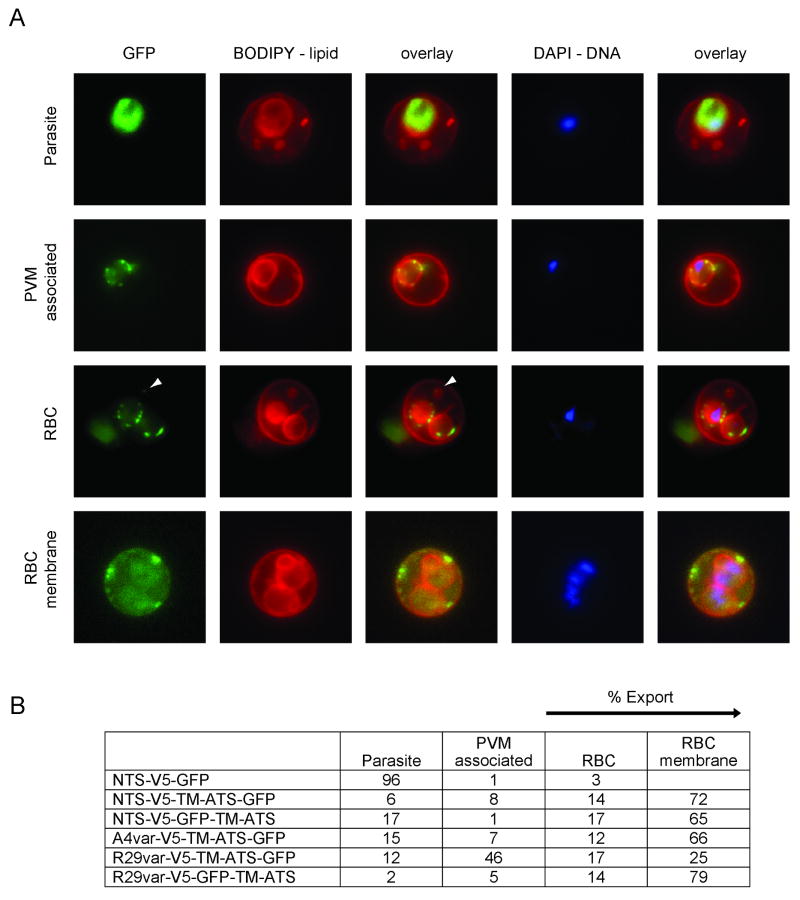
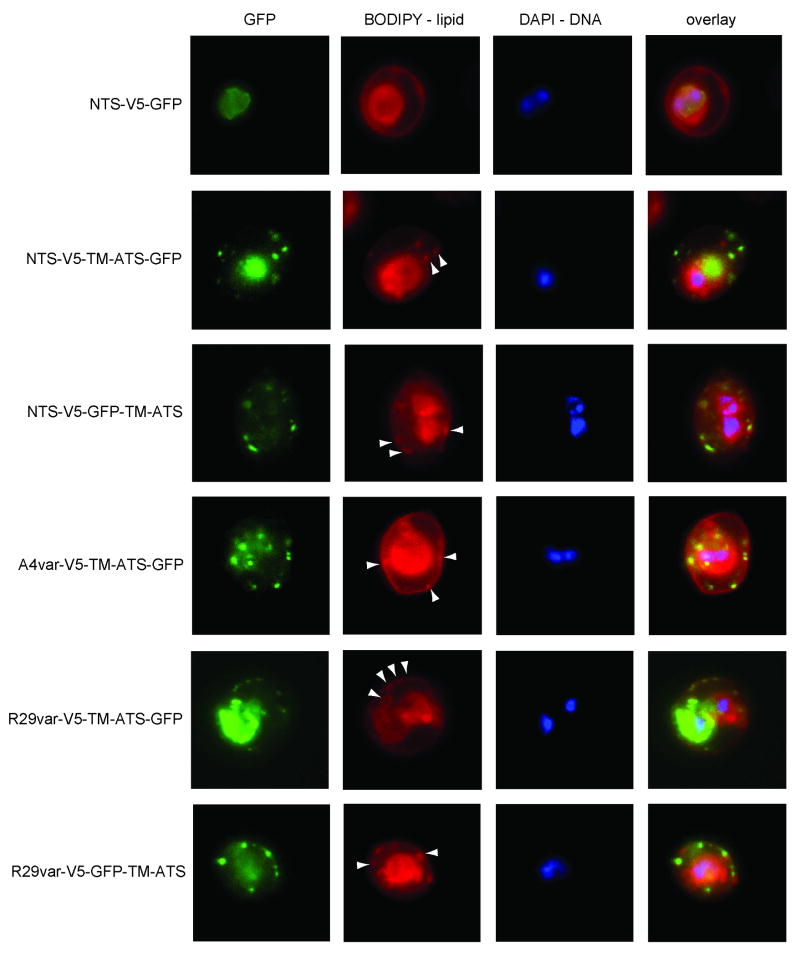
Patterns of miniPfEMP1 export into and within the infected erythrocyte
To analyze miniPfEMP1 transport, BODIPY lipid and DAPI DNA staining were employed to distinguish between the parasite and RBC compartments by fluorescence microscopy of trophozoite-infected RBCs (Tamez et al., 2008). BODIPY stained several lipid-containing structures including the RBC membrane, lipid structures in the RBC cytoplasm, and the parasite membrane/PVM. The presence of DNA staining marked the intracellular parasite. In combination with these staining methods, GFP fluorescence allowed us to identify several patterns of miniPfEMP1 trafficking: retention within the parasite (“Parasite”), retention at the PVM (“PVM associated”), export into the RBC cytoplasm (“RBC”), or localization at or near the RBC membrane (“RBC membrane”) (Fig. 4A). The percentage of each pattern was determined by analyzing at least 50 individual infected RBCs from each line (Fig. 4B). As predicted by the absence of an N-terminal hydrophobic sequence, the construct containing only the NTS was insufficient to mediate export to the RBC, with “export” defined as fluorescence within the RBC cytoplasm or at the RBC membrane (“RBC” and “RBC membrane”). 96% of infected RBCs retained the NTS-V5-GFP miniPfEMP1 within the parasite (Fig. 4B).
Fig. 4. Localization of miniPfEMP1 proteins in the infected red blood cells by live cell fluorescence imaging.
A. Characteristic fluorescence patterns observed in the miniPfEMP1-expressing parasite lines: 1st row: localization to the intracellular parasite; 2nd row: association with the parasite membrane/parasitophorous vacuole membrane (PVM); 3rd row: export into the RBC; 4th row: export to or near the RBC membrane. White arrowheads indicate GFP signal in lipid-enclosed vesicles in the RBC cytoplasm.
B. Percentage of each fluorescence pattern observed in each miniPfEMP1-expressing parasite line, as determined by cell counts of fluorescence patterns of at least 50 infected RBC per line.
In contrast, the placement of the TM domain and the cytosolic tail downstream of the NTS, in the lines NTS-V5-TM-ATS-GFP and NTS-V5-GFP-TM-ATS, increased export to greater than 80% of observed cases. Furthermore, the miniPfEMP1 transgenes expressed in the A4var-V5-TM-ATS-GFP and R29var-V5-GFP-TM-ATS lines were exported in 78% and 93% of observed cases, respectively (Fig. 4B).
For most constructs, placement of the GFP tag within the predicted extracellular or intracellular portion of the miniPfEMP1 polypeptide did not significantly modify export efficiency beyond the PVM. However, the R29var-V5-TM-ATS-GFP construct was less well exported than other constructs with ∼46% of the infected RBC exhibiting some protein retention at the parasite membrane and/or PVM (Fig. 4B). Retained R29var-V5-TM-ATS-GFP proteins were found in discrete foci in the membrane surrounding the parasite (Fig. 4A), in a pattern reminiscent of the one reported for the protein translocon machinery (de Koning-Ward et al., 2009). In the majority of cases, export of miniPfEMP1 proteins beyond the PVM led to localization near or at the RBC membrane. In addition, all five miniPfEMP1 constructs that were exported to the RBC cytoplasm gave a punctate staining pattern. In virtually all observed cases, the miniPfEMP1 proteins that were localized within the RBC cytoplasm appeared to be associated with BODIPY-labeled structures (Fig. 5). These might correspond to Maurer's clefts or other vesicular structures in the RBC cytoplasm.
Fig. 5. Export of miniPfEMP1 proteins to the red blood cell cytoplasm.
Representative fluorescence images showing the localization of miniPfEMP1-GFP fusion proteins. Parasite lines are indicated on the left. The first column of images represents GFP fluorescence, the second column BODIPY lipid staining, the third column DAPI staining of parasite DNA, and the fourth column an overlay of the three images. The NTS-V5-GFP construct was confined to the parasite and other miniPfEMP1 constructs were typically exported to the RBC cytoplasm. Exported proteins had a punctate staining pattern and GFP signal generally colocalized with BODIPY-labeled structures in the RBC cytoplasm (white arrowheads).
MiniPfEMP1 proteins containing a complete head structure are exposed at the RBC surface in the correct orientation
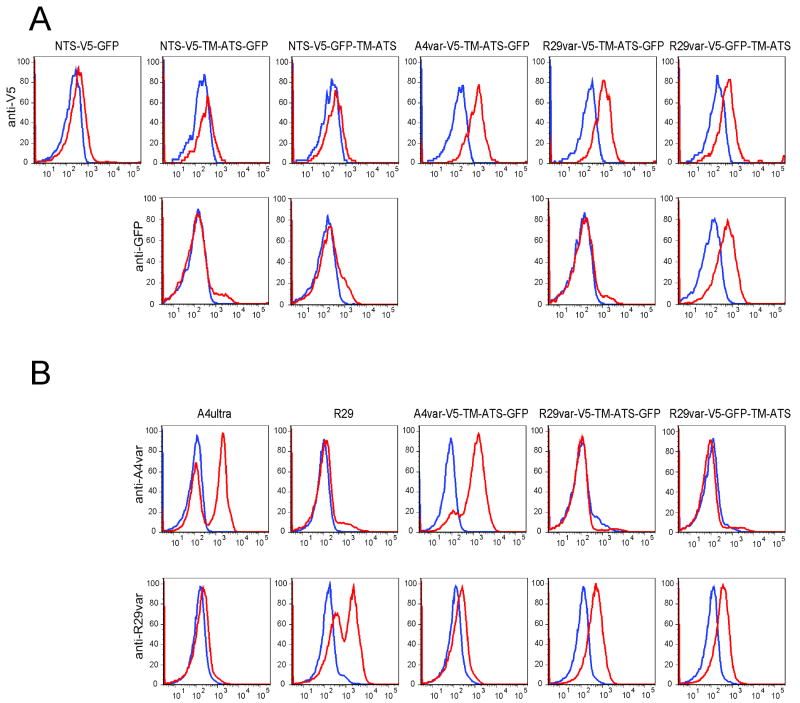
To examine whether miniPfEMP1 proteins were transported to the RBC surface in the proper membrane orientation, we performed flow cytometric analysis on live trophozoite-stage infected RBCs using antibodies directed against either the V5 epitope or GFP tags (Fig. 6A). Whereas miniPfEMP1 proteins containing the NTS in combination with the TM region and cytoplasmic tail, namely NTS-V5-TM-ATS-GFP and NTS-V5-GFP-TM-ATS, were efficiently exported to the RBC and localized near the RBC membrane (Fig. 5), we were unable to detect recombinant PfEMP1 surface staining in either line (Fig. 6A, left panels). This suggests a possible defect in the translocation and presentation of these miniPfEMP1 proteins on the RBC surface. In contrast, all of the miniPfEMP1 proteins that encoded complete semi-conserved head structures with full-length DBL and CIDR domains (namely A4var-V5-TM-ATS-GFP, R29var-V5-TM-ATS-GFP and R29var-V5-GFP-TM-ATS; Fig. 6A) were surface positive with anti-V5 monoclonal antibody. Moreover, anti-GFP polyclonal sera reacted with infected RBC in which the GFP label was positioned in the extracellular region of the miniPfEMP1 protein (R29var-V5-GFP-TM-ATS) and not within the luminal side of the RBC membrane (R29var-V5-TM-ATS-GFP) (Fig. 6A).
Fig. 6. Red blood cell surface exposure of miniPfEMP1 proteins.
A. FACS histograms of gated infected RBCs stained with antibodies directed against either the V5 epitope (top line) or GFP (bottom line). Cells stained with antibodies are in red, controls without primary antibodies are in blue.
B. FACS histograms of gated infected RBCs. Control parasite lines expressing native PfEMP1 are shown on the left (A4ultra for A4var and R29 for R29var), and recombinant miniPfEMP1 lines are shown on the right. Antibody stainings were against A4var CIDR1 (top line) or R29var DBL1 domains (bottom line). Color key as per A.
Surface exposure was further confirmed using specific antisera to the A4var CIDR1 or the R29var DBL1 domains and controlled by non-transfected parasite lines expressing native A4var or R29var PfEMP1 proteins. We detected similar levels of anti-A4var CIDR1 antibody binding at the RBC surface of lines expressing native A4var PfEMP1 and the A4var-V5-TM-ATS-GFP miniPfEMP1 fusion protein (Fig. 6B, top panels). Similarly, anti-R29var DBL1 antibodies labeled the surface of infected RBCs expressing native R29var or recombinant R29var-V5-TM-ATS-GFP and R29var-V5-GFP-TM-ATS (Fig. 6B, bottom panels). Antibody binding was specific, as shown by the lack of cross-reactivity between parasite lines expressing either A4var or R29var PfEMP1 domains. In these three parasite lines, the complete population of infected RBCs displayed miniPfEMP1 at the surface, including the R29var-V5-TM-ATS-GFP line that showed partial retention at the PVM. Thus, miniPfEMP1 proteins containing semi-conserved head structures possessed all the necessary signals for transport and integration in the correct membrane orientation at the RBC surface.
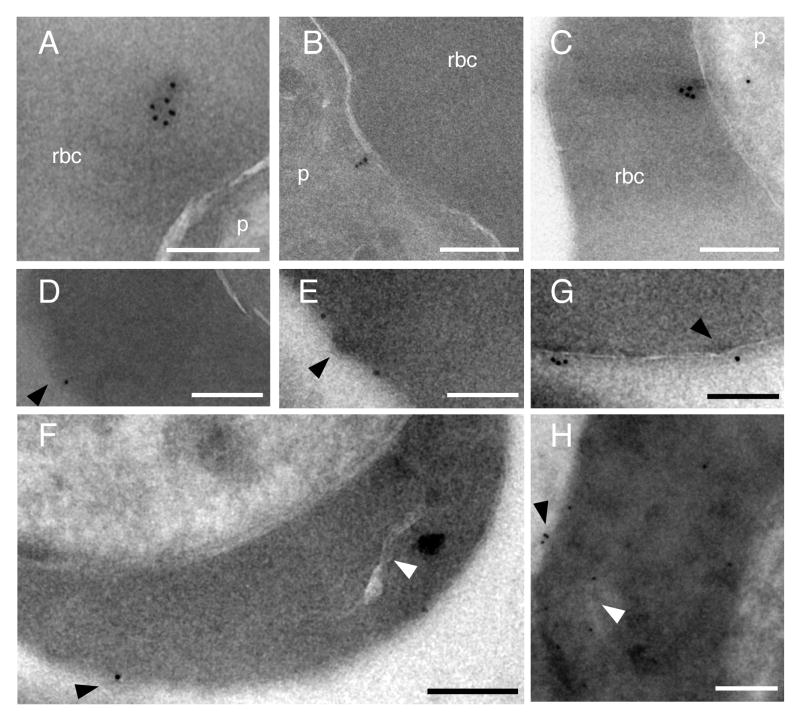
To investigate miniPfEMP1 localization at the ultrastructural level, immuno-electron microscopy (immuno-EM) was performed with anti-GFP antibodies. With the NTS-V5-TM-ATS-GFP line, we observed immunogold particles in the RBC cytoplasm, with evidence of association with electron dense material (Fig. 7A). In this line we also observed particles in the parasite cytoplasm, in some cases located adjacent to the parasite plasma membrane (Fig. 7B). Consistent with the flow cytometry data, surface labeling of infected RBCs was never observed in 40 images taken of this line, although the anti-GFP antibody reacted with this protein in the RBC cytoplasm (Fig. 7A). Electron tomogram studies with the R29var-V5-TM-ATS-GFP line confirmed export of this miniPfEMP1 fusion protein into the RBC cytoplasm (Supplemental Movie S1).
Fig. 7. Ultrastructural analysis of miniPfEMP1 transport.
The NTS-V5-TM-ATS-GFP or R29var-V5-GFP-TM-ATS parasite lines were analyzed by immunogold electron microscopy. A. NTS-V5-TM-ATS-GFP, immunogold particles were observed in association with an electron dense structure in the RBC cytoplasm.
B. NTS-V5-TM-ATS-GFP localized close to the parasite plasma membrane.
C. R29var-V5-GFP-TM-ATS immunogold particles were observed close to the PVM. D–H. R29var-V5-GFP-TM-ATS parasites were labeled with anti-GFP, revealing miniPfEMP1 fusion proteins at the RBC surface at or near knob structures, as well as near a Maurer's cleft in the RBC cytoplasm (H). Anti-GFP immunogold label is found at the red blood cell surface or in close proximity to Maurer's clefts (H). Labeling with 10-nm immunogold was performed after cryosectioning (A to F and H) or prior to fixation with 2% glutaraldehyde (G). White arrowheads, Maurer's clefts; black arrowheads, surface knobs of infected RBCs. Scale bars 200 nm, p parasite, rbc red blood cell cytoplasm.
Immuno-EM studies were also performed with the R29var-V5-GFP-TM-ATS line that efficiently exported PfEMP1 into the host RBC (Fig. 7C–H). These identified miniPfEMP1 proteins in the RBC cytoplasm (Fig. 7C), and at sites associated with or adjacent to knob-like structures in the RBC membrane (Fig. 7D–H). Some parasites also showed evidence of physical proximity to Maurer's clefts (Fig. 7H). These patterns are reminiscent of images observed with a previously reported KAHRP-PfEMP1 chimera (Knuepfer et al., 2005). Taken together, our studies suggest that miniPfEMP1 proteins encoding the PfEMP1 semi-conserved head structure and cytoplasmic tail were capable of being transported to the RBC surface where they were often located at or near parasite-induced knob structures.
MiniPfEMP1 proteins can be surface exposed simultaneously with endogenous PfEMP1
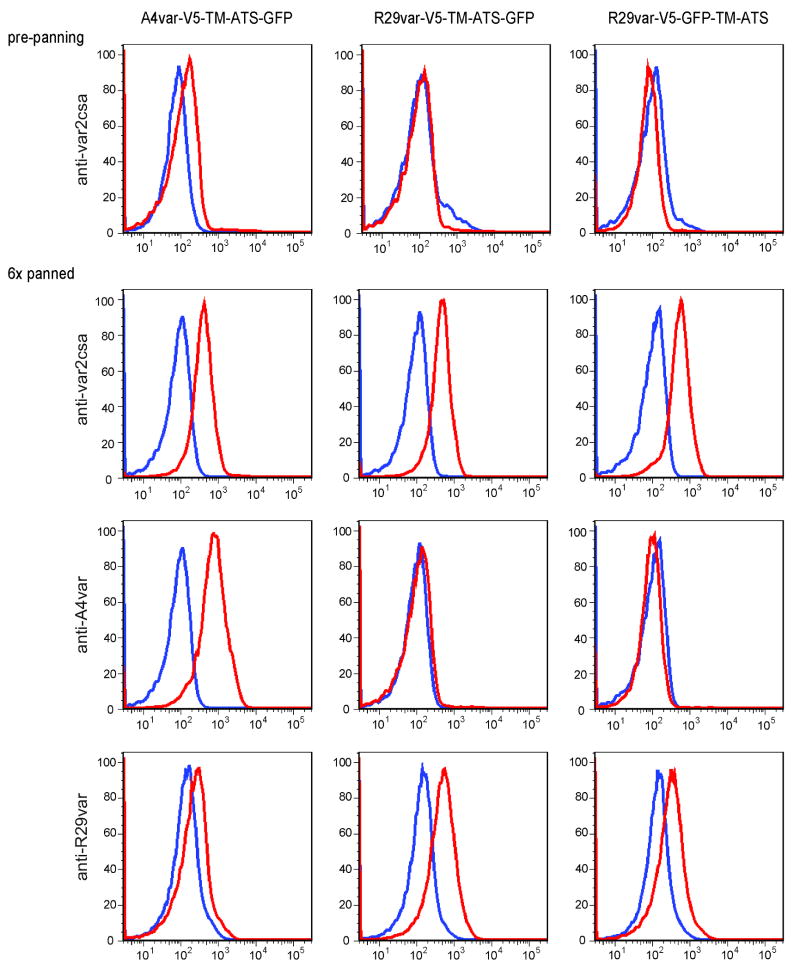
From our flow cytometry data, we observed that whereas the A4var miniPfEMP1 protein was expressed at approximately the same level as native A4var PfEMP1 in non-transfected parasites, the R29var miniPfEMP1 proteins were expressed at slightly lower levels than their native counterparts (Fig. 6B). Because miniPfEMP1 proteins are expressed independent of var gene regulatory mechanisms, this raises the possibility of competition between miniPfEMP1 and native PfEMP1 proteins for their association with knob structures or surface display. To determine whether two different PfEMP1 variants can be simultaneously displayed at the RBC surface, parasite lines expressing the A4var and R29var miniPfEMP1 proteins were selected on the placental adhesion receptor CSA to enrich for a specific PfEMP1 variant (Salanti et al., 2003). After 6 rounds of panning on CSA, all of the parasite lines had switched to VAR2CSA as the endogenous PfEMP1 protein, as evidenced by anti-VAR2CSA antibody labeling. Notably, these also retained expression of miniPfEMP1 protein on the infected RBC surface (Fig. 8), demonstrating that a single infected RBC can express two different PfEMP1 proteins at the RBC surface. We note that levels of endogenous VAR2CSA PfEMP1 were comparable between the A4var and the two R29var lines, despite differences in the degrees to which they exported their miniPfEMP1 out to the RBC cytoplasm (Fig. 4B).
Fig. 8. Simultaneous surface exposure of miniPfEMP1 proteins with endogenous PfEMP1.
FACS histograms of gated infected RBCs pre- or post- CSA-panning. Specific antibody stainings are as indicated on the left. Cell populations labeled with specific antibodies are shown in red, and control cell populations without primary antibodies are shown in blue.
Having selected for VAR2CSA as the endogenous PfEMP1 variant, we also investigated whether miniPfEMP1 proteins were able to confer new binding properties on infected RBCs. VAR2CSA is known to bind to the host receptor CSA, but does not mediate infected erythrocyte rosetting or CD36 binding. Our studies were predicated on earlier reports that these binding properties reside within the R29var and A4var head structures, respectively (Rowe et al., 1997; Smith et al., 1998). As expected, all of the panned lines bound to CSA above background (Supplementary Fig. S2). However, we could not detect binding of the A4var miniPfEMP1 to CD36, and infected RBCs expressing the R29var miniPfEMP1 were insufficient to mediate rosetting (data not shown). As the larger VAR2CSA protein may have sterically hindered binding, we also tried limited protease digestion of infected erythrocytes to selectively remove the native PfEMP1. The miniPfEMP1 proteins were found to be highly sensitive to low trypsin concentrations, ranging from 0.1-1.0 mg/ml (Supplementary Fig. S3), supporting their surface localization. However, we were unable to define protease conditions that would digest the VAR2CSA protein and leave the miniPfEMP1 protein intact (Supplementary Fig. S3). Thus, miniPfEMP1 proteins containing the semi-conserved head structure possessed sufficient structural integrity for surface transport and presentation, but not for binding or rosetting activity, suggesting that additional domains might be required to mediate adhesion.
Discussion
P. falciparum causes infected RBC sequestration in peripheral blood vessels through the exportation of PfEMP1 virulence factors to the host cell surface. In this study, we provide the first model system to study the trafficking of a PfEMP1 protein to the RBC surface, by using the Bxb1 integrase system to rapidly integrate mini-var constructs into the cytoadherent A4 parasite line. We demonstrate that this system of transgene expression is highly efficient, and does not require drug selection on the attP plasmid. Integration is stable, because attB × attP recombination yields the attL and attR sites that are resistant to excision by Bxb1 integrase. This should permit integration of larger full-length PfEMP1 transgenes through the removal of the blasticidin S-deaminase (bsd) selection, thereby allowing the cloning and propagation of larger transgenes with smaller plasmid backbones.
Two signals are typically required to allow protein export to the RBC, namely an N-terminal hydrophobic sequence for entry into the parasite ER, and a PEXEL/VTS motif for translocation across the PVM (Hiller et al., 2004; Marti et al., 2004; de Koning-Ward et al., 2009). However, PfEMP1 proteins differ from most exported proteins because they lack a defined N-terminal signal sequence, with the PfEMP1 TM domain appearing to direct the protein to the parasite ER instead (Knuepfer et al., 2005). Additionally, two different non-canonical PEXEL-like motifs have been proposed in PfEMP1 proteins. Both motifs were shown to mediate export to the host cell when combined with the TM and ATS domains (Hiller et al., 2004; Marti et al., 2004). However, while the first is located within the NTS domain, the second motif is found in all DBL domains, including several EBL genes that localize to microneme organelles within the parasite; therefore its function has been called into question (Templeton et al., 2005). In this study, we show that the first motif is sufficient for export of miniPfEMP1 proteins into the RBC, but that additional information in the protein head structure is required for RBC surface display. This presumably reflects the existence of additional control mechanisms that operate post-PVM export.
Our study reveals that the PfEMP1 semi-conserved head structure in cooperation with the TM region and ATS domain encodes sufficient information for RBC surface transport of both a Group A (R29var) and a Group B (A4var) miniPfEMP1 protein. These miniPfEMP1 proteins associate with knob-like protrusions at the infected RBC surface in the correct membrane orientation. This is notable because the head structure is the most conserved extracellular region between PfEMP1 proteins (Smith et al., 2000b) and was previously known to be important only for host receptor binding (Baruch et al., 1997; Rowe et al., 1997; Chen et al., 1998; Robinson et al., 2003). Our results provide compelling evidence to suggest that this region is also under selection for PfEMP1 protein translocation to the cell surface.
In contrast, miniPfEMP1 proteins with truncated head structures were exported to the RBC cytoplasm but could not be detected at the host cell surface. Several explanations are possible, including that a truncated head structure might not fold properly or be unable to interact with an accessory protein(s) required for the final stage of protein transport from Maurer's clefts to the infected RBC surface. Indeed, a number of accessory proteins required for PfEMP1 transport have been described, including those that are located within the Maurer's cleft lumen or that are associated with Maurer's clefts in the RBC cytoplasm (Cooke et al., 2006; Maier et al., 2006; Maier et al., 2007). Another possibility would be a requirement for an additional signal in the PfEMP1 head structure that is necessary for the final stages of transport. While a prior study observed transport to the RBC surface with a fusion protein of the first 120 amino acids of KAHRP (containing a PEXEL motif and an N-terminal hydrophobic sequence) that were coupled with PfEMP1 TM and ATS domains (Knuepfer et al., 2005), those findings do not exclude a role for additional features within the PfEMP1 head structure. The fact that there exist different final destinations for P. falciparum proteins that are exported to the infected RBC might even argue for the necessity of additional signals in these proteins, especially if the Maurer's cleft serves as a sorting point.
This analysis also shows the absence of any mechanistic barriers to having two different PfEMP1 proteins (mini and endogenous) expressed at the RBC surface. Such a scenario could occur in multiply infected RBC, potentially modifying their binding properties and endothelial binding tropism. Previous work had suggested that infected RBC might gain adhesion properties upon disruption of var gene silencing mechanisms (Tonkin et al., 2009). However, the current report would appear to be the first to directly show simultaneous expression of two PfEMP1 variants at the RBC surface by antibody labeling.
Interestingly, we did not observe adhesion of the A4var miniPfEMP1 to CD36, an initially unexpected result given that CD36 binding was previously mapped in vitro to the A4var CIDR1 domain encoded within our transgene construct (Smith et al., 1998). Nor did we find the R29var miniPfEMP1 to be sufficient to mediate infected RBC rosetting, even though the DBL1 domain, present within this miniPfEMP1, binds complement receptor 1 on RBCs when expressed as a fusion protein from heterologous cells (Rowe et al., 1997). Neither of those former studies on adhesive properties of individual domains, however, addressed binding activity at the surface of infected RBCs, and conceivably the assembly of PfEMP1 proteins within the knob structure might have influenced miniPfEMP1 binding properties. To assess whether binding regions in miniPfEMP1 proteins could have been buried or rendered sterically inaccessible, perhaps hidden by the larger co-expressed VAR2CSA protein, we also attempted partial protease digestion. However, we were unable to define protease conditions that would cleave the VAR2CSA protein while leaving miniPfEMP1 intact, thus making it impossible to test for steric hindrance. We note that most PfEMP1 proteins have four or more extracellular domains, and it is possible that internal domains may be required to maintain the head structure at a proper distance from the RBC membrane to enable CIDR1-CD36 interactions.
Another interpretation of our data is that single domains were insufficient to mediate binding because they do not reconstitute the structural requirements for adhesion. This would infer that multiple domains would be required for binding. This interpretation is consistent with two recent reports showing that recombinant VAR2CSA protein expressing the complete extracellular domain (containing six DBL domains) had at least 1,000-fold greater affinity for binding CSA than that displayed by individual domains, implying that multiple binding domains may contribute to higher binding affinity or that ligand binding required higher-order structure of multiple domains involving inter-domain interactions (Khunrae et al., 2010; Srivastava et al., 2010).
With the expression of larger miniPfEMP1 proteins using the Bxb1 integrase system, it should prove possible in future studies to identify the minimal requirements for infected RBC binding and investigate cooperation between domains as they pertain to binding, in the native context at the RBC cell surface. This would represent a significant advance from the limitations of the current technology, based on recombinant PfEMP1 domain expression in heterologous eukaryotic cells. This system also provides an in vivo tool for the characterization of molecular interactions between the PfEMP1 cytoplasmic tail and knob structures. In conclusion, the miniPfEMP1 system developed herein provides a useful system to analyze transport of this virulence determinant to the RBC surface and suggests the PfEMP1 head structure is under selection for both binding and protein transport.
Experimental procedures
Plasmid construction
Plasmids pINT, pLN-GFP and pCBD-P were described previously (Nkrumah et al., 2006; Muhle et al., 2009). The construction of transfection and intermediary shuttle plasmids is detailed in the Supplementary Experimental Procedures. Table S1 lists all plasmids created during this study. This includes pDC-attB-var3(HA), which was generated by introducing an attB element upstream of the HA-tagged IT4var3 exon 1 and inserting this cassette into a plasmid expressing the human dhfr selectable marker. The attP containing plasmids were generated from pDC-attB-var3(HA) by replacing the truncated IT4var3 open reading frame with mini-var transgene elements, and exchanging the human dhfr cassette with a bsd cassette. All fragments were PCR amplified from genomic DNA of the P. falciparum IT4/25/5 clone A4 (Roberts et al., 1992), unless otherwise noted. Plasmids were verified by sequencing prior to transfection. Sequences of primers used for cloning or PCR analysis are listed in Supplementary Table S2. Representative mini-var plasmid maps are presented in Supplementary Fig. S4.
Parasite propagation and transfections
All parasite lines described in this study were generated from the IT4/25/5 clone A4 (Roberts et al., 1992), and were cultured under standard conditions (Trager and Jensen, 1976) in RPMI-1640 medium supplemented with 10% pooled heat-inactivated AB+ serum (Interstate Blood Bank, Memphis, TN), 0.5% w/v Albumax II (Invitrogen, Carlsbad, CA) and Lymphoprep (Axis-Shield)-purified human RBCs at 3 to 5% hematocrit. Cultures were frequently subjected to gelatin flotation (Waterkeyn et al., 2001) to maintain selection for cytoadherence. The A4attB parasite line was propagated in the presence of 0.4 nM WR99210 (Jacobus Pharmaceuticals, Princeton, NJ). MiniPfEMP1 lines were cultured in the presence of 1.0-2.5 μg/mL blasticidin hydrochloride (BSD) (Invitrogen).
Transfections to incorporate the attB element were performed using ring-stage parasites at approximately 5% parasitemia. Cells were electroporated with 100 μg of plasmid in 0.4 cm cuvettes using low voltage (0.31 kV) and high capacitance (950 μF) (Fidock and Wellems, 1997), and aliquoted to triplicate 2 mL wells. Transfected lines were cloned by limiting dilution on day 104 post-transfection. Site-specific recombination of attP plasmids was accomplished by co-transfecting A4attB parasites with 50 μg of attP plasmid and 50 μg of pINT plasmid (Nkrumah et al., 2006) as described above. To select for transfected parasites, 100 μg/mL of G418 sulfate (NEO) (Cellgro) ± 1.0 μg/mL BSD was added on day 2 post-transfection. Integration events were verified by sequence-specific PCR screening and Southern blot analysis, as documented below.
DNA analysis
P. falciparum genomic DNA was obtained from trophozoite-stage infected RBCs by saponin lysis and purified using DNeasy Tissue Kits (Qiagen). Integration of plasmid pDC-attB-var3(HA) was detected with primers 1093 to the 5′untranslated region (UTR) and 1096 to exon 2 of the IT4var3 locus, primers 1094 and 1095 to the HA epitope tag, primer 1091 to the ef-1α 5′UTR, and primer M13R to the plasmid backbone of pDClink (described in Supplementary Experimental Procedures and Table S1). To detect integrase-mediated attB × attP site-specific integration of mini-var transgenes, the attL junction was detected using primers 1091 and 1090 (to the hrp3 5′ untranslated region of pLN-GFP), the attR junction using primers 594 (specific to bsd cassette) and 1094, and plasmid was detected with primers 594 and 1090 (primer locations depicted in Fig. 2A). For Southern blot analysis, 1.5 μg of DNA were digested with enzyme(s) as indicated, electrophoresed on a 0.8% polyacrylamide gel, and transferred onto nylon membranes overnight. A [32P]-radiolabeled probe (made from a ZraI-BstBI fragment isolated from plasmid pDC-attB-var3(HA)) was hybridized to the membranes overnight at 56°C.
Antibody production
To produce antibodies specific to R29var-encoded PfEMP1, we produced the recombinant domain encompassing residues 1-481 as a soluble protein in Escherichia coli using a baculovirus codon-adjusted version (to decrease the A-T richness of the sequence) and the pMAL-c2X (New England Biolabs) vector. This expressed domain was cleaved from the maltose binding protein fusion by treatment with the protease Factor Xa. Purified R29var protein was used to immunize BALB/c mice (10 μg per injection in Freund's complete adjuvant for the first injection and Freund's incomplete adjuvant for two subsequent boosts at three-week intervals). A pool of sera was obtained by mixing serum from five animals bled eight days after the final immunization (Vigan-Womas et al., in preparation). A4var antibodies were produced by immunizing rabbits with a plasmid DNA vector (VR1051) expressing the A4-CIDR1 region (residues 402 to 846), as described previously (Avril et al., 2008).
Western blotting
Western blotting was performed on SDS-detergent extracts of mature infected cells harvested after MACS cell sorting (Miltenyi biotec). Cell extracts were electrophoresed on a 6% SDS-PAGE gel and transferred to polyvinylidene difluoride membranes. Primary incubation was performed with mouse anti-GFP monoclonal antibody (Clontech), followed by secondary incubation with horseradish peroxidase-conjugated donkey anti-rabbit IgG (Amersham Biosciences). Bands were visualized by chemiluminescence using the ECL Western blot analysis system (Amersham Biosciences). The blot was stripped and reprobed with anti-BiP antibodies (Kumar et al., 1991) followed by secondary antibody detection with anti-rabbit IgG as described above.
Flow cytometry analysis
Trophozoite stage infected RBCs were incubated for one hour at room temperature with a variety of primary antibodies: mouse anti-V5 (Invitrogen) (diluted 1:50), rabbit anti-GFP (Millipore) (1:50), rabbit anti-A4var CIDR1 (1:10), rabbit anti-VAR2CSA DBL1 (1:10) (Avril et al., 2008), or mouse anti-R29var DBL1 (1:500). The rabbit antibodies were further incubated with mouse anti-rabbit IgG-R-Phycoerythrin (SouthernBiotech) (1:50) and then goat anti-mouse IgG-R-Phycoerythrin (Sigma) (1:20) for 30 minutes each. The secondary antibodies used for anti-R29var were goat anti-mouse IgG-R-Phycoerythrin (Sigma) (1:20) for 30 minutes. Anti-V5 was further incubated with unlabelled rabbit anti-mouse IgG (H+L) (Southern Biotech) (1:50), mouse anti-rabbit IgG-R-Phycoerythrin (Southern Biotech) (1:50) and then goat anti-mouse IgG-R-Phycoerythrin (Sigma) (1:20) for 30 minutes each. SYTO 61 DNA dye (Invitrogen) (1:1000) was added to the last antibody incubation in each case. Stained cells were washed in PBS and analyzed on an LSRII FACS machine (BD Biosciences). Analysis was performed using FlowJo 8 (Tree Star, Inc).
Immunogold electron microscopy
Immuno-electron microscopy was performed according to methodology described in (Tokuyasu, 1973). Cells were purified over a SuperMACS column of magnetic beads, fixed in 80 mM phosphate buffer containing 4% paraformaldehyde (EMS) and 0.1% glutaraldehyde, embedded in 2% gelatin, and cryo-preserved in 2.3 M sucrose. Gelatin blocks were frozen in liquid nitrogen and cut in an ultramicrotome (Leica) at -110°C to achieve 90 nm thin sections. Immunolabeling was done in 0.8% BSA and 1% fish gelatin in PBS with rabbit polyclonal anti-GFP primary antibodies (diluted 1:30; Invitrogen) and 10 nm immunogold-coupled anti-rabbit IgG secondary antibodies (diluted 1:40; Electron Microscopy Sciences). For immunolabeling of cells prior to fixation, anti-GFP labeling was done in 2% BSA in PBS followed by fixation with 2% glutaraldehyde in 80 mM phosphate buffer. After immunolabeling, the sections were stained at 4°C in an aqueous solution of 0.3% uranyl acetate and 1.8% methylcellulose. Images were taken at room temperature in a Tecnai F20 FEG at 200keV at 14500 × magnification and processed with ImageJ (NIH). Tilt series for tomographic reconstruction were obtained at room temperature at 11500 × magnification. The sample was rotated ±60° in maximally 2° increments following the Saxton scheme (Saxton et al., 1984). The final tomograms were calculated with the “protomo” software package (Taylor et al., 1999) using a marker-free alignment and weighted backprojection. The supplementary movie S1, which shows tomographic reconstruction of R29var-V5-TM-ATS-GFP parasites stained with anti-GFP antibodies, can be viewed with H.264 capable movie players, including the open-source platform-independent “vlc player” (http://www.videolan.org/videolan).
Panning of infected RBCs on CSA and infected RBC binding assays
A sterile culture flask was coated with 0.1 mg/ml CSA overnight at 4°C and blocked with 2% BSA in PBS for 1 hour at 37°C. Infected RBCs were enriched by gelatin flotation and incubated in the coated flask for 2 hours. Non-adherent RBCs were removed by gentle washing with warm culture medium. Adherent (panned) RBCs were washed off with medium and cultured. Infected RBCs were panned a total of six times on CSA with periods of parasite replication between rounds of panning. Assessments of infected RBC binding to CD36 or CSA or infected rosetting were performed according to standard methodologies ((Rowe et al., 1997; Avril et al., 2008); see Supplementary Experimental Procedures for further details).
Live cell imaging
Infected erythrocytes at the trophozoite stage were enriched by gelatin flotation. Cells were incubated with BODIPY-TR-ceramide (diluted 1:500; Invitrogen) in culture medium for one hour at 37°C, followed by incubation with 1 μg/mL DAPI in PBS for 15 minutes. Cells were subsequently washed with PBS. A drop of cell suspension in PBS was then put on a slide, covered by a coverslip and pictures were taken on a Nikon Eclipse E600 microscope. Picture analyses and overlays were performed using Metamorph (Molecular Devices).
Supplementary Material
Acknowledgments
We extend our deep graduate to the members of the New York Structural Biology Center, in particular William Rice and KD Kerr, for their assistance with electron microscopy studies. The Center is a STAR center supported by the New York State Office of Science, Technology, and Academic Research. R.A.M. gratefully acknowledges support from the Cellular and Molecular Biology Training Grant (NIGMS T32 07491; PI. Dr. Pamela Stanley) and the Albert Einstein College of Medicine Medical Scientist Training Program (Director: Dr. Myles Akabas). We would like to also thank Sophie Adjalley, Marcus Lee, Micheline Guillotte and Alexandre Juillerat for their expert help with this work. Funding for this work was provided in part by the NIH (R01 AI47953; PI: J. Smith) and startup funds from the Albert Einstein College of Medicine and Columbia University (to D. Fidock).
References
- Andrews KT, Pirrit LA, Przyborski JM, Sanchez CP, Sterkers Y, Ricken S, et al. Recovery of adhesion to chondroitin-4-sulphate in Plasmodium falciparum varCSA disruption mutants by antigenically similar PfEMP1 variants. Mol Microbiol. 2003;49:655–669. doi: 10.1046/j.1365-2958.2003.03595.x. [DOI] [PubMed] [Google Scholar]
- Avril M, Kulasekara BR, Gose SO, Rowe C, Dahlback M, Duffy PE, et al. Evidence for globally shared, cross-reacting polymorphic epitopes in the pregnancy-associated malaria vaccine candidate VAR2CSA. Infect Immun. 2008;76:1791–1800. doi: 10.1128/IAI.01470-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Baruch DI, Ma XC, Singh HB, Bi X, Pasloske BL, Howard RJ. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- Boddey JA, Moritz RL, Simpson RJ, Cowman AF. Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic. 2009;10:285–299. doi: 10.1111/j.1600-0854.2008.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Falick AM, Carlton PM, Sedat JW, DeRisi JL, Marletta MA. N-terminal processing of proteins exported by malaria parasites. Mol Biochem Parasitol. 2008;160:107–115. doi: 10.1016/j.molbiopara.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Barragan A, Fernandez V, Sundstrom A, Schlichtherle M, Sahlen A, et al. Identification of Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) as the rosetting ligand of the malaria parasite P. falciparum. J Exp Med. 1998;187:15–23. doi: 10.1084/jem.187.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke BM, Buckingham DW, Glenister FK, Fernandez KM, Bannister LH, Marti M, et al. A Maurer's cleft-associated protein is essential for expression of the major malaria virulence antigen on the surface of infected red blood cells. J Cell Biol. 2006;172:899–908. doi: 10.1083/jcb.200509122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward TF, Gilson PR, Boddey JA, Rug M, Smith BJ, Papenfuss AT, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmendorf HG, Bangs JD, Haldar K. Synthesis and secretion of proteins by released malarial parasites. Mol Biochem Parasitol. 1992;52:215–230. doi: 10.1016/0166-6851(92)90054-n. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc Natl Acad Sci U S A. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock DA, Nomura T, Wellems TE. Cycloguanil and its parent compound proguanil demonstrate distinct activities against Plasmodium falciparum malaria parasites transformed with human dihydrofolate reductase. Mol Pharmacol. 1998;54:1140–1147. doi: 10.1124/mol.54.6.1140. [DOI] [PubMed] [Google Scholar]
- Frank M, Deitsch K. Activation, silencing and mutually exclusive expression within the var gene family of Plasmodium falciparum. Int J Parasitol. 2006;36:975–985. doi: 10.1016/j.ijpara.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehde N, Hinrichs C, Montilla I, Charpian S, Lingelbach K, Przyborski JM. Protein unfolding is an essential requirement for transport across the parasitophorous vacuolar membrane of Plasmodium falciparum. Mol Microbiol. 2009;71:613–628. doi: 10.1111/j.1365-2958.2008.06552.x. [DOI] [PubMed] [Google Scholar]
- Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estrano C, Haldar K. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- Horrocks P, Pinches R, Christodoulou Z, Kyes SA, Newbold CI. Variable var transition rates underlie antigenic variation in malaria. Proc Natl Acad Sci U S A. 2004;101:11129–11134. doi: 10.1073/pnas.0402347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khunrae P, Dahlback M, Nielsen MA, Andersen G, Ditlev SB, Resende M, et al. Full-length recombinant Plasmodium falciparum VAR2CSA binds specifically to CSPG and induces potent parasite adhesion-blocking antibodies. J Mol Biol. 2010;397:826–834. doi: 10.1016/j.jmb.2010.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilejian A, Rashid MA, Aikawa M, Aji T, Yang YF. Selective association of a fragment of the knob protein with spectrin, actin and the red cell membrane. Mol Biochem Parasitol. 1991;44:175–181. doi: 10.1016/0166-6851(91)90003-o. [DOI] [PubMed] [Google Scholar]
- Knuepfer E, Rug M, Klonis N, Tilley L, Cowman AF. Trafficking of the major virulence factor to the surface of transfected P. falciparum-infected erythrocytes. Blood. 2005;105:4078–4087. doi: 10.1182/blood-2004-12-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr Opin Microbiol. 2006;9:374–380. doi: 10.1016/j.mib.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, Christodoulou Z, et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriek N, Tilley L, Horrocks P, Pinches R, Elford BC, Ferguson DJ, et al. Characterization of the pathway for transport of the cytoadherence-mediating protein, PfEMP1, to the host cell surface in malaria parasite-infected erythrocytes. Mol Microbiol. 2003;50:1215–1227. doi: 10.1046/j.1365-2958.2003.03784.x. [DOI] [PubMed] [Google Scholar]
- Kumar N, Koski G, Harada M, Aikawa M, Zheng H. Induction and localization of Plasmodium falciparum stress proteins related to the heat shock protein 70 family. Mol Biochem Parasitol. 1991;48:47–58. doi: 10.1016/0166-6851(91)90163-z. [DOI] [PubMed] [Google Scholar]
- Lingelbach K, Przyborski JM. The long and winding road: protein trafficking mechanisms in the Plasmodium falciparum infected erythrocyte. Mol Biochem Parasitol. 2006;147:1–8. doi: 10.1016/j.molbiopara.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Lopez-Estrano C, Bhattacharjee S, Harrison T, Haldar K. Cooperative domains define a unique host cell-targeting signal in Plasmodium falciparum-infected erythrocytes. Proc Natl Acad Sci U S A. 2003;100:12402–12407. doi: 10.1073/pnas.2133080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AG, Braks JA, Waters AP, Cowman AF. Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol Biochem Parasitol. 2006;150:118–121. doi: 10.1016/j.molbiopara.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Maier AG, Rug M, O'Neill MT, Beeson JG, Marti M, Reeder J, Cowman AF. Skeleton-binding protein 1 functions at the parasitophorous vacuole membrane to traffic PfEMP1 to the Plasmodium falciparum-infected erythrocyte surface. Blood. 2007;109:1289–1297. doi: 10.1182/blood-2006-08-043364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Good RT, Rug M, Knuepfer E, Cowman AF. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- Marti M, Baum J, Rug M, Tilley L, Cowman AF. Signal-mediated export of proteins from the malaria parasite to the host erythrocyte. J Cell Biol. 2005;171:587–592. doi: 10.1083/jcb.200508051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- Muhle RA, Adjalley S, Falkard B, Nkrumah LJ, Muhle ME, Fidock DA. A var gene promoter implicated in severe malaria nucleates silencing and is regulated by 3′ untranslated region and intronic cis-elements. Int J Parasitol. 2009;39:1425–1439. doi: 10.1016/j.ijpara.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkrumah LJ, Muhle RA, Moura PA, Ghosh P, Hatfull GF, Jacobs WR, Jr, Fidock DA. Efficient site-specific integration in Plasmodium falciparum chromosomes mediated by mycobacteriophage Bxb1 integrase. Nature Methods. 2006;3:615–621. doi: 10.1038/nmeth904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SS, Voigt S, Fisher D, Yi SJ, LeRoy PJ, Derick LH, et al. Plasmodium falciparum erythrocyte membrane protein 1 is anchored to the actin-spectrin junction and knob-associated histidine-rich protein in the erythrocyte skeleton. Mol Biochem Parasitol. 2000;108:237–247. doi: 10.1016/s0166-6851(00)00227-9. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, Newbold CI. Rapid switching to multiple antigenic and adhesive phenotypes in malaria. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BA, Welch TL, Smith JD. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol Microbiol. 2003;47:1265–1278. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Salanti A, Dahlback M, Turner L, Nielsen MA, Barfod L, Magistrado P, et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W, Hahn M. Three-dimensional reconstruction of imperfect two-dimensional crystals. Ultramicroscopy. 1984;13:57–70. doi: 10.1016/0304-3991(84)90057-3. [DOI] [PubMed] [Google Scholar]
- Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, et al. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. Embo J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Kyes S, Craig AG, Fagan T, Hudson-Taylor D, Miller LH, et al. Analysis of adhesive domains from the A4VAR Plasmodium falciparum erythrocyte membrane protein-1 identifies a CD36 binding domain. Mol Biochem Parasitol. 1998;97:133–148. doi: 10.1016/s0166-6851(98)00145-5. [DOI] [PubMed] [Google Scholar]
- Smith JD, Craig AG, Kriek N, Hudson-Taylor D, Kyes S, Fagen T, et al. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proc Natl Acad Sci U S A. 2000a;97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol Biochem Parasitol. 2000b;110:293–310. doi: 10.1016/s0166-6851(00)00279-6. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Gangnard S, Round A, Dechavanne S, Juillerat A, Raynal B, et al. Full-length extracellular region of the var2CSA variant of PfEMP1 is required for specific, high-affinity binding to CSA. Proc Natl Acad Sci U S A. 2010;107:4884–4889. doi: 10.1073/pnas.1000951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Tamez PA, Bhattacharjee S, van Ooij C, Hiller NL, Llinas M, Balu B, et al. An erythrocyte vesicle protein exported by the malaria parasite promotes tubovesicular lipid import from the host cell surface. PLoS Pathog. 2008;4:e1000118. doi: 10.1371/journal.ppat.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KA, Schmitz H, Reedy MC, Goldman YE, Franzini-Armstrong C, Sasaki H, et al. Tomographic 3D reconstruction of quick-frozen, Ca2+-activated contracting insect flight muscle. Cell. 1999;99:421–431. doi: 10.1016/s0092-8674(00)81528-7. [DOI] [PubMed] [Google Scholar]
- Templeton TJ, Deitsch KW. Targeting malaria parasite proteins to the erythrocyte. Trends Parasitol. 2005;21:399–402. doi: 10.1016/j.pt.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin CJ, Carret CK, Duraisingh MT, Voss TS, Ralph SA, Hommel M, et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009;7:e84. doi: 10.1371/journal.pbio.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Trelka DP, Schneider TG, Reeder JC, Taraschi TF. Evidence for vesicle-mediated trafficking of parasite proteins to the host cell cytosol and erythrocyte surface membrane in Plasmodium falciparum infected erythrocytes. Mol Biochem Parasitol. 2000;106:131–145. doi: 10.1016/s0166-6851(99)00207-8. [DOI] [PubMed] [Google Scholar]
- Viebig NK, Gamain B, Scheidig C, Lepolard C, Przyborski J, Lanzer M, et al. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO Rep. 2005;6:775–781. doi: 10.1038/sj.embor.7400466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller KL, Cooke BM, Nunomura W, Mohandas N, Coppel RL. Mapping the binding domains involved in the interaction between the Plasmodium falciparum knob-associated histidine-rich protein (KAHRP) and the cytoadherence ligand P. falciparum erythrocyte membrane protein 1 (PfEMP1) J Biol Chem. 1999;274:23808–23813. doi: 10.1074/jbc.274.34.23808. [DOI] [PubMed] [Google Scholar]
- Waller KL, Nunomura W, Cooke BM, Mohandas N, Coppel RL. Mapping the domains of the cytoadherence ligand Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) that bind to the knob-associated histidine-rich protein (KAHRP) Mol Biochem Parasitol. 2002;119:125–129. doi: 10.1016/s0166-6851(01)00395-4. [DOI] [PubMed] [Google Scholar]
- Waterkeyn JG, Cowman AF, Cooke BM. Plasmodium falciparum: gelatin enrichment selects for parasites with full-length chromosome 2. implications for cytoadhesion assays. Exp Parasitol. 2001;97:115–118. doi: 10.1006/expr.2000.4593. [DOI] [PubMed] [Google Scholar]
- Wickert H, Wissing F, Andrews KT, Stich A, Krohne G, Lanzer M. Evidence for trafficking of PfEMP1 to the surface of P. falciparum-infected erythrocytes via a complex membrane network. Eur J Cell Biol. 2003;82:271–284. doi: 10.1078/0171-9335-00319. [DOI] [PubMed] [Google Scholar]
- Wickham ME, Rug M, Ralph SA, Klonis N, McFadden GI, Tilley L, Cowman AF. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 2001;20:5636–5649. doi: 10.1093/emboj/20.20.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.