Abstract
Background and aims
Occult hepatitis B virus infection (OBI) poses a challenge to the safety of blood donation. The prevalence of OBI is not well documented in Indonesia, although this information in such an endemic country is needed. This study was aimed to evaluate the prevalence of occult hepatitis B in blood donors from two cities of Indonesia, and to study the genetic variation and its effect on the predicted antigenicity of HBsAg.
Methods
Serum samples of 309 regular blood donors negative for HBsAg were tested for anti-HBs and anti-HBc. Hepatitis B virus (HBV) DNA isolated from anti-HBc-positive samples were analyzed by polymerase chain reaction, cloned and sequenced. Antigenic properties of identified HBsAg mutants were predicted by calculation of the antigenic index.
Results
Of the 309 HBsAg-negative samples, anti-HBc was positive in 134 (43.4%) and HBV DNA was detected in 25 (8.1%). Seven of the viremic samples had nucleotide substitutions (A521G, A551T, C582T, and A562G) in the S gene, causing amino acid mutations (T123A, M133L, and T143M) in the ‘a’ determinant of HBsAg that resulted in changes in the predicted antigenicity.
Conclusions
OBI was detected in blood donors’ samples in Indonesia. Anti-HBc was shown to be a better screening parameter than HBsAg, however, it might result in the loss of donors particularly in endemic countries. HBsAg detection failure in this study might be due to mutations altering the protein antigenicity and/or the low-level carriage of HBV.
Keywords: Hepatitis B, Hepatitis B virus, HBV, Occult hepatitis B, OBI, Antigenic index, Antigenic property, Blood donor
Background
Chronic hepatitis B virus (HBV) infection continues to be a global public health problem that affects an estimated 360 million individuals [1]. Two-thirds of these HBV carriers live in Asia–Pacific region where hepatitis B is the leading cause of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) [2]. It is of particular concern that Indonesia is a region with an intermediate-to-high level of hepatitis B endemicity [1].
One important mode of HBV transmission is through contaminated blood transfusion. The safety of blood transfusion has become an important issue, since occult HBV infection (OBI) has been detected and could be transmitted to the recipients [3, 4]. OBI is defined as the presence of HBV DNA in serum and/or liver without detectable HBsAg [5]. It is found in several conditions [5, 6]: (a) recovery from past infection defined by the presence of anti-HBs, (b) chronic hepatitis with surface gene escape mutants that are not, or poorly, recognized by current assays, (c) chronic carriage without any marker of HBV infection other than HBV DNA, referred to as seronegative, and (d) chronic carriage with HBsAg too low to be detected and recognized solely by the presence of anti-HBc.
The occurrence of OBI to a large extent depends on the prevalence of HBV infection in the general population. It is most common in regions where HBV infection is endemic [7, 8]. Since the first evidence of OBI was reported in 1979, there has been continuous increase in the number of publications on OBI covering various areas of bio-medical and public health aspects [9]. Most of the publications came from countries with low endemicity. The prevalence of occult hepatitis B in blood donors from two cities in Indonesia, to analyze the genetic characteristics of HBV, and to study the effect of the genetic alteration of HBV DNA on the predicted antigenicity of HBsAg.
Materials and methods
Study samples
A total of 309 serum samples of regular blood donors negative for HBsAg, anti-HCV and anti-HIV (aged 17–56, mean 28.97 ± 8.81 years; male/female 273/36) were used for this study. The samples were obtained from a serological surveillance for the main transfusion-transmitted infections including hepatitis B, conducted by the Indonesian Red Cross Blood Transfusion Unit in two cities of Indonesia, Solo in Java and Medan in Sumatra islands. Informed consent for participation in this study was obtained from each blood donor. All samples were collected in the year 2004–2005 and stored at −70°C until use. The study protocol was in accordance with and approved by the Eijkman Institute Research Ethics Commission (EIREC No. 24/2007).
Serological detection of HBV
Prior to this study, all regular blood donors from the two cities were tested by two immunoassay procedures, namely Murex HBsAg Version 3 (Abbott/Murex Biotech Ltd) for screening and Auzyme® Monoclonal (Abbott Laboratories) for confirmation. The 309 HBsAg-negative samples were considered for this study and examined by enzyme immunoassays for anti-HBs (AUSAB EIA®, Abbott Laboratories) and total anti-HBc (HBV Core Antigen CORZYME®, Abbott Laboratories) according to the manufacturer’s instruction. The quantification of anti-HBs was performed using Ausab Anti-HBs Quantitation Panel (Abbott Laboratories). Anti-HBs concentration equal to or greater than 10 mIU/mL was considered positive [1].
HBV DNA extraction and polymerase chain reaction
HBV DNA was isolated from 100 μL of HBsAg-negative and anti-HBc-positive sera by proteinase-K digestion and phenol–chloroform–isoamyl alcohol extraction [10]. The resulting precipitate was resuspended in 20 μL of double-distilled water and stored at −20°C. Nested polymerase chain reaction (PCR) was performed targeting a segment within the S gene that codes for the ‘a’ determinant of HBsAg. The outer primers were S2–1 and S1–2, while the inner primers were S088 and S2–2 [11, 12]. Sequences of the oligonucleotide primers are shown in Table 1. Denaturizing, annealing and extension were done at 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, respectively, for both rounds (35 cycles for the first and 25 for the second steps of PCR).
Table 1.
HBV DNA-specific primers used in nested PCR, clone analysis, and sequencing
| Primer | Nucleotide sequence (5′ → 3′) | Nucleotide position# | Polarity | Ref. |
|---|---|---|---|---|
| S2–1 | CAAGGTATGTTGCCCGTTTG | 455–474 | Sense | [28] |
| S088 | TGTTGCCCGTTTGTCCTCTA | 462–471 | Sense | [29] |
| S1–2 | GCCATTTGTTCAGTGGTTCG | 685–704 | Antisense | [28] |
| S2–2 | TGGCTCAGTTTACTAGTGCC | 668–687 | Antisense | [28] |
| PS8–1 | GTCACCATATTCTTGGGAAC | 2817–2836 | Sense | [30] |
| HS6–2 | GCCAAGTGTTTGCTGACGCA | 1175–1194 | Antisense | |
| HS4–2 | CCTATTGATTGGAAGGTGTG | 970–989 | Antisense | [30] |
| T728 | GGAATCAAACCTTATTATCC | 2688–2707 | Sense | [30] |
| T703 | CAGAGTCTAGACTCGTGGTG | 242–261 | Antisense | [30] |
| PS5–2 | CTCGTGTTACAGGCGGGGTT | 191–210 | Antisense | |
| M13F | GCCAGGGTTTTCCCAGTCACGAC | 2949–2972 | Sense | |
| M13R | GTCATAGCTGTTTCCTGTGTGA | 176–197 | Antisense |
Number based on EcoRI site numbering
To confirm the detection of HBV DNA and mutations in the ‘a’ determinant of the S gene, semi-nested PCR was also performed to amplify part of the overlapping P gene using primers PS8–1 and HS6–2 for the first round, and PS8–1 and HS4–2 for the second round (Table 1) [13]. To determine the presence of mutations in the S promoter that could affect the transcription of the S gene, a fragment covering the PreS1 region of the S gene was amplified by semi-nested PCR using primer set T728/T703 for the first round, and T728/PS5–2 for the second round, with primer sequences as shown in Table 1 [13].
The amplification products were visualized on ethidium bromide-stained 2% agarose gel under ultraviolet light. Kwok and Higuchi [14] rules were followed strictly in all experimental steps. The nucleotide positions of the primers used in this study are based on EcoRI site. Positive PCR products were purified using QIAquick™ PCR Purification kit (QIAGEN, Hilden, Germany).
Cloning and sequence analysis of S gene
Purified PCR products of S gene were ligated to pGEM®T Easy vector (Promega Co., Madison, WI, USA) and transformed into E. coli JM109. Transformed bacteria were selected by plating on Luria–Bertoni agar in the presence of ampicillin at 100 μg/mL and screened by PCR using primers S088 and S2–2. Six clones of each sample containing the HBV insert were selected and grown overnight in Luria-Bertoni broth containing 50 μg/mL ampicillin. Recombinant plasmids were recovered by standard alkaline lysis miniprep procedure and sequenced using the Bigdye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA, USA) on an automatic sequencer (Applied Biosystems 337 DNA, Perkin Elmer). Primers M13F and M13R were used as the sequencing primers (Table 1). Each clone was sequenced bidirectionally by two independent reactions. The nucleotide sequences comprising 226 bp fragment of the S gene were aligned and compared using the software BioEdit Sequence Alignment Editor Ver. 7.0.5.2 [15] with two wild-type HBV sequences, M54923 (Genotype B, adw) and AP011097 (Genotype C, adr) retrieved from GenBank [16, 17].
Direct sequencing of P gene and PreS1 region of S gene
Direct sequencing of purified PCR products of the P gene and PreS1 region of the S gene was also performed using the same methods. Each sample was also sequenced bidirectionally using PCR products from two independent reactions. The sequences obtained were aligned and compared with that of M54923 retrieved from Genbank.
Calculation of antigenicity and secondary structure
To determine whether the changes in amino acid sequence alter the antigenicity of HBsAg, the study of Jameson-Wolf Antigenic Index Prediction was performed using Lasergene Protean v8.1 program (DNASTAR Inc., Madison, WI, USA). The antigenicity index prediction study combines information from the primary amino acid sequence hydrophilicity (Hoop-Wood and Kyte Doolittle method), surface probability (Emini method), and backbone flexibility (Karplus–Schultz method) predictions, together with the secondary structure predictions (Chou-Fasman and Robson-Garner) [18, 19].
Results
Detection of occult HBV in blood donors
Of 309 HBsAg-negative blood donor samples, 134 (43.4%) were positive for total anti-HBc, referred to hereafter as anti-HBc, and therefore had serological evidence of prior and/or ongoing HBV infection. The remaining samples were negative for anti-HBc and excluded from this study. Of the 134 samples with anti-HBc, 68 (50.7%) had detectable anti-HBs and 66 (49.3%) were anti-HBs negative (referred to as isolated anti-HBc). HBV DNA was detected in 25 (18.7%) of all anti-HBc-positive samples, including 6 (8.8%) from the anti-HBs positive/anti-HBc positive and 19 (28.8%) from the isolated anti-HBc groups. Thus, the overall prevalence of OBI in 309 blood donors was 25 (8.1%). Of these, six (24%) were anti-HBs positive and two samples had antibody titer >100 mIU/mL. Demographic and serologic data of donors with HBV DNA are shown in Table 2.
Table 2.
Serological markers of HBV DNA-positive blood donors and mutation pattern of HBsAg
| No. | Donor ID | Age (years) | Sex (M/F) | HBsAg | Anti-HBc | Anti-HBs | Anti-HBs titer (mIU/mL) | HBV-DNA | HBsAg mutation |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2013 | 45 | L | – | + | + | 346 | + | T143M |
| 2 | 2509 | 41 | L | – | + | + | 103 | + | wt |
| 3 | 2314 | 20 | L | – | + | + | 59 | + | wt |
| 4 | 2096 | 45 | P | – | + | + | 46 | + | wt |
| 5 | 2050 | 22 | L | – | + | + | 39 | + | wt |
| 6 | 2542 | 38 | L | – | + | – | 10 | + | wt |
| 7 | 2350 | 25 | L | – | + | – | 9 | + | M133L |
| 8 | 2411 | 22 | L | – | + | – | 9 | + | wt |
| 9 | 2028 | 23 | L | – | + | – | 8 | + | wt |
| 10 | 2054 | 25 | L | – | + | – | 8 | + | wt |
| 11 | 2072 | 20 | L | – | + | – | 8 | + | T143M |
| 12 | 2362 | 35 | L | – | + | – | 8 | + | wt |
| 13 | 2407 | 27 | L | – | + | – | 8 | + | wt |
| 14 | 2412 | 23 | L | – | + | – | 8 | + | wt |
| 15 | 2537 | 29 | L | – | + | – | 8 | + | wt |
| 16 | 2083 | 28 | L | – | + | – | 7 | + | T143M |
| 17 | 2361 | 37 | L | – | + | – | 7 | + | T123A |
| 18 | 2414 | 21 | L | – | + | – | 7 | + | wt |
| 19 | 2427 | 30 | L | – | + | – | 7 | + | wt |
| 20 | 2182 | 32 | L | – | + | – | 6 | + | T143M |
| 21 | 2392 | 35 | L | – | + | – | 5 | + | wt |
| 22 | 2524 | 26 | L | – | + | – | 5 | + | wt |
| 23 | 2357 | 25 | L | – | + | – | 4 | + | wt |
| 24 | 2133 | 28 | L | – | + | – | 3 | + | T143M |
| 25 | 2351 | 23 | L | – | + | – | 3 | + | wt |
A level of anti-HBs equal to or higher than 10 mIU/mL was considered positive [1]
The sensitivity of the nested PCR was validated using a panel of sera with various HBV DNA titers tested by COBAS TaqMan 48 Real-Time PCR (Roche Molecular System, Branchburg, NJ, USA). The method was capable of detecting HBV DNA at titers lower than the detection limit of the COBAS TaqMan 48 Real-Time PCR (6 IU/mL). Thus, it met the sensitivity requirement for detection of occult hepatitis B of less than 10 IU/mL [5].
Analysis of nucleotide sequences and protein products
Of the 150 clones derived from 25 HBV DNA-positive samples, 42 (28%) from 7 samples had nucleotide substitutions: 6 clones from 1 sample exhibiting A521G substitution, 6 clones from 1 sample with A551T and A562G substitutions, and the other 30 clones from 5 samples with C582T substitution.
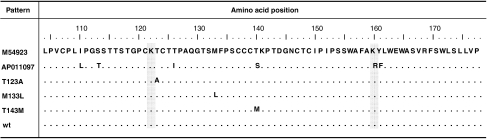
Three of these four substitution patterns, A521G, A551T, and C582T, caused mutations within the ‘a’ determinant: T123A in one sample, M133L in one sample, and T143M in five samples, respectively, whereas pattern A562G caused silent mutation. The HBsAg mutation patterns identified in these HBV DNA-positive samples are shown in Table 2 and Fig. 1. As a consequence of HBV overlapping open reading frames, the nucleotide changes in the S gene (A521G, A551T, and A562G) were associated with amino acid alterations in the reverse-transcriptase domain of the HBV polymerase protein: rtN131S, rtY141F, and rtM145V, respectively, while C582T caused silent mutation [20]. The remaining 108 (72%) clones from 18 samples had identical nucleotides to that of the M54923 sequence, even when the screening was extended to 10 additional clones of each sample. The 42 nucleotide sequences from 7 samples with substitution and representations from each of the 18 samples with wild-type HBV were deposited at GenBank (AN EF507434–EF507475 and HM116516–HM116533). Analysis of PreS1 sequences generated from direct sequencing showed wild-type HBV in all samples.
Fig. 1.
Mutation pattern of the hepatitis B surface antigen (HBsAg) of 25 blood donor samples with HBV DNA. M54923 (Genotype B, adw) and AP011097C (C, adr) obtained from GenBank are used as reference sequences. Dots represent amino acids identical to those in the M54923. Amino acids that determine HBsAg serotype are shaded. Three substitution patterns were observed: T123A (in 1 donor), M133L (in 1 donor), and T143M (in 5 donors). wt represents sequences with no amino acid substitution in 18 donors
Alteration in predicted antigenicity of HBsAg
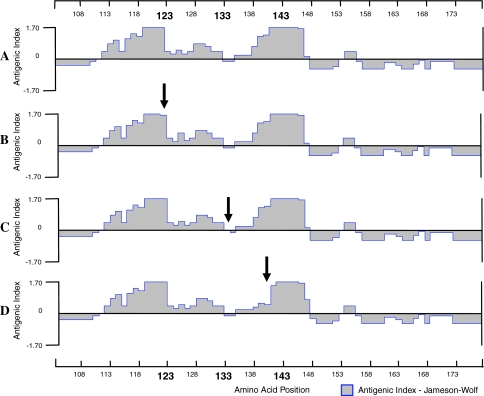
To determine the effect of amino acid changes to HBV surface protein antigenicity, Jameson and Wolf antigenic index prediction was conducted [19]. All mutation patterns found in this study showed changes in the secondary structure and the predicted antigenicity of HBsAg (Fig. 2). Sequence with T123A pattern, a substitution located one residue upstream of the first HBsAg loop, only experienced relatively minor and localized change in the antigenicity index at position 123 and its close proximity compared to the wild type. In M133L pattern, with substitution in the first loop of HBsAg in the secondary structure prediction, the effect was also slight and localized (−0.05 instead of −0.2 at residue 133). However, sequences with T143M pattern, the most prevalent variant found in this study, showed significant decrease in the pattern of antigenicity of the second HBsAg loop, which was observed in a larger area surrounding the amino acid substitution (Fig. 2).
Fig. 2.
Antigenicity plots based on amino acid sequences of the ‘a’ determinant region of HBsAg. Antigenicity plots of the reference sequence (M54923) (a) and of HBV mutants isolated from blood donors found in this study, with amino acid changes T123A (b), M133L (c), and T143M (d). The change in each antigenic index is indicated by an arrow, with the most significant alteration observed in the T143M substitution, while T123A and M133L show minor antigenicity changes
Discussion
The implications of HBV infection involve several clinical aspects, varying from chronic asymptomatic carriers to complicated liver diseases including liver cirrhosis and HCC [1, 2]. OBI has appeared to have similar infectivity and pathogenicity in the development of fulminant hepatitis, liver cirrhosis and HCC [7], and would possibly affect the safety of blood transfusion [21].
OBI is also related to the endemicity of HBV infection. It is most commonly reported in high endemic areas and infrequently detected in low endemic areas [7, 8]. In this study, OBI was detected in 8.1% of regular blood donors’ samples, nearly equaling the average 9.4% HBsAg prevalence in Indonesia [22]. It was higher than those found elsewhere (7% in Taiwan and 1.4% in Ghana) [21, 23]. The evidence that OBI in blood donors with negative HBsAg status is not negligible, alarms us.
The detection of anti-HBc in HBsAg-negative individuals has been considered a marker of past HBV exposure and/or of resolved infection. However, the application of molecular biology techniques has shown that HBV viremia are detectable in 1.33–38% of HBsAg-negative/anti-HBc-positive donors [24, 25]. In this study, HBV DNA was detected in 25 (18.7%) of anti-HBc-positive regular blood donors with higher frequency of HBV DNA in isolated anti-HBc subjects than in those with anti-HBc and anti-HBs. This finding highlights the importance of anti-HBc compared to other serological HBV markers for predicting latent HBV infection in apparently healthy individuals, and reiterates that the implementation of anti-HBc screening would improve the safety of blood supply [25–27]. However, in highly endemic regions including Indonesia, anti-HBc screening would be impractical due to the high loss of potential donors (approximately, 70% of isolated anti-HBc donors). This study could give support to the potential use of molecular detection as an alternative once it is widely available at a lower cost for public health.
The presence of anti-HBs and anti-HBc is usually indicative of immunity after infection. In some countries such as Germany, Austria and Japan, blood units with anti-HBs levels greater than 100 IU/L is considered to be safe [26]. However, there was evidence that transmission of HBV from occult hepatitis B subjects occurred in the presence of concurrent neutralizing anti-HBs in the same specimen [28]. Detection of HBV DNA in some anti-HBs-positive samples in this study indicates that the absence of HBsAg and the presence of anti-HBs do not totally reflect the safety of blood donations. A similar report from Italy also supports the notion that some blood donors with anti-HBs titer over 100 IU/L still had detectable HBV DNA [8]. Overall, these results raise several important public health issues: the absence of HBsAg as HBV infection marker does not exclude the possibility of viral transmission; anti-HBc-positive sera potentially contain HBV; and the presence of anti-HBs is not a sign of total HBV eradication.
In this study, although the frequently emerged variant G145R was not found, some other variants showing single amino acid substitution patterns of the ‘a’ determinant, which had been reported previously, were observed [29–31]. All isolates showing T123A and M133L substitutions, together with 4 of the T143M isolates, were obtained from isolated anti-HBc samples; while one of the T143M isolates was found in samples with anti-HBc and anti-HBs. This finding might suggest that mutation in the ‘a’ determinant was more frequently observed in the isolated anti-HBc samples (Table 2).
Conformationally-dependent antigenic determinant might be affected by changes of its amino acid residues [32, 33]. While M133L substitution did not show significant alteration, both T123A and T143M mutations demonstrated results that should be taken into account; pattern T123A resided in close proximity to the cysteine residues at 121 and 124, which form disulfide bonds that are important for maintaining the ‘a’ determinant’s conformation [30, 33]. This close proximity substitution might cause alteration in the steric hindrance that would disturb the disulfide bonds, and hence might affect HBsAg conformation and its detection. The other substitution, T143M, caused marked alterations demonstrated by extensive changes of antigenic index of the mutated amino acid and its surroundings. This pattern affected the second loop of the ‘a’ determinant, which was more antigenic than the first [32, 34]. Thus, mutations in the second loop would more significantly disrupt the HBsAg antigenicity. These substitutions might partly explain the detection failure of HBsAg in this study.
It is acknowledged that this conclusion is based on mathematical modeling and may not reflect actual changes in antibody recognition. Further protein model prediction based on these amino acid substitution patterns might explain the conformational changes of HBsAg, and assays to confirm reduction in binding affinity of the altered epitopes to monoclonal anti-HBs are suggested.
As a consequence of gene overlapping, the nucleotide substitutions also caused amino acid mutations in the HBV polymerase: rtN131S, rtY141F, and rtM145V. These mutations lay between domain A and B of the reverse-transcriptase region of polymerase protein, which is crucial for its function in the HBV replication process [35, 36]. One or several of the identified mutations could be responsible for diminished rate of replication causing the detection failure of HBsAg.
In this study, 72% of the viremic donors had HBV DNA with the wild-type S gene. This finding could indicate that there were other factors besides antigenic property that might cause negative HBsAg status. Since all the samples were negative for anti-HCV and had HBV of the same genotype and serotype, the low viral load titer would provide alternative explanation for the wild-type HBV undetectable by HBsAg serological assay. This is supported by the fact that HBV DNA in these samples was detectable only in the second-round of nested PCR.
In conclusion, occult Hepatitis B was detected in samples of regular blood donors from Indonesia. Failure of HBV detection in these cases demonstrated that anti-HBc seemed to be a better screening parameter than HBsAg. Amino acid mutation in the S gene, which alters HBsAg antigenic property, may in part be the molecular background of the failure of HBsAg detection. Another factor that contributes to the insensitivity of the assay could be the low titer of viral load. Further experimental studies are needed to confirm the changes in antigenicity of these HBsAg variants. Studies involving more samples from various regions in Indonesia are important for investigating the magnitude of occult hepatitis B infection and the characteristics of occult HBV strains among blood donors in Indonesia.
Footnotes
An erratum to this article can be found at http://dx.doi.org/10.1007/s12072-010-9212-4
References
- 1.World Health Organization. Hepatitis B vaccines: weekly epidemiological record. WHO annual report. Switzerland; 2009, 84:405–420
- 2.Liaw YF, Leung N, Kao JH, Piratvisuth T, Gane E, Han KH, Guan R, Lau GKK, Locarnini S. Asian–Pacific consensus statement on the management of chronic hepatitis B: a 2008 update. Hepatol Int. 2008;2:263–283. doi: 10.1007/s12072-008-9080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoofnagle JH, Seeff LB, Bales ZB, Zimmerman HJ. Type B hepatitis after transfusion with blood containing antibody to hepatitis B core antigen. N Engl J Med. 1978;298:1379–1383. doi: 10.1056/NEJM197806222982502. [DOI] [PubMed] [Google Scholar]
- 4.Levicnik-Stezinar S, Rahne-Potokar U, Candotti D, Lelie N, Allain JP. Anti-HBs positive occult hepatitis B virus carrier blood infectious in two transfusion recipients. J Hepatol. 2008;48:1022–1025. doi: 10.1016/j.jhep.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 5.Raimondo G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxi A, Donato F, Ferrari C, Gaeta GB, Gerlich WH, Levrero M, Locarnini S, Michalak T, Mondelli MU, Pawlotsky JM, Pollicino T, Prati D, Puoti M, Samuel D, Shouval D, Smedile A, Squadrito G, Trepo C, Villa E, Will H, Zanetti AR, Zoulim F. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J Hepatol. 2008;49:652–657. doi: 10.1016/j.jhep.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Allain JP. Occult hepatitis B virus infection: implications in transfusion. Vox Sang. 2004;86:83–91. doi: 10.1111/j.0042-9007.2004.00406.x. [DOI] [PubMed] [Google Scholar]
- 7.Torbenson M, Thomas DL. Occult hepatitis B. Lancet Infect Dis. 2002;2:479–486. doi: 10.1016/S1473-3099(02)00345-6. [DOI] [PubMed] [Google Scholar]
- 8.Manzini P, Girotto M, Borsotti R, Giachino O, Guaschino R, Lanteri M, Testa D, Ghiazza P, Vacchini M, Danielle F, Pizzi A, Valpreda C, Castagno F, Curti F, Magistroni P, Abate ML, Smedile A, Rizzetto M. Italian blood donors with anti-HBc and occult hepatitis B infection. Haematologica. 2007;92:1664–1670. doi: 10.3324/haematol.11224. [DOI] [PubMed] [Google Scholar]
- 9.Chemin I, Trepo C. Clinical impact of occult HBV infections. J Clin Virol. 2005;34(Suppl 1):S15–S21. doi: 10.1016/S1386-6532(05)80005-8. [DOI] [PubMed] [Google Scholar]
- 10.Okamoto H, Yano K, Nozaki Y, Matsui A, Miyazaki H, Yamamoyo K, Tsuda F, Machida A, Mishiro S. Mutation within the S gene of hepatitis B virus transmitted from mothers to babies immunized with hepatitis B immunoglobulin and vaccine. Pediatric Res. 1992;32:264–268. doi: 10.1203/00006450-199209000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Iizuka H, Ohmura K, Ishijima A, Satoh K, Tanaka T, Tsuda F, Okamoto H, Miyakawa Y, Mayumi M. Correlation between anti-HBc titers and HBV DNA in blood units without detectable HBsAg. Vox Sang. 1992;63:107–111. doi: 10.1111/j.1423-0410.1992.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto H, Nishizawa T. Non-B Non-C Non-G hepatitis virus gene, polynucleotide, polypeptide, virion, method for separating virion, and method for detecting virus; 1992. http://www.freepatentsonline.com/EP1010759.html (with permission)
- 13.Takahashi K, Akahane Y, Hino K, Ohta Y, Mishiro S. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: comparative analysis of 40 full-length isolates. Arch Virol. 1998;143:2313–2326. doi: 10.1007/s007050050463. [DOI] [PubMed] [Google Scholar]
- 14.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp. 1999;41:95–98. [Google Scholar]
- 16.Sastrosoewignjo RI, Omi S, Okamoto H, Mayumi M, Rustam M, Sujudi T. The complete nucleotide sequence of HBV DNA clone of subtype adw (pMND122) from Menado in Sulawesi Island, Indonesia. ICMR Ann 1987;7:51–60.
- 17.Mulyanto, Depamede SN, Surayah K, Tsuda F, Ichiyama K, Takahashi M, Okamoto H. A nationwide molecular epidemiological study on hepatitis B virus in Indonesia: identification of two novel subgenotypes, B8 and C7. Arch Virol. 2009;154(7):1047–1059. doi: 10.1007/s00705-009-0406-9. [DOI] [PubMed] [Google Scholar]
- 18.Jameson BA, Wolf H. The antigenic index: a novel algorithm for predicting antigenic determinants. Cabios. 1988;4:181–186. doi: 10.1093/bioinformatics/4.1.181. [DOI] [PubMed] [Google Scholar]
- 19.Ghany MG, Ayola B, Villamil FG, Gish RG, Rojter S, Vierling JM, Lok AS. Hepatitis B Virus mutants in liver transplant recipients who were reinfected despite Hepatitis B Immune Globulin prophylaxis. Hepatology. 1998;27:213–221. doi: 10.1002/hep.510270133. [DOI] [PubMed] [Google Scholar]
- 20.Stuvyer LJ, Locarnini SA, Lok A, Richman DD, Carman WF, Dienstag JL, Schinazi RF. The HEP DART IC. Nomenclature for antiviral-resistant human Hepatitis B Virus mutations in the polymerase region. Hepatology. 2001;33:751–757. doi: 10.1053/jhep.2001.22166. [DOI] [PubMed] [Google Scholar]
- 21.Wang JT, Lee CZ, Chen PJ, Wang TH, Chen DS. Transfusion-transmitted HBV infection in an endemic area: the necessity of more sensitive screening for HBV carriers. Transfusion. 2002;42:1592–1597. doi: 10.1046/j.1537-2995.2002.00274.x. [DOI] [PubMed] [Google Scholar]
- 22.Khan M, Dong JJ, Acharya SK, Dhagwahdorj Y, Abbas Z, Jafri SMW, Mulyono DH, Tozun N, Sarin SK. Hepatology issues in Asia: perspectives from regional leaders. J Gastroenterol Hepatol. 2004;19:S419–S430. doi: 10.1111/j.1440-1746.2004.03728.x. [DOI] [Google Scholar]
- 23.Owusu-Ofori S, Temple J, Sarkodie F, Anokwa M, Candotti D, Allain JP. Predonation screening of blood donors with rapid test: implementation and efficacy of a novel approach to blood safety in resource-poor settings. Transfusion. 2005;45:133–140. doi: 10.1111/j.1537-2995.2004.04279.x. [DOI] [PubMed] [Google Scholar]
- 24.Yotsuyanagi H, Yasuda K, Moriya K, Shintani Y, Fujie H, Tsutsumi T, Nojiri N, Juji T, Hoshino H, Shimoda K, Hino K, Kimura S, Iino S, Koike K. Frequent presence of HBV in the sera of HBsAg-negative, anti-HBc-positive blood donors. Transfusion. 2001;41(9):1093–1099. doi: 10.1046/j.1537-2995.2001.41091093.x. [DOI] [PubMed] [Google Scholar]
- 25.El-Sherif AM, Abou-Shady MA, Al-Hiatmy MA, Al-Bahrawy AM, Motawea EA. Screening for hepatitis B virus infection in Egyptian blood donors negative for hepatitis B surface antigen. Hepatol Int. 2007;1:469–470. doi: 10.1007/s12072-007-9017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollinger FB. Hepatitis B virus infection and transfusion medicine: science and the occult. Transfusion. 2008;48:1001–1026. doi: 10.1111/j.1537-2995.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 27.Niederhauser C, Taleghani BM, Graziani M, Stolz M, Tinguely C, Schneider P. Blood donor screening: how to decrease the risk of transfusion-transmitted hepatitis B virus? Swiss Med Wkly. 2008;138(9–10):134–141. doi: 10.4414/smw.2008.12001. [DOI] [PubMed] [Google Scholar]
- 28.Matsumoto C, Tadokoro K, Fujimura K, Hirakawa S, Mitsunaga S, Juji T. Analysis of HBV infection after blood transfusion in Japan through investigation of a comprehensive donor specimen repository. Transfusion 2001;41:878–884. [DOI] [PubMed]
- 29.Ho MS, Mau YC, Lu CF, Huang SF, Hsu LC, Lin SR, Hsu HM. Patterns of circulating hepatitis B surface antigen variants among vaccinated children born to hepatitis B surface antigen carrier and non-carrier mothers: a population-based comparative study. J Biomed Sci. 1998;5:355–362. doi: 10.1007/BF02253445. [DOI] [PubMed] [Google Scholar]
- 30.Hou JL, Wang ZH, Cheng JJ, Lin YL, Lau GKK, Sun J, Zhou FY, Waters J, Karayiannis P, Luo KX. Prevalence of naturally occuring surface gene variants of hepatitis B virus in non-immunized surface antigen-negative Chinese carriers. Hepatology. 2001;34:1027–1034. doi: 10.1053/jhep.2001.28708. [DOI] [PubMed] [Google Scholar]
- 31.Thakur V, Kazim SN, Guptan RC, Hasnain SE, Bartholomeusz A, Malhotra V, Sarin SK. Transmission of G145R mutant of HBV to an unrelated contact. J Med Virol. 2005;76:40–46. doi: 10.1002/jmv.20321. [DOI] [PubMed] [Google Scholar]
- 32.Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-A. [DOI] [PubMed] [Google Scholar]
- 33.Seddigh-Tonekaboni S, Waters JA, Jeffers S, Gehrke R, Ofenloch B, Horsch A, Hess G, Thomas HC, Karayiannis P. Effect of variation in the common “a” determinant on the antigenicity of hepatitis B surface antigen. J Med Virol. 2000;60:113–121. doi: 10.1002/(SICI)1096-9071(200002)60:2<113::AID-JMV2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 34.Zuckerman AJ. Effect of hepatitis B virus mutants on efficacy of vaccination. Lancet. 2000;355:1382–1383. doi: 10.1016/S0140-6736(00)02132-2. [DOI] [PubMed] [Google Scholar]
- 35.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64(2):613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torresi J, Earnest-Silveira L, Civitico G, Walters TE, Lewin SR, Fyfe J, Locarnini SA, Manns M, Trautwein C, Bock TC. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the “Fingers” subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology. 2002;299:88–99. doi: 10.1006/viro.2002.1448. [DOI] [PubMed] [Google Scholar]