SUMMARY
Huntington’s disease (HD) is an incurable neurodegenerative disease caused by neuronal accumulation of the mutant protein huntingtin. Improving clearance of the mutant protein is expected to prevent cellular dysfunction and neurodegeneration in HD. We report here that such clearance can be achieved by posttranslational modification of the mutant Huntingtin (Htt) by acetylation at lysine residue 444 (K444). Increased acetylation at K444 facilitates trafficking of mutant Htt into autophagosomes, significantly improves clearance of the mutant protein by macroautophagy, and reverses the toxic effects of mutant huntingtin in primary striatal and cortical neurons and in a transgenic C. elegans model of HD. In contrast, mutant Htt that is rendered resistant to acetylation dramatically accumulates and leads to neurodegeneration in cultured neurons and in mouse brain. These studies identify acetylation as a mechanism for removing accumulated protein in HD, and more broadly for actively targeting proteins for degradation by autophagy.
INTRODUCTION
Accumulation and aggregation of mutant proteins is a hallmark of several neurodegenerative disorders such as Parkinson’s, Alzheimer’s, and Huntington’s disease (HD) (Ross and Poirier, 2004). One of the major therapeutic challenges in the field of neurodegeneration has been to improve the degradation of accumulated mutant proteins. While the ubiquitin-proteosome system (UPS) represents an important defense against abnormal protein accumulation, aggregation-prone proteins appear to be poor substrates for proteosomal degradation and better targets for autophagic-lysosomal degradation (Levine and Kroemer, 2008). In terms of the mode of cargo delivery to the lysosome, three forms of autophagic degradation have been described so far—microautophagy, chaperone-mediated autophagy, and macroautophagy (Klionsky, 2007). This latter form, whereby cytosolic constituents and organelles are engulfed by multilamellar vesicles which then fuse to the lysosome, has been implicated in a wide array of neurological disorders including HD (Cuervo, 2004; Nixon, 2005; Levine and Kroemer, 2008).
Huntington’s disease is a devastating neurodegenerative disorder characterized by progressive and severe motor and cognitive impairment; death ensues about 15 years after the onset of symptoms (Vonsattel and DiFiglia, 1998). The mutation is inherited as autosomal dominant and causes expansion of a stretch of glutamines near the N terminus of huntingtin, a protein of uncertain function whose mutant form accumulates as nuclear and cytoplasmic inclusions in HD brain (DiFiglia et al., 1997). In a conditional mouse model of HD, it was found that elimination of mutant Huntingtin (Htt) expression not only halted symptomatic progression but also led to regression of the disease-like symptoms (Yamamoto et al., 2000). Initial experiments in human HD brain found aberrant accumulation of huntingtin in late endosomal structures, suggesting dependence on autophagy (Sapp et al., 1997). Recent findings showed that activation of autophagy by systemic administration of rapamycin may be sufficient to partially ameliorate symptoms in an HD mouse model (Ravikumar et al., 2004). While these and other studies demonstrate neuroprotection by the inhibition of the ubiquitous protein kinase mTOR and nonspecific activation of autophagy, it remains unclear whether autophagy can be selectively activated in order to remove disease proteins of interest.
In this study, we demonstrate a link between acetylation of a nonhistone protein and targeted degradation by autophagy. Modification of mutant huntingtin by acetylation promotes its targeting into autophagosomes and facilitates specific degradation of the mutant protein by the autophagiclysosomal pathway. Furthermore, we show that acetylation and clearance of mutant huntingtin leads to neuroprotection in primary neurons and a transgenic C. elegans model of HD, highlighting the importance of selective targeting of disease proteins to autophagosomes for degradation.
RESULTS
Mutant Huntingtin Is Acetylated at Lysine 444
Recent studies demonstrate that mutant Htt interacts directly with the histone acetyltransferase (HAT) domain of CREB-binding protein (CBP) (Steffan et al., 2001), suggesting possible mutant Htt acetylation by CBP. To determine whether Htt gets acetylated, COS-7 cells were transiently transfected with mutant Htt containing the N-terminal 480 amino acids and 68 glutamines (Htt480-68Q), treated with a combination of HDAC inhibitors trichostatin A (TSA) and nicotinamide (NAM), and subjected to tandem mass spectrometry (MS). Using three different mass spectrometers at separate facilities, we repeatedly identified a single acetyl-lysine-containing peptide (GKAcVLLGEEEA LEDDSESR) mapping the acetylated lysine (K) to position 444 of human Htt (K444; Figure 1A). Protein sequence alignment of Htt homologs from different species demonstrated conservation of human K444 residue in mouse, rat, zebrafish, and pufferfish (Figure S1A available online). Interestingly, K444 was identified within the caspase 6 fragment of mutant Htt (586aa) that represents the required cleavage step for neurodegeneration due to mutant Htt (Graham et al., 2006a).
Figure 1. Mutant Huntingtin Is Acetylated at Lysine 444.
(A) LC-MS/MS spectrum of acetylated peptide GKAcVLLGEEEALEDDSESR obtained from Htt480-68Q. The unfragmented peptide as well as a series of b-ions displayed a mass change of +42 Da, indicative of acetylation.
(B) Specificity of the Ac-K444 antibody was determined in COS-7 cells transfected with Htt590-97Q or Htt590-97Q-KR, treated with TSA and NAM, and analyzed by western blotting.
(C) Htt480-68Q was transfected into COS-7 cells together with indicated HATs. Levels of total and acetylated Htt are shown. β-tubulin was used as loading control.
(D) CBP-HAT increases acetylation of Htt. COS-7 cells were transfected with Htt480-68Q and CBP-HAT domain, or HAT-deficient CBP-HAT-DY and Htt detected with MAB5490 and AcK444 antibodies. CBP-HAT levels were analyzed by HA antibody.
(E) Htt is deacetylated by HDAC1. Neuro2a cells were transfected with Htt480-68Q along with CBP-HAT, vector, or HDACs 1–5 and levels of total and acetyl-Htt determined by western blotting. β-tubulin was used as loading control. Equal expression of all HAT and HDAC constructs was confirmed by western blotting (not shown). All data are representative of at least three independent experiments.
(F) Treatment with HDAC inhibitors increased acetylation of Htt at K444. Neuro2a cells were transfected with Htt480-17Q or Htt480-68Q and treated with TSA and NAM. Htt was immunoprecipitated with anti-Htt antibody (MAB5490) and probed with AcK444 and MAB5490 antibodies. All data are representative of at least three independent experiments.
(G) Full-length mutant Htt is acetylated in knock-in mouse brains. Brain samples of 140Q knock-in mice were analyzed with Htt antibody (MAB 5490; upper panel) and acetyl-Htt antibody (lower panel). Comparison of wild-type (HdhQ7/Q7), heterozygous (HdhQ7/Q140), and homozygous (HdhQ140/Q140) animals revealed acetylation of only full-length mutant Htt (lower panel). At least three independent samples were examined and a representative blot is presented.
(H) Homogenates from 111Q knock-in mouse model were analyzed as in (G). The comparison of wild-type (HdhQ7/Q7) and heterozygous (HdhQ7/Q111) littermates confirmed that only mutant Htt is acetylated in vivo (lower panels).
(I) Mutant huntingtin is acetylated in postmortem human HD brain. Htt was immunoprecipitated from postmortem brain tissues of presymptomatic heterozygous HD subjects (grades 0–1) and advanced HD subjects (grades 3–4) (Figure S3A) and analyzed as in (G). The upper band representing mutant huntingtin was acetylated at K444.
In order to further characterize acetylation of Htt at K444, a specific antibody against acetylated K444 (AcK444) was generated. Using a dot blot assay, we demonstrated that the antibody specifically reacted to K444-acetylated peptide but not to the native peptide (Figure S1B). To further assess the specificity of the antibody, lysine 444 was mutated to arginine (R) in an expression vector encoding the N-terminal domain of Htt with 97 glutamines (Htt590-97Q), generating an Htt590-97Q-KR mutant construct. When overexpressed in COS-7 cells in the presence of HDAC inhibitors, Htt590-97Q was detected by the AcK444 antibody, but the K444R mutation completely abrogated reactivity of AcK444 to Htt, demonstrating specificity of AcK444 antibody for acetylated K444 (Figure 1B).
Next, we sought to identify the histone acetyltransferase (HAT) and deacetylase (HDAC) responsible for acetylation/deacetylation of Htt at K444. A number of known HATs and HDACs were examined by cotransfection together with mutant Htt (Htt480-68Q). While p300, P/CAF, Tip60, HAT1, or HBO1 and a number of HDACs did not alter Htt acetylation, CBP and HDAC1 strongly increased and decreased the acetylation of mutant Htt, respectively (Figures 1C and 1E). Consistent with a previous finding that the glutamine-rich domain of CBP (CBPΔQ) is not required for the CBP/Htt interaction (Steffan et al., 2001), CBPΔQ also efficiently acetylated mutant Htt (Figure 1C). To determine if the HAT domain of CBP was sufficient for Htt acetylation, CBP-HAT was subcloned into a mammalian expression vector (Figure S2). While CBP-HAT potently induced acetylation of Htt, mutagenesis of conserved aspartic acid residue (D1435) to tyrosine (Y) in the HAT catalytic domain (CBP-HAT-DY) that eliminates HAT activity of CBP completely abolished the ability of CBP-HAT to acetylate Htt at K444 (Figure 1D). Together, these findings demonstrate the requirement of the functional HAT domain in CBP for acetylation of Htt and suggest that a specific subset of HATs and HDACs modulates Htt acetylation.
Mutant Huntingtin Is Acetylated at Lys444 in Heterozygous Mouse and Human HD Brain
Using the specific AcK444 antibody we next examined the acetylation status of a 480 aa fragment of wild-type huntingtin (Htt480-17Q) and mutant huntingtin (Htt480-68Q) in Neuro2a cells. As shown in Figure 1F, treatment with HDAC inhibitors primarily augmented the acetylation of mutant huntingtin while acetylation of wild-type huntingtin was barely detectable despite higher levels of the wild-type protein expression.
In order to examine acetylation of endogenous huntingtin in the absence of overexpression, we tested brain samples from wild-type, homozygous, or heterozygous knock-in mouse models of HD that carry 111Q or 140Q polyglutamine stretches knocked into the murine HD gene homolog (Menalled et al., 2003; Wheeler et al., 1999). Surprisingly, probing with the AcK444 antibody revealed that only mutant Htt was acetylated in both the 140Q HD mouse brain (Figure 1G) and the 111Q HD mouse brain (Figure 1H). Similarly, in heterozygous human HD brain samples, only acetylation of the mutant but not the wild-type form of Htt protein was detected in early or advanced stages of disease (Figures 1I and S3A). These in vivo results were consistent with the preferential acetylation of mutant Htt observed in cultured cells (Figure 1F).
To examine, as a proof-of-principle, whether Htt acetylation can be increased in mouse brains, heterozygous knock-in HD mice (HdH7Q/140Q) were treated with a combination of TSA and NAM for 10 days. Consistent with the results in cultured cells (Figure 1F), we found that HDAC inhibitors increased acetylation of the mutant but not the wild-type form of Htt (Figure S3B). Since only the mutant protein was acetylated, we used the ratio of mutant Htt over wild-type Htt to determine the effects of acetylation on total huntingtin protein levels. This approach revealed a decrease in mutant but not the wild-type Htt levels in treated animals (Figure S3B), suggesting that preferential acetylation of mutant Htt in vivo may affect the stability of the mutant protein.
Acetylation-Resistant Mutant Htt Accumulates in Cultured Neurons and in Mouse Brain
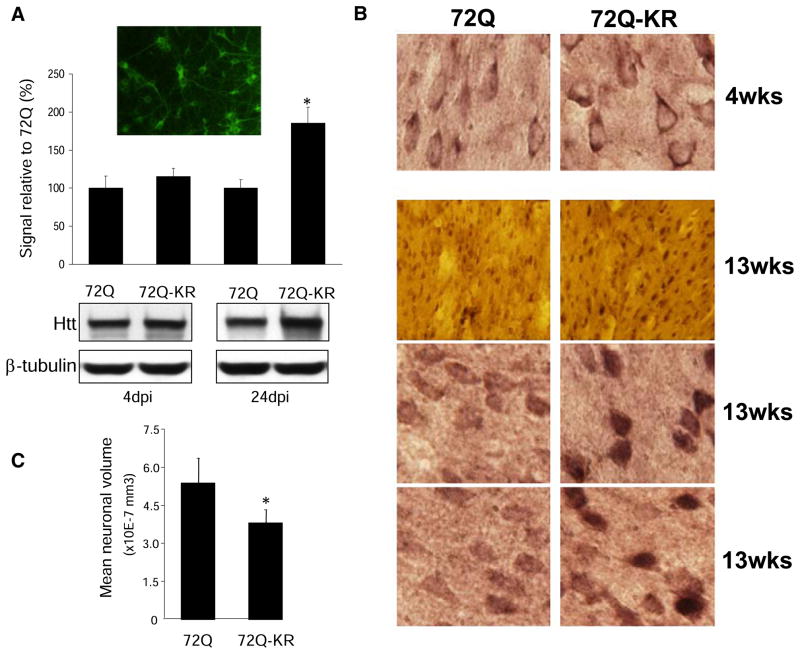
In order to examine if acetylation of K444 affects the stability of mutant Htt in neurons, a lentiviral-mediated delivery of mutant huntingtin was employed (Régulier et al., 2003). Analysis of various lentiviral promoters revealed that the mouse phosphoglycerate kinase (PGK) promoter resulted in easily detectable expression of Htt in more than 90% of neurons at nontoxic titers (Figure 2A, inset). Primary neuronal cultures were transduced with acetylated (lenti-Htt571-72Q) or acetylation-resistant Htt (lenti-Htt571-72Q-KR) and the expression monitored over a period of 24 days. These experiments revealed significant accumulation of lenti-Htt571-72Q-KR compared to lenti-Htt571-72Q (Figure 2A), suggesting increased stability and impaired clearance of acetylation-resistant mutant Htt. In order to validate these observations in vivo, brains of wild-type mice were stereotaxically injected with lentiviral vectors expressing lenti-Htt571-72Q or lenti-Htt571-72Q-KR. While no significant accumulation was noted 4 weeks after injection, a dramatic accumulation of lenti-Htt571-72Q-KR compared to lenti-Htt571-72Q was observed 13 weeks after the injection (Figure 2B). The accumulation of lenti-Htt571-72Q-KR led to increased neurodegeneration as determined by a significant decrease in the mean neuronal volumes of infected neurons (Figure 2C). These findings in mice are in agreement with our studies in primary neurons demonstrating increased accumulation of acetylation-resistant mutant Htt.
Figure 2. Acetylation-Resistant Mutant Htt Accumulates in Cultured Neurons and in Mouse Brains.
(A) Rat primary cortical neurons were transduced with acetylated (lenti-Htt571-72Q) or acetylation-resistant mutant Htt (lenti-Htt571-72Q-KR) on DIV2. More than 90% of neurons expressed lenti-Htt (Ab1 antibody) (inset). Twenty-four days post-infection lenti-Htt571-72Q-KR significantly accumulated compared to lenti-Htt571-72Q. *p < 0.01 for three independent experiments.
(B) Lentiviral delivery of mutant Htt in mouse cortex and striatum. Expression of lenti-Htt571-72Q and lenti-Htt571-72Q-KR Htt in contralateral brain sections was examined at 4 and 13 weeks after injection by immunostaining with EM48 antibody against mutant Htt. Serial sections from at least ten mice in each group were examined. Representative sections show predominantly cytoplasmic neuronal expression of lenti-Htt571-72Q and cytoplasmic and nuclear expression of lenti-Htt571-72Q-KR. Magnification 40× and 100×.
(C) Neuronal volumes were significantly decreased in lenti-Htt571-72Q-KR compared to lenti-Htt571-72Q injected mice. Unbiased stereological analysis (Micro-BrightField) was performed 13 weeks after injection. Graphs represent means ± standard error of the mean (SEM) of five animals per group; *p < 0.01 compared to 72Q.
Acetylation at Lys444 Is Required for Neuroprotection from Mutant Htt Toxicity in Primary Neurons and Transgenic C. elegans Model of HD
To further establish that acetylation at K444 affects neuronal toxicity, we found that primary neurons transduced with acetylation-resistant mutant Htt (lenti-Htt571-72Q-KR) showed significantly higher toxicity compared to acetylated (lenti-Htt571-72Q) mutant protein (Figure 3A). Based on these results, we hypothesized that increased acetylation of mutant Htt at K444 by CBP may lead to neuroprotection. Previous studies demonstrated that depletion of CBP enhanced toxicity whereas overexpression of CBP suppressed toxicity by mutant Htt (Bates et al. 2006; Nucifora et al., 2001). As expected from the previous work, transfection of CBP significantly reduced Htt590-97Q-induced cell death (Figure 3B). In contrast, neuronal toxicity mediated by the acetylation-resistant mutant Htt (Htt590-97Q-KR) was not protected by CBP (Figure 3B), suggesting that intact lysine at position 444 is required for the CBP-mediated rescue.
Figure 3. Acetylation of Mutant Htt at K444 Leads to Neuroprotection.
(A) Lentiviral expression of acetylation-resistant Htt (lenti-Htt571-72Q-KR) in primary corticostriatal cultures leads to increased toxicity compared to acetylated Htt (lenti-Htt571-72Q), as determined by caspase 3/7 activation. At least three independent experiments were performed; *p < 0.01.
(B) Protection from Htt toxicity by CBP requires acetylation of Htt at K444. Rat primary corticostriatal neurons were transfected with Htt590-97Q or Htt590-97Q-KR together with CBP-HAT or vector, and toxicity scored for at least 150 neurons per sample. Results of three independent experiments are expressed as means + SEM (p = 0.001).
(C) Acetylation of mutant Htt is protective in vivo in C. elegans. In animals expressing acetylated mutant Htt (Htt564-150Q) and CBP-HAT, the intact GFP-expressing ASH neuron (arrow, left upper panel) takes up the red fluorescent dye DiD (left middle and lower panels); no degeneration is detected. In contrast, in most animals expressing acetylation-resistant mutant Htt (Htt564-150Q-KR) and CBP-HAT, ASH neurons are GFP positive (arrow, right upper panel) and DiD negative (right middle and lower panels); neurodegeneration increases with K444R mutation. Consistent with previous studies, the ASI neurons (arrowheads) express less Htt protein, are less consistently affected by polyglutamine toxicity, and, hence, were not scored.
(D) Affected ASH neurons were scored and compared for Htt564-150Q + CBP and Htt564-150Q-KR + CBP. Average percentage degenerated ± SEM is shown for multiple independent strains (p = 0.0012, see Experimental Procedures for details).
To examine whether acetylation of mutant Htt can be neuroprotective in vivo, we studied the effects of Htt acetylation in C. elegans, a well-established model organism for polyglutamine toxicity (Faber et al., 1999; Voisine and Hart, 2004). Recent work showed that CBP protects neurons from mutant Htt toxicity in the C. elegans model (Bates et al., 2006). Transgenic C. elegans lines expressing CBP-HAT together with the N-terminal 564 amino acids of human Htt with intact K444 (Htt564-150Q) or mutated K444 (Htt564-150Q-KR) were generated (Figure S4). The transgenes were coexpressed with GFP in a subset of neurons including the ASH sensory neurons. Consistent with previous studies, the expanded Htt polyglutamine tract caused neuronal degeneration and cell death (Faber et al., 1999). Degeneration is revealed as process retraction, which prevents uptake of the fluorescent dye DiD from the environment via the exposed sensory endings of ASH neurons, and polyglutamine-mediated cell death is assessed using GFP expression (Hart et al., 1999). Intact ASH neurons express GFP and are DiD positive (Figure 3C). Degenerating ASH neurons fail to take up DiD and are detected as GFP-positive, DiD-negative cells (Figure 3C). ASH neurons that have undergone cell death are both GFP and DiD negative. While mutant Htt resulted in degeneration of 58% of neurons in the absence of CBP (not shown), coexpression of CBP was neuroprotective with only 14% of neurons affected (Figure 3D). Importantly, when CBP was expressed along with the acetylation-resistant mutant Htt (Htt564-150Q-KR), almost 60% of the ASH neurons degenerated (Figure 3D), suggesting that CBP-mediated neuroprotection in C. elegans requires intact K444. These results are consistent with the data obtained in primary neurons (Figure 3B) and provide further in vivo evidence that acetylation of mutant Htt at K444 is required for neuroprotection from mutant Htt toxicity.
Increased Acetylation at Lys444 Facilitates Clearance of Mutant Huntingtin
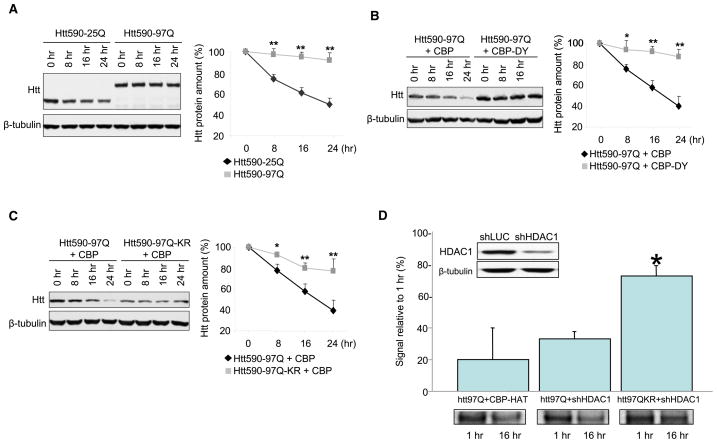
Since the above experiments demonstrated increased accumulation of acetylation-resistant mutant Htt, we directly investigated whether stability of Htt protein is modulated by acetylation of K444. Neuro2a cells were transfected with mutant or wild-type Htt, treated with the protein synthesis inhibitor cycloheximide (CHX), then monitored for the clearance of Htt. Previous studies have suggested that lysosomal protein clearance may be impaired by CHX treatment (Lawrence and Brown, 1993); however, we provide evidence that lysosomal protein degradation remains intact in our experimental system (Figure S5). Although wild-type Htt (Htt590-25Q) showed rapid clearance, mutant Htt (Htt590-97Q) clearance was significantly slower at baseline (Figure 4A). Cotransfection with CBP-HAT, but not with mutant CBP with inactive HAT activity (CBP-HAT-DY), significantly enhanced the clearance of mutant Htt (Figure 4B), further suggesting that increased acetylation of mutant Htt promotes its clearance. Importantly, no cellular toxicity was observed in these experiments, which enabled the assessment of protein clearance in the absence of cell death (Figure S6). While cotransfection of CBP-HAT led to progressive clearance of Htt590-97Q, the level of Htt590-97Q-KR remained comparatively unaltered (Figure 4C), further implicating K444 as the required residue that modulates the stability of Htt protein.
Figure 4. Acetylation of K444 Promotes Clearance of Mutant Htt.
Neuro2a cells were transfected with Htt and CBP-HAT, treated with cycloheximide (CHX), and harvested at the indicated time points. Western blots were analyzed by densitometry and values normalized to the amount of Htt at the time of cycloheximide treatment (100%). Values represent means of at least four independent experiments + SEM; *p < 0.05; **p < 0.01 compared to Htt590-25Q (A), Htt590-97Q + CBP-HAT (B), and Htt590-97Q + CBP-HAT (C), respectively.
(A) Mutant Htt590-97Q has a longer half-life than wild-type Htt590-25Q.
(B) Cotransfection of CBP-HAT, but not the HAT-deficient CBP-HAT-DY, resulted in increased clearance of mutant Htt.
(C) Acetylation-induced clearance of mutant Htt depends on K444. Coexpression of CBP-HAT led to increased clearance of Htt590-97Q, whereas mutation of Htt K444 to R prevented this effect of CBP-HAT.
(D) Enhanced clearance of Htt590-97Q by HDAC1 knockdown requires residue K444. Efficient knockdown of HDAC1 was achieved 3 days after transfection of pSUPER-HDAC1 compared to transfection of pSUPER-luciferase in Neuro2a cells. β-tubulin was used as loading control (inset). Levels of Htt590-97Q were monitored by radiolabeled pulse-chase. Knockdown of HDAC1 led to significant clearance of Htt590-97Q (32.6% ± 4.9%), comparable to levels achieved with coexpression of CBP-HAT (20.0% ± 16.9%). Mutagenesis of K444 to R (K444R) led to a significant decrease of this clearance (73.0% ± 8.5%). Densitometry measurements are expressed as % of signal after a 1 hr chase. Three independent experiments were performed and are represented as mean + SEM. *p < 0.001 compared to Htt97Q+shHDAC1.
Having demonstrated that CBP overexpression leads to increased acetylation and clearance of mutant Htt, we then examined the effects of HDAC1 knockdown on Htt clearance. These experiments revealed that knockdown of endogenous HDAC1 by shRNA significantly increased acetylation of mutant Htt (Figure S7A) and enhanced its clearance, as determined by radiolabeled pulse-chase. In contrast, mutagenesis of K444 to R (K444R) led to a significant decrease of this clearance (Figure 4D). No effect was observed with a shRNA construct targeting luciferase (Figure S7B). Taken together, these results demonstrate that modulation of Htt acetylation at K444 by CBP overexpression or HDAC1 knockdown facilitates clearance of the mutant protein.
Clearance of Acetylated Mutant Huntingtin by Autophagic-Lysosomal Pathway
To examine whether the lysosomal or the proteasomal pathway is responsible for clearance of acetylated Htt, Neuro2a cells were transfected with mutant Htt and treated with inhibitors of proteasome or lysosome. While the proteasome inhibitors epoxomicin and ALLN did not impede the clearance of mutant Htt (Figure S8A), treatment of Neuro2A cells with the lysosomal cathepsin L inhibitor leupeptin, cathepsin B, H, and L inhibitor E-64d, lysosomal proton pump inhibitor bafilomycin A1 (Baf A1), and the macroautophagy inhibitor 3-MA led to accumulation of acetylated mutant Htt (Figures 5C and S8B), suggesting that autophagy and lysosomal degradation play an important role in its clearance. To further examine the contribution of autophagy to acetyl-Htt clearance, we transfected mutant Htt into wild-type mouse embryo fibroblasts (MEFs) and Atg5-deficient MEFs (Atg5 KO) that exhibited defective autophagosome formation (Hara et al., 2006). When compared with the CBP-HAT-transfected wild-type MEFs, clearance of mutant Htt was greatly diminished in Atg5 KO MEFs transfected with CBP-HAT (Figure S8C), confirming that autophagy represents the key pathway for clearance of acetylated mutant Htt.
Figure 5. Acetylation of Mutant Htt Enhances Its Clearance by Autophagy.
(A) Acetylation of mutant Htt at lysine 444 leads to increased recruitment of LC3 to autophagic vacuoles. RFP-Htt480-68Q and RFP-Htt480-68Q-KR were transfected into COS-7 cells together with GFP-LC3 and CFP-CBP-HAT. Live cells were sequentially scanned to detect distribution of Htt (red), LC3 (green), and CBP-HAT (blue). Scale bar, 10 μm. (a) Coexpression of RFP-Htt480-68Q with LC3 and CBP-HAT led to accumulation of LC3 puncta. (b) Cells transfected with RFP-Htt480-68Q-KR, LC3, and CBP-HAT displayed a marked decrease in the LC3 puncta. (c) LC3-G120A mutant does not form punctate structure. (d) The percentage of cells containing >5 puncta was scored in 150 cells per experiment. Values are expressed as means + SEM of three independent experiments.
(B) HeLa cell line, stably expressing YFP-LC3, was transfected with Htt590-97Q or Htt590-97Q-KR plus CFP-CBP-HAT or inactive CBP-HAT-DY. ANOVA analysis of the percentage of transfected cells with Htt puncta that colocalized with LC3 puncta revealed a significant effect of the presence of CBP-HAT (p < 0.001) and the requirement of K444 (p < 0.001).
(C) Accumulation of mutant acetyl-Htt after treatment with lysosomal inhibitor leupeptin (200 μM). β-tubulin was used as loading control; p < 0.001.
(D) Clearance of acetylated mutant Htt is impeded by inhibition of lysosomal enzymes. Levels of Htt590-97Q, monitored by radiolabeled pulse-chase, were determined after 1, 8, 16, and 24 hr by immunoprecipitation for Htt590-97Q using an N-terminal FLAG antibody. Coexpression of CBP-HAT significantly enhanced clearance of Htt590-97Q (filled square) compared to inactive CBP-HAT-DY (open square) (p < 0.05). This clearance required K444 (open diamond) (p < 0.005) and was impeded by 200 μM leupeptin (filled diamond) (p < 0.05). Significant degradation was achieved after a chase period of 16 hr and 24 hr (*p < 0.001). Densitometry measurements are expressed as % of signal after a 1 hr chase. Representative radiograms are shown. Three independent experiments were performed and are represented as mean + SEM. ANOVA reveals a significant difference across chase time (F(3,23) = 61.779, p < 0.0001), across group (F(3,23) = 19.656, p < 0.0001), and across an interaction between chase time and group (F(9,23) = 5.479, p = 0.005).
(E) Acetyl-Htt is preferentially localized to the cytoplasm. Levels of cytoplasmic and nuclear acetyl-Htt and CBP-HAT were determined by subcellular fractionation and western blotting. HDAC1 and β-tubulin were used as nuclear and cytoplasmic markers, respectively. Results are representative of three independent experiments.
Next, we examined clearance of acetylated mutant Htt by monitoring autophagic activity in live cells by LC3, a mammalian homolog of yeast Atg8 that gets incorporated into autophagosomal (AV) membrane (Kabeya et al., 2000; Tanida et al., 2005). LC3, when fused to fluorescent proteins, can thus be used to monitor the formation of AV in live cells by the appearance of LC3-positive puncta (Mizushima, 2004). We found that COS-7 cells expressing Htt480-68Q displayed significantly more LC3-positive vacuoles compared to cells expressing Htt480-68Q-KR (Figures 5A, S9A, and S9B). Transfections of LC3 mutant, GFP-LC3-G120A, that does not get incorporated into AV membranes (Kabeya et al., 2000) led to a dramatic decrease in the number of LC3 puncta (Figure 5A), suggesting that the observed LC3 puncta represent AV rather than nonspecific GFP-LC3 aggregates.
To quantify the frequency with which Htt-positive puncta colocalized with LC3-positive puncta, Htt590-97Q or Htt590-97Q-KR mutant Htt was cotransfected together with CBP-HAT into HeLa cells that stably express YFP-tagged LC3. The number of Htt-containing puncta per cell and the percentage of transfected cells exhibiting this colocalization event were determined (Figure 5B). Correlative with the increased degradation of acetylated mutant Htt, there was a significantly higher probability of finding Htt puncta colocalizing with LC3 in cells transfected with acetylated Htt (Figure 5B). In contrast, mutating the lysine 444 to an arginine eliminated the ability of CBP-HAT to augment the colocalization of LC3 and Htt (Figure 5B), suggesting preferential targeting of acetylated mutant Htt to autophagosomes.
Next, we sought to directly investigate whether targeting of acetylated Htt to autophagosomes results in increased clearance of the mutant protein. Similar to our observation using CHX, coexpression of CBP-HAT but not inactive CBP-HAT-DY led to significantly enhanced clearance of Htt590-97Q (Figure 5D) as determined by radiolabeled pulse-chase experiments. This clearance required K444 (◇, Figure 5D) and was indeed impeded by lysosomal inhibitor leupeptin (◆, Figure 5D), further suggesting that acetylation of Htt at K444 promotes autophagic and lysosomal degradation of the mutant protein.
CBP is a resident nuclear protein that is known to acetylate histone proteins (Bannister and Kouzarides, 1996). Since CBP presumably acetylates Htt in the nucleus and clearance by autophagy takes place in the cytoplasm, we examined if acetylated Htt is indeed in the correct cellular compartment to be a substrate for autophagy. While CBP was primarily localized to the nucleus, acetylated Htt was predominantly found in the cytoplasmic compartment (Figure 5E), suggesting that, upon acetylation by CBP in the nucleus, mutant Htt becomes available for autophagic degradation in the cytoplasm.
Acetylated Mutant Huntingtin Is Preferentially Trafficked into Autophagosomes
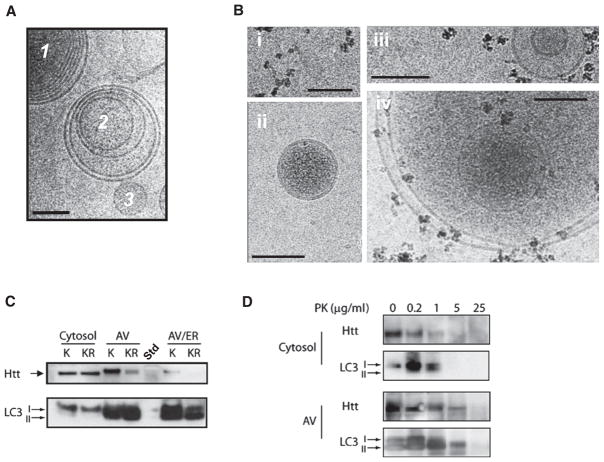
To directly examine whether acetylated mutant huntingtin can be targeted to autophagosomes, the distribution of mutant Htt (Htt590-97Q) was examined in autophagosomes isolated from Neuro2a cells by modified versions of two previously published protocols (Marzella et al., 1982; Stromhaug et al., 1998). Neuro2a cells were cotransfected with acetylated (Htt590-97Q) or acetylation-resistant (Htt590-97Q-KR) mutant Htt and CBP-HAT and autophagosomes isolated in the absence of chemical modifiers of autophagosomal maturation. Cryoelectron micoscopy (cryo-EM) of gradient flotation autophagosomal fractions revealed a variety of membranous structures consistent with previous publications (Reggiori and Klionsky, 2002). These fractions were enriched in multivesicular bodies that are morphologically consistent with autophagosomes, large multivesicular endosomes and/or autolysosomes and unilamellar structures like transport vesicles (Figure 6A). Immuno-EM using an antibody against LC3 revealed that only the multilamellar structures were positive for LC3 (Figure 6B). Autophagosome-enriched fractions (AV) were also confirmed by probing for the membrane-bound form of LC3, LC3-II (Figures 6C and S10). Whereas both acetylated and acetylation-resistant mutant Htt were detected at similar levels in the cytosolic fraction, acetylated Htt was preferentially detected in the AV fraction (Figure 6C). To determine if the acetylated Htt detected in the AV fraction is indeed within a membranous structure, a protease protection assay was performed. In the cytosolic fraction, both Htt590-97Q and the non-membrane-bound form of LC3 (LC3-I) were mostly degraded after exposure to 1 μg/ml proteinase K. In contrast, despite degradation of LC3-I at 1 μg/ml proteinase K in the AV fraction, Htt590-97Q did not degrade until 25 μg/ml proteinase K when the membrane-bound form of LC3 (LC3-II) was also degraded (Figure 6D). Taken together, these results further suggest that acetylation at K444 facilitates trafficking of the mutant protein into autophagosomes for degradation.
Figure 6. Acetylated Mutant Htt Is Trafficked to Autophagosomes.
(A) Purification of autophagosomes (AV). Neuro2A cells were fractionated and AV, lysosomes, and endoplasmic reticulum (ER) isolated using two distinct protocols, as described in Experimental Procedures. Cryo-EM revealed fractions enriched in multivesicular bodies that are morphologically consistent with AV (1, 2) and unilamellar structures like transport vesicles (3).
(B) To determine whether LC3 was associated with membrane structures, AV fractions were incubated with an antibody against LC3 followed by iron-rich magnetic beads conjugated to Protein A. (i) Beads imaged alone. (ii) Unilamellar transport vesicles were not labeled by beads. (iii and iv) Multilamellar autophagosomal-like particles are labeled by beads.
(C) Preferential trafficking of acetylated Htt to autophagosomes. Cells were fractionated and AV isolated, run on SDS-PAGE, and probed with anti-Htt antibody or LC3 antibody. Htt590-97Q was preferentially detected in the AV-enriched fractions compared to Htt590-97QKR. The cytosolic fractions predominantly contain the unbound form of LC3 (LC3-I), while the AV-enriched fractions contain the membrane-bound form of LC3 (LC3-II). Fraction “AV/ER” differs from “AV” as it is also enriched for ER markers such as calnexin (not shown). Std, protein standard.
(D) Mutant Htt is detected within the AV membrane fraction. Protease protection assay was performed with 20 μg of protein from a cytosol fraction or AV fraction collected from (C), subjected to increasing amounts of proteinase K (PK) and examined by SDS-PAGE. In the cytosolic fraction, both Htt590-97Q and the LC3-I were degraded after exposure to 1 μg/ml PK. In the AV fraction, Htt590-97Q did not degrade until 25 μg/ml PK when the LC3-II was also degraded. Htt590-97Q was detected using MAB5492 (Chemicon) and LC3 with rabbit polyclonal antibody (Abcam). A representative image from three independent experiments is shown.
p62-Dependent Clearance of Acetylated Mutant Huntingtin
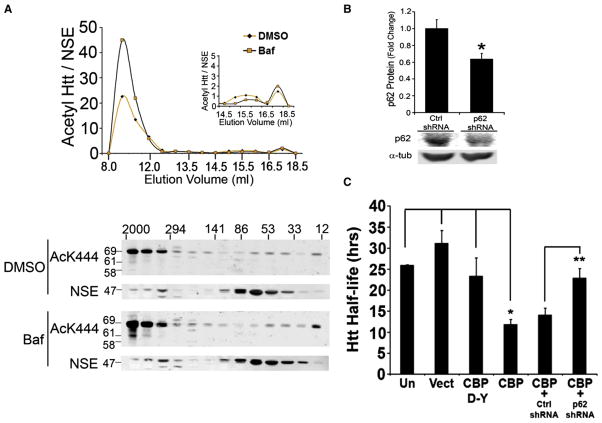
In order to further examine mechanisms of clearance of acetylated mutant Htt, we evaluated the biochemical state of Htt upon acetylation. Extracts from Htt480-68Q/CBP-HAT cotransfected Neu-ro2a cells were analyzed by native size-exclusion chromatography (SEC) followed by western blot analysis of collected fractions. Although the expected molecular mass of the monomeric Htt480-68Q is 59 kDa, the majority of acetylated Htt eluted off as a high molecular weight (HMW) species corresponding to 2000–294 kDa in size (Figure 7A). Further analysis of this fraction by denaturing SDS-PAGE revealed that the HMW species contained a 69 kDa Ac-Htt species (Figure 7A). Treatment of cells with the lysosomal inhibitor Baf A1 enhanced the amount of HMWAc-Htt species, whereas no changeinthe53 kDa monomeric Ac-Htt species was observed (HMW/monomer ratios: DMSO = 16; Baf A1 = 45) (Figure 7A). These results suggest that acetylation of mutant Htt at K444 leads to formation of a larger complex of proteins that may serve as a substrate for lysosomal degradation.
Figure 7. p62-Mediated Clearance of Mutant Htt.
(A) Acetyl-Htt exists as a HMW species and accumulates upon lysosomal inhibition. Neuro2a cells were cotransfected with Htt480-68Q and CBP-HAT, in the presence of bafilomycin A1 (Baf A1, 100 nM) or DMSO. Triton-soluble lysates were analyzed by size-exclusion chromatography (SEC) and western blot using the anti-acetyl K444 antibody (bottom). Neuron-specific enolase (NSE) was used to monitor gel filtration column performance and loading. The horizontal MW marker (kDa) represents the MW obtained by SEC analysis as determined by the elution of globular protein standards. The vertical marker (kDa) represents the MW obtained by SDS-PAGE analysis of fractions. Western blots shown were analyzed by densitometric analysis from at least three independent experiments and normalized to NSE (top). Inset, the amount of monomeric Ac-Htt is shown at a smaller y axis.
(B) Knockdown of p62 in inducible HN10 cell lines expressing Htt573-72Q. Values are the mean ± SEM (n = 5); *p = 0.01 for p62 shRNA compared to control (Ctrl) shRNA, normalized to α-tubulin.
(C) Clearance of Ac-Htt is mediated by p62 in HN10 cells. Mutant Htt half-life was determined in untransfected (Un) or cells transfected with empty vector (Vect), CBP D-Y, CBP-HAT, control shRNA, or p62 shRNA. Half-life values of Htt573-72Q were determined as described in Experimental Procedures. The values represent the mean ± SEM (n = 5). *p < 0.01 compared to Un and Vect; *p < 0.05 compared to CBP D-Y; **p < 0.01 compared to Ctrl-shRNA.
Next, we examined the role of p62/SQSTM1 (also called sequestosome 1) that interacts with LC3 and has been suggested to have a role in autophagic clearance of protein aggregates (Komatsu et al., 2007). Recent data suggest that reduction of p62 protein levels or interference with p62 function significantly increases mutant Htt-mediated cell death (Bjorkoy et al., 2005). To determine if p62 plays a role in the clearance of Ac-Htt, we used the HN10 neuronal cell line (Weiss et al., 2009) to generate conditional expression of a 573 aa fragment of mutant Htt (573-72Q). Initial characterization of this cell line revealed a half-life of mutant Htt of about 25 hr (Figure S11). Consistent with our findings in CHX-based experiments (Figure 4A), the half-life of the wild-type (WT) Htt (573-25Q) in HN10 cells was significantly shorter (12.5 hr) compared to the mutant protein (data not shown). As expected, we observed a significant decrease in half-life of Htt573-72Q upon CBP-mediated acetylation, compared to vector- or CBP-DY-transfected cells (Figure 7C). Interestingly, knockdown of endogenous p62 in HN10 cells by approximately 40% significantly hindered the clearance of Ac-Htt (Figures 7B and 7C). Moreover, p62 preferentially interacts with Ac-Htt (Figure S13), further suggesting that p62 plays a role in clearance of acetylated mutant Htt.
DISCUSSION
We demonstrate that in tissue culture models of HD as well as in mouse and human HD brain, there is baseline acetylation of mutant Htt at K444. Acetylation of mutant Htt at this residue can be increased by CBP-mediated acetylation or the inhibition of HDAC1, which in turn leads to an increased clearance of mutant Htt and neuroprotection in vitro and in vivo. Previous studies showed that full-length and truncated Htt are found in the nucleus where Htt can interact with the transcriptional machinery to disrupt gene expression (Kegel et al., 2002; Dunah et al., 2002; Zhai et al., 2005; Cui et al., 2006). Our results suggest that as mutant Htt accumulates in the nucleus, it can become acetylated by CBP and then exported to the cytoplasm for autophagic degradation. Our finding of increased clearance of acetylated mutant Htt is in agreement with the observation that acetylation-resistant mutant Htt dramatically accumulates and causes neurodegeneration in cultured neurons and in mouse brains (Figures 2 and 3). Interestingly, we detect baseline acetylation of only mutant but not wild-type Htt in both cultured cells and mouse or human HD brain. Since we observe relatively rapid clearance of wild-type Htt under baseline conditions when compared to the mutant protein, wild-type protein may also be acetylated but degraded too rapidly to permit detection. The accumulation of mutant Htt in HD patients, despite acetylation, is clearly an indication that even this attempt by the neuron is not sufficient to evade disease onset; however since our data in mice and cultured neurons indicate that the acetylation-resistant form of mutant Htt accumulates even more, it can be concluded that some benefit arises from baseline acetylation. Moreover, our data suggest that by increasing the rate of acetylation at K444, we can enhance degradation of mutant huntingtin that results in neuroprotection, suggesting that acetylation of K444 represents a means of regulated clearance of mutant Htt. From a standpoint of therapy for human HD, a specific decrease in mutant but not wild-type Htt would be expected to delay disease onset and progression (Yamamoto et al., 2000; Arrasate et al. 2004; Graham et al., 2006b). While CBP and the existing HDAC inhibitors are not sufficiently specific as therapeutic agents, we describe a pathway that should allow for development of more selective compounds to specifically promote acetylation and clearance of mutant Htt in HD.
We find that acetylation of mutant Htt at K444 promotes clearance of the mutant protein via an autophagiclysosomal pathway. Recent work has revealed an important role for autophagy in general control of protein turnover in the central nervous system (Hara et al., 2006; Komatsu et al., 2006). In HD, autophagy is also emerging as an important cellular defense mechanism (Kegel et al., 2000; Ravikumar et al., 2004; Yamamoto et al., 2006). In these previous studies, shorter Htt fragments that rapidly aggregate were studied. Unlike the short fragments, the longer Htt fragments used in our study did not form visible aggregates, a phenomenon often seen with longer fragments of Htt (Hackam et al., 1998; Lunkes and Mandel, 1998; Menalled et al., 2003). We showed the enrichment of acetylated, but not acetylation-resistant, 590 aa Htt fragment in purified autophagosomes, further suggesting that these longer Htt fragments can be trafficked to autophagosomes. Biochemical analysis indicated that acetyl-Htt is present mainly as a HMW species, with <10% in the monomeric form. The inherent property of polyQ-containing proteins to self-aggregate has been well established (Perutz, 1999; Scherzinger et al., 1997), suggesting that Ac-Htt detected in the HMW fractions may exist, in part, as a self-associated oligomeric species. Lysosomal inhibition caused a specific accumulation of HMW Ac-Htt, whereas the monomeric form was unchanged suggesting that a soluble oligomeric Ac-Htt is likely the substrate for autophagic degradation. This is further supported by the fact that accelerated clearance of Ac-Htt is affected by levels of p62, a protein known to co-localize to mutant huntingtin inclusions (Nagaoka et al., 2004), and has been previously implicated in modulating levels of mutant exon1-Htt (Bjorkoy et al., 2005). While our results suggest that the HMW species of acetylated Htt may serve as a substrate for p62-mediated degradation of mutant Htt, further studies will be required to elucidate the precise nature of the interaction between p62 and acetylated Htt.
Importantly, current approaches to modulate autophagy inhibit mTOR (i.e., rapamycin) and in turn result in a global and nonspecific activation of autophagy and other cellular pathways that could have deleterious consequences in neurons (Levine and Kroemer, 2008). Our study showed that a direct modification of a target protein promotes its specific clearance and provides a link between protein acetylation and targeted degradation by autophagy. Since acetylation of nonhistone proteins has only recently emerged as an important cellular mechanism (Kouzarides, 2000), it will be of interest to further examine whether protein acetylation represents a more general regulatory mechanism for selective targeting of proteins for lysosomal degradation.
EXPERIMENTAL PROCEDURES
Cell Culture, Live-Cell Imaging, Lentiviral Transductions
Neuro2a, COS-7, HeLa, MEFs, and primary neurons were transfected using Lipofectamine 2000. Toxicity was measured using a ToxiLight nondestructive cytotoxicity bioassay kit (Cambrex) according to the manufacturer’s protocol. For primary cultures, cortical neurons were isolated from Sprague-Dawley rats at E19. Cell death was monitored as described previously (Cui et al., 2006). For live-cell imaging COS-7 were analyzed on a Zeiss LSM 510 confocal microscope (Carl Zeiss, Jena, Germany). Cells were kept at 37°C and 5% CO2 on the microscope stage and images were acquired sequentially in red, green, and blue channels. To create YFP-LC3 stable cell lines, Tet-off HeLa cell lines were cotransfected with YFP-LC3 and pTk-Hygro (10:1) with Lipofectamine (Invitrogen) per manufacturer’s instructions. For studies of Htt half-life, HN10 cells were stably transfected with a 573-72Qaa fragment of Htt using the Rheoswitch-inducible expression system (New England Biolabs, Ipswich, MA, USA) and expression induced with 500 nM Rheoswitch ligand (RSL1, New England Biolabs). The first order rate constant (k) of Htt clearance curve was obtained by an exponential decay fit, and protein half-life was determined by the equation Htt t1/2 = ln(2)/k. Rat embryonic primary corticostriatal neurons were infected with lentiviral vectors on DIV2. For the in vivo experiments, concentrated lentiviral stocks expressing Htt571-72Q or Htt571-72Q-KR were injected in the left or the right striatum of pentobarbital anesthetized C57BL/6 mice using stereotaxic coordinates of 0.4 mm rostral to bregma, 1.8 mm lateral to midline, and 3.5 mm ventral from the dural surface. The brain was perfused and processed as described (Cui et al., 2006).
Mass Spectrometry
Htt480-68Q was immunoprecipitated with Htt antibody (MAB5492) from transfected COS-7 cells treated with TSA (1 μM) and NAM (5 mM). Protein bands were stained with Simply Blue Safe Stain (Invitrogen), digested with trypsin, and analyzed by mass spectrometry (LC-MS/MS). MS/MS data were analyzed by the SEQUEST algorithm. MS was performed at Taplin Biological Mass Spectrometry Facility (Thermo Electron 7-T LTQ FT), Partners HealthCare Center for Genetics and Genomics Proteomic Facility (ThermoFinnigan DECA LCQ), and the LNT/NIMH/NIH (Finnigan LCQ Classic).
Purification of Autophagosomes and Electron Microscopy
To collect the lysosomal/AV fraction supernatants, Neuro2a cells were fractionated using a discontinuous Nycodenz (Stromhaug et al., 1998) or 50% metrizamide gradient (Marzella et al., 1982). AV-enriched fractions from both methods were collected and examined by electron cryomicroscopy and by immunoblotting. For immunolabeling of vesicles, anti-LC3 antibody (Nanotools, Germany) was incubated at 1:250 with AV fractions. For cryo-EM, organelles were imaged in vitreous ice over open holes on a perforated carbon support to preserve all details in the image. Samples were imaged on a Tecnai 20 FEG electron microscope operating at 200 kV, with magnifications from 25,000 to 50,000 and a typical defocus of −3.5 μm. For protease protection, 20 μg of protein from the cytosolic or AV fraction generated from the Marzella protocol were subject to 0, 0.2, 1, 5, or 25 μg/ml of proteinase K.
Radiolabeled Pulse-Chase Experiments
Radioactive isotope was purchased from ICN Pharmaceuticals. Neuro2a cells were transfected using Lipofectamine 2000 per manufacturer’s instructions. GFP-CBP-HAT was transfected in a 1:8 ratio with Htt590 constructs, and pSUPER-HDAC1 or pSUPER-Luciferase were transfected in 1:1 ratio with Htt590 constructs. Cells were incubated in serum-free, methionine-free DMEM, followed by a 3 hr pulse with [35S]methionine/cysteine (0.03 mCi/ml).
C. elegans Assays
Transgenic animals were generated by injecting pqe-1(rt13) unc-32(e189) pha-1(e2123) animals with osm-10::GFP (pHA#29, 40 ng/μl), osm-10::CBP-HAT (CBP-HAT, 10 ng/μl), and osm-10::Htn564-150Q with or without the K444R mutation (100 ng/μl). pJM#67 (elt-2::GFP) was used as a selection marker. ASH neuron degeneration was assessed in aged animals using GFP expression and DiD staining as described previously (Bates et al., 2006). No difference was observed in ASH neuronal degeneration and death in pqe-1;HtnQ150 animals with (7% degenerated and 89% dead) or without (5% degenerated and 94% dead) the unc-32(e189) mutation (Figure S4).
Size-Exclusion Chromatography
Neuro2a cells were transiently cotransfected with Htt 480-68Q and CBP-HAT, treated with either 100 nM Baf A1 (Calbiochem) or DMSO, and lysate resolved on a Superdex 200 HR10/30 column (GE Healthcare BioSciences, Uppsala, Sweden) as described previously (Mazzulli et al., 2006). The column was calibrated using a mixture of globular proteins (HMW standards, GE Healthcare BioSciences) and the void volume determined by the elution of blue dextran (2000 kDa). Normalized Ac-Htt signal was graphed according to elution volume of globular protein standards, and chromatographic peaks were integrated using GraphPad Prism software, V4 (GraphPad Software, Inc. LaJolla, CA, USA). Integrated peak values were used to calculate oligomer (pk at 1280 kDa)/monomer (pk at 53 kDa) ratios.
Supplementary Material
Acknowledgments
We are grateful to Drs. K. Kegel, M. DiFiglia, C.D. Allis, A. Basu, H. Park, and A.B. Young and members of the Krainc laboratory for helpful suggestions and comments on the manuscript. We also thank Drs. D. Sarracino, B. Krastins, J. Kowalak, S. Markey, and A. Makusky for help with mass spectrometry; B. Hyman, W. Stoothoff, and C. Lill for help with live-cell imaging; E. Regulier for help with lentiviral transductions; A. Weiss and A. Roscic for help with HN10 cells; H. Ischiropoulos for use of HPLC system; N. Mizushima for Atg5-deficient cells; A. Kazantsev for CBP and CBPΔQ constructs; Y. Yoshimori for GFP-LC3; G.A. Fitzgerald for p300 and P/CAF; P.W. Faber for osm::Htt564-150Q; and the New York Structural Biology Center for the EM facilities. This work was supported by R01NS050352 and R01 NS051303 (to D.K.) and R01 NS050199 (to A.Y.). F.T. is a fellow of the DFG.
Footnotes
Supplemental Data include Supplemental Experimental Procedures and thirteen figures and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(09)00317-1.
Author contributions: H.J. and F.T. contributed equally to this work.
References
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy: many paths to the same end. Mol Cell Biochem. 2004;263:55–72. doi: 10.1023/B:MCBI.0000041848.57020.57. [DOI] [PubMed] [Google Scholar]
- Cui L, Jeong H, Borovecki F, Parkhurst C, Tanese N, Krainc D. Repression of PGC-1alpha gene transcription by mutant huntingtin leads to mitochondrial dysfunction in Huntington’s disease. Cell. 2006;126:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dunah AW, Jeong H, Griffin A, Kim MJ, Standaert DG, Hersch SM, Mouradian MM, Young AB, Tanese N, Krainc D. Sp1 and TAF130 transcriptional activity disrupted in early Huntington’s Disease. Science. 2002;296:2238. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci USA. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006a;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Graham RK, Slow EJ, Deng Y, Bissada N, Lu G, Pearson J, Shehadeh J, Leavitt BR, Raymond LA, Hayden MR. Levels of mutant huntingtin influence the phenotypic severity of Huntington disease in YAC128 mouse models. Neurobiol Dis. 2006b;21:444–455. doi: 10.1016/j.nbd.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T, Kalchman M, Hayden MR. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol. 1998;141:1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hart AC, Kass J, Shapiro JE, Kaplan JM. Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci. 1999;19:1952–1958. doi: 10.1523/JNEUROSCI.19-06-01952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Meloni AR, Yi Y, Kim YJ, Doyle E, Cuiffo BG, Sapp E, Wang Y, Qin ZH, Chen JD, et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal Binding Protein, and represses transcription. J Biol Chem. 2002;277:7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou Y, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence BP, Brown WJ. Inhibition of protein synthesis separates autophagic sequestration from the delivery of lysosomal enzymes. J Cell Sci. 1993;105:473–480. doi: 10.1242/jcs.105.2.473. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunkes A, Mandel JL. A cellular model that recapitulates major pathogenic steps of Huntington’s disease. Hum Mol Genet. 1998;7:1355–1361. doi: 10.1093/hmg/7.9.1355. [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzulli JR, Mishizen AJ, Giasson BI, Lynch DR, Thomas SA, Nakashima A, Nagatsu T, Ota A, Ischiropoulos H. Cytosolic catechols inhibit alpha-synuclein aggregation and facilitate the formation of intracellular soluble oligomeric intermediates. J Neurosci. 2006;26:10068–10078. doi: 10.1523/JNEUROSCI.0896-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J Comp Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Nagaoka U, Kim K, Jana NR, Doi H, Maruyama M, Mitsui K, Oyama F, Nukina N. Increased expression of p62 in expanded polyglutamine-expressing cells and its association with polyglutamine inclusions. J Neurochem. 2004;91:57–68. doi: 10.1111/j.1471-4159.2004.02692.x. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Nucifora FC, Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- Perutz MF. Glutamine repeats and nueodegenerative diseases: molecular aspects. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régulier E, Trottier Y, Perrin V, Aebischer P, Déglon N. Early and reversible neuropathology induced by tetracycline-regulated overexpression of mutant huntingtin in rat striatum. Hum Mol Genet. 2003;12:2827–2836. doi: 10.1093/hmg/ddg305. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Sapp E, Schwarz C, Chase K, Bhide PG, Young AB, Penney J, Vonsattel JP, Aronin N, DiFiglia M. Huntingtin localization in brains of normal and Huntington’s disease patients. Ann Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335:217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Voisine C, Hart AC. Caenorhabditis elegans as a model system for triplet repeat diseases. Methods Mol Biol. 2004;277:141–160. doi: 10.1385/1-59259-804-8:141. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Weiss A, Roscic A, Paganetti P. Inducible mutant huntingtin expression in HN10 cells reproduces Huntington’s disease-like neuronal dysfunction. Mol Neurodegener. 2009;9:11–23. doi: 10.1186/1750-1326-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler VC, Auerbach W, White JK, Srinidhi J, Auerbach A, Ryan A, Duyao MP, Vrbanac V, Weaver M, Gusella JF, et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum Mol Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington’s disease. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Jeong H, Cui L, Krainc D, Tjian R. In vitro analysis of huntingtin mediated transcriptional repression reveals novel target and mechanism. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.