Abstract
Citron-N (citron) binds activated Rho and concentrates at excitatory postsynaptic densities on the smooth dendritic shafts of hippocampal GABAergic neurons. Since little is known about how cytoplasmic proteins become targeted and retained at synapses, we asked how citron attains this discrete distribution. We first sought to determine whether the synaptic localization machinery is unique to GABAergic interneurons, but in neurons cultured from either cerebral cortex or hippocampus low levels of citron are found in a small population of glutamatergic neurons concentrated at the tips of dendritic spines in addition to shaft synapses. These data suggest that the targeting domains in citron can be utilized appropriately by any neuron, and consistent with this idea, tagged, exogenous, full-length citron becomes concentrated at postsynaptic sites in all hippocampal neurons in which it is introduced. Citron contains multiple sites for protein-protein interaction, including coiled-coil, pleckstrin homology, cysteine rich zinc binding, and CMG domains, as well as a proline rich region and a PDZ binding tail that is known to interact with PSD-95/SAP90. Using citron deletion mutants, we find that the coiled-coil (CC) domain, located in the N-terminal half of the protein, mediates somatodendritic targeting, self-oligomerization and puncta formation independent of the Rho binding domain, while regions in the C-terminal half of citron are responsible for precise alignment of postsynaptic citron clusters at sites apposing presynaptic boutons. Rho binding and activation regulate membrane targeting and the number of synapses containing citron. Thus, citron’s clustering at postsynaptic membranes is mediated by multiple domains, and its synaptic distribution is modified in response to Rho activation.
Introduction
All CNS synapses possess certain common elements that can be seen by electron microscopy. These include synaptic vesicles and pre- and postsynaptic membrane thickenings. Not surprisingly then there are certain key structural proteins that appear to be common to most or all synapses (Benson et al., 2001, Ziv and Garner, 2004). Interwoven among the common elements are a variety of proteins that impart structural and functional distinctions, and these have been used to define particular synapse types. Some of the most common examples include proteins composing the postsynaptic density (PSD) which is an electron dense, proteinaceous network unique to excitatory synapses. The majority of PSDs are found in dendritic spines on glutamatergic neurons where they function partly as anchoring structures, but perhaps more importantly as organizers of signal transduction machinery at postsynaptic sites (Walikonis et al., 2000, Kim and Sheng, 2004). However, PSDs also form at excitatory synapses on smooth dendritic shafts of GABAergic interneurons. By light microscopy, such PSDs appear a bit longer and straighter than they do in spines and some evidence suggests that the protein composition is distinct. Such synapses lack alpha-actinin (Rao et al., 1998), they can interact with the pentraxin, Narp (O'Brien et al., 1999) and their PSDs can contain high levels of the protein citron-N (Zhang et al., 1999). The mechanisms that target and anchor cytoplasmic proteins like citron-N are poorly understood.
Citron-N (citron), a brain specific isoform of citron-kinase (citron-K), is a major component of isolated PSDs (Zhang et al., 1999). Along with NMDA receptors, PSD-95/SAP90, CaMKIIα and a few other proteins, it is part of the “core” PSD that remains after strong detergent extraction (Walikonis et al., 2000, Peng et al., 2004). It binds to the PDZ2 and 3 domains of the scaffolding protein, PSD-95/SAP90 through its C-terminal tail (Furuyashiki et al., 1999, Zhang et al., 1999). Citron can bind to activated Rho, suggesting it is a Rho effector (Madaule et al., 1995), but it does not contain an active kinase domain making its effector role unclear (Madaule et al., 1998, Furuyashiki et al., 1999). Recent work suggests that citron is important for the localization of the Golgi apparatus within neuronal cell bodies (Camera et al, 2004), but how citron becomes concentrated at synapses is unknown.
In this paper we asked how citron becomes selectively targeted and clustered at postsynaptic densities and whether the machinery required to read the targeting signals is unique to GABAergic interneurons. Additionally we assessed the effects of Rho activation on citron distribution. We find that any neuron can target citron postsynaptically, that distinct citron domains dictate homo-oligomerization, postsynaptic targeting and synapse localization, and that Rho activity alters both citron clustering and synaptic distribution. Based on these findings, we propose that citron may act to cluster interacting molecules at postsynaptic sites by virtue of its homo-oligomerization in a process that is regulated by Rho GTPases.
Methods
Dissociated neuronal cultures were prepared from hippocampi or cortices of embryonic day 18 rats according to the procedure described in Goslin and Banker, 1991. Neurons were fixed in 4% paraformaldehyde for immunocytochemistry using antibodies against synaptophysin (monoclonal, Boehringer Mannheim; and G95, gift of P. DeCamilli), PSD-95 family (monoclonal, Upstate Biotechnology, Waltham, MA), GluR1 (Upstate Biotechnology), GM130 (BD Transduction Laboratories) and lysosomal glycoprotein 120 (anti-lgp120, LY1C6, gift of Bettina Winckler). Neurons were fixed in −20°C methanol for immunocytochemistry when the following antibodies were used: citron (mouse and rabbit polyclonal, Zhang et al. 1999), CaMKII-alpha (monoclonal, Invitrogen, Carlsbad, CA) and PSD-95 (monoclonal 6g6, BioAffinity Reagents, Golden, CO).
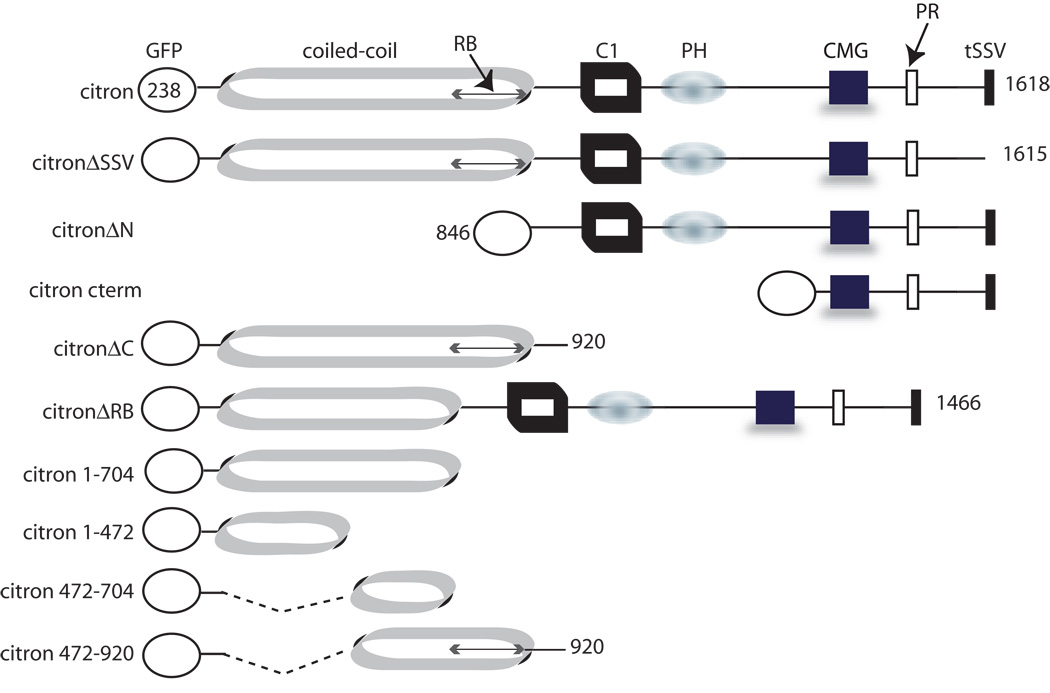
Rat cDNAs encoding citron (NCBI AF039218) were cloned into pcDNA3 for expression of full length, untagged citron-N. cDNAs encoding citron and its mutants were inserted in pEGFP-C2 (Clontech, Palo Alto, CA) to generate GFP-citron (GFP-CT) and other constructs described below. GFP-CTΔssv has a seven amino acid deletion at C-terminus. GFP-CTΔN, GFP-CTΔC, and GFP-CTΔRB have deletions of N-terminal coiled-coil domain (residue 1–846), C-terminal half of the protein (residue 920 to C terminal end), and Rho binding region (residue 704–846), respectively. GFP-cterm is a fusion protein of GFP with citron c-terminal region residue 1133 to the c-terminal end, containing citron homology, proline rich and PDZ binding domains. Four other constructs GFP-CT1–704, GFP-CT1–472, GFP-CT472–704, GFP-CT472–920 are GFP fusions with truncated citron coiled-coil domains with the included regions indicated by the amino acid numbers. For 2myc-RhoA constructs, green fluorescence protein cDNA in GFP-Myc-RhoA and GFP-Myc-RhoAG14V plasmid (kind gift of H. Udo and E. R. Kandel, (Udo and Kandel, 1996) was deleted and replaced by a Myc tag.
For crosslinking reactions, HEK293 cells were transfected with citron GFP fusion constructs using effectene (Qiagen, Valencia, CA), and cells were harvested 36–48 hours later in lysis buffer (25mM HEPES, 150mM NaCl, 2.5mM MgCl2, 0.5% Nonidet P 40, 1mM DTT, 5% glycerol, 0.23mM PMSF, 77 mM aprotinin, with 1X protease inhibitor cocktail: 8 uM benzamidine, 10uM iodoacetamide, 0.01 uM leupeptin, 0.007uM pepstatin A). The nuclei and cell debris were cleared by centrifugation at 10,000g for 10 minutes at 4 C. Cleared cell extracts were incubated with BS3 (Pierce Biotechnology, Rockford, IL) at final concentration of 0.2mM for 30 min on ice, loaded to 7% SDS-PAGE and immunoblotted by anti-GFP (1:1000, monoclonal, Clontech, Palo Alto, CA).
For fusion protein expression, HEK293 cells or 10–12 days old hippocampal neurons in culture were transfected with GFP-citron or 2myc-RhoA constructs using Effectene (Qiagen) or Lipofectamine 2000 (Gibco BRL), and fixed after 24 hours in paraformaldehyde. Expression of GFP fusion proteins is detected by green fluorescence, and expression of 2xmyc-RhoA proteins was detected by anti-Myc immunocytochemistry.
Fluorescence images were acquired using confocal microscopy (Zeiss LSM410) or a cooled CCD (SPOT camera, Diagnostic Instruments, Sterling Heights, MI) mounted on a Zeiss Axiophot. Images were overlayed using Adobe Photoshop. Density and alignment of individual punta were assessed with the aid of Neurolucida software (Microbrightfield, Williston, VT), numbers were imported into Excel and differences between groups were determined using JMP statistical software (Cary, North Carolina).
Results
Synaptic localization of citron in hippocampal and cortical neurons
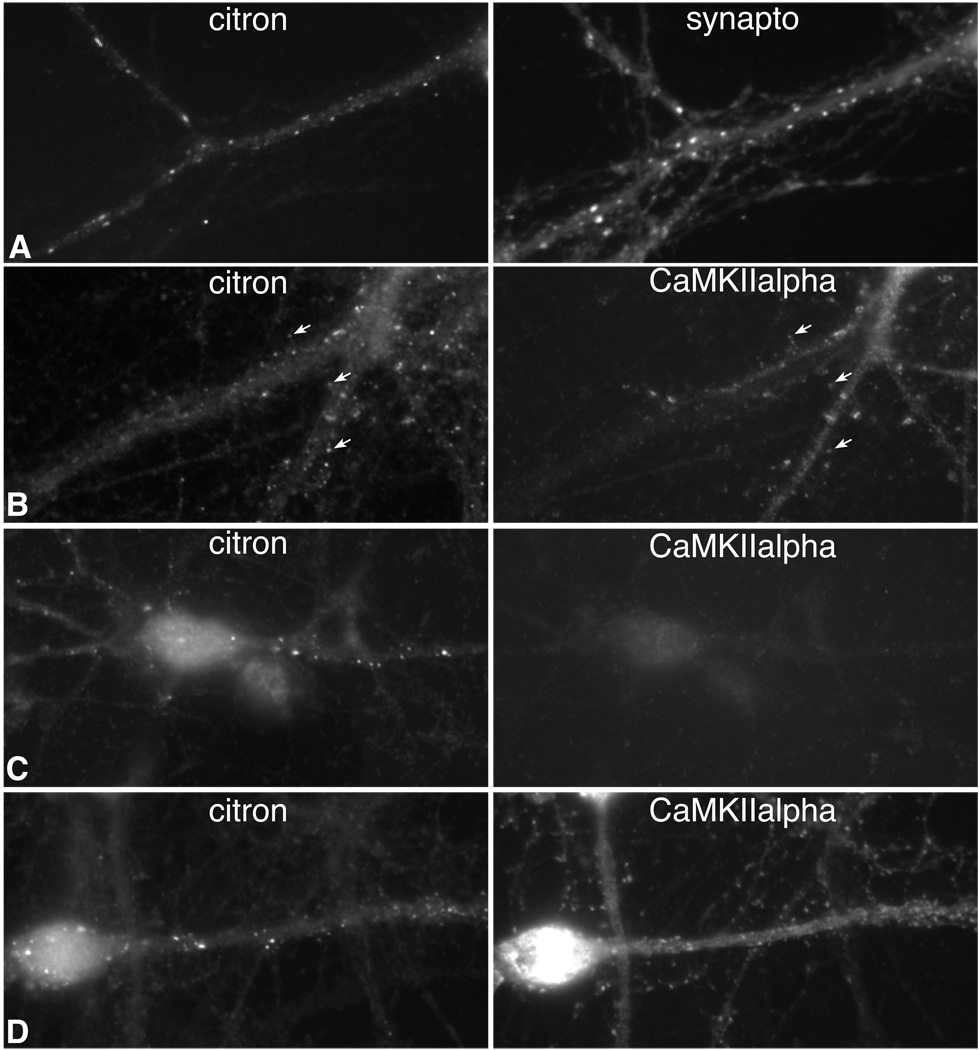
In hippocampal neurons in vivo and in culture, citron concentrates in postsynaptic densities clustered on the smooth dendrites of GABAergic neurons (Figure 1A), and it is excluded from GABAergic synapses (Zhang et al., 1999). This localization pattern suggests that citron targeting to synapses may be a property unique to GABAergic neurons. Since immunohistochemical experiments on tissue sections show that some excitatory neurons in the cerebral cortex express citron (Zhang et al., 1999), we sought to determine whether citron concentrates at shaft or spine synapses in neurons cultured from cortex where subcellular localization could be assessed unequivocally. Similar to hippocampus, we find citron to be expressed in a small number of neurons, but the cortical population is far more heterogeneous, containing both glutamatergic and GABAergic neurons and encompassing a variety of morphologies (Fig. 1B–D). In all cases, citron is discretely localized to puncta that in excitatory neurons overlap with labeling for CaMKII alpha which is the major postsynaptic density protein (Fig. 1B,D), or it apposes labeling for synaptophysin, a synaptic vesicle protein (data not shown). Some puncta are displaced from the dendritic shaft, strongly suggesting citron can also be targeted to dendritic spines (Fig. 1B, arrows). These data suggest that citron function is not limited to excitatory synapses on interneurons, but can also be executed at excitatory postsynaptic sites in a heterogeneous subpopulation of neurons throughout the forebrain.
Figure 1.
Fluorescence images of immunolabeled neurons show that citron clusters at synapses in a small, heterogeneous population of excitatory as well as inhibitory neurons having different morphologies. Cultured hippocampal neurons (A) illustrate the prominent pattern with citron clusters along dendrite shafts aligning with synaptophysin (synapto) labeling. An excitatory, CaMKII alpha labeled cortical neuron (B) shows citron co-clustering at CaMKIIalpha labeled spines. Other glutamatergic (D) and nonglutamatergic (C, CaMKII negative) cortical neurons of varying morphologies also show clustered citron labeling. Bar = 10 µm.
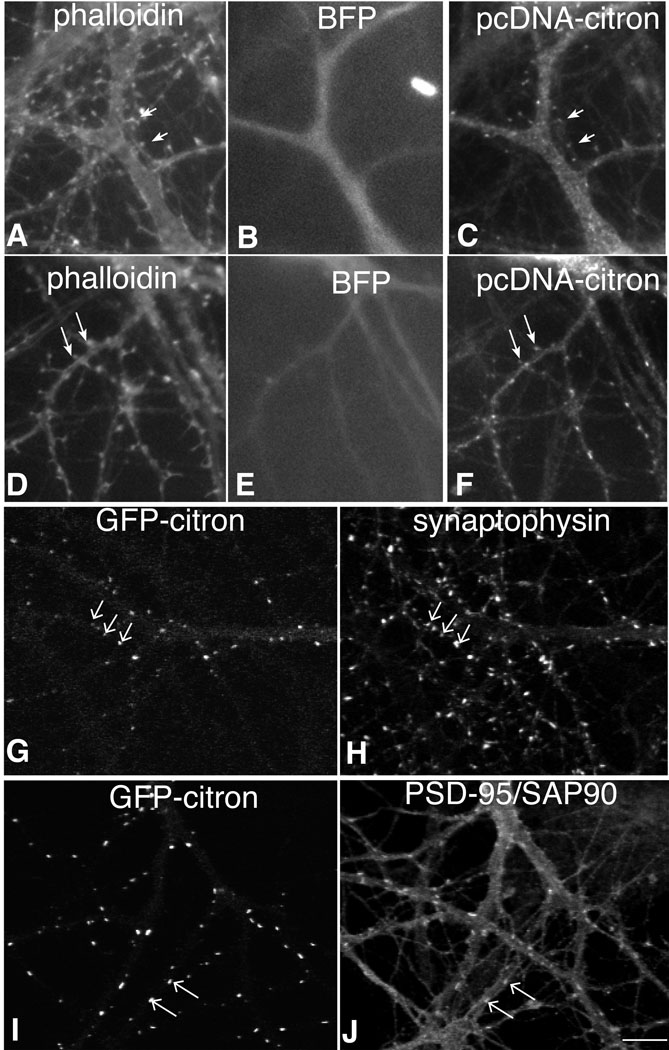
Since citron appears to concentrate at postsynaptic sites in both excitatory and inhibitory cortical neurons, it seems likely that the postsynaptic targeting machinery driving its localization will be common to most neurons. To further investigate this hypothesis, we co-transfected 10–12 day old hippocampal neurons with full length citron (pcDNA3-citron) and blue fluorescent protein (BFP) and 24 h later examined citron distribution in pyramidal cells which could be identified by the presence of F-actin labeled dendritic spines (Fischer et al., 1998). Similar to its expression pattern in hippocampal GABAergic interneurons, citron concentrates in puncta at discrete sites in dendritic shafts, but is also often found concentrated at the tips of dendritic spines (Fig. 2A, arrows). GFP-tagged, full length citron, shows a similar distribution concentrating at sites labeled for the excitatory postsynaptic density protein PSD-95/SAP90 (Fig. 2G–J). Neither spiny nor smooth neurons showed any obvious change in cell morphology. These data suggest that citron is targeted to excitatory postsynaptic sites regardless of the neuron (excitatory or inhibitory) or excitatory synapse (shaft or spine) type. Consistent with these observations, close examination of native citron expression in hippocampal neurons carried out at high magnification reveals a small number of pyramidal neurons that express low levels of citron at spine synapses (data not shown).
Figure 2.
Citron is targeted to synapses when expressed in hippocampal pyramidal neurons.
When untagged, full length citron (C, F) is introduced into hippocampal pyramidal cells along with blue fluorescent protein (B, E), immunolabeled citron (antibody 1b) clusters at the tips of phalloidin labeled (A, D) dendritic spines (arrows). GFP-tagged citron (G, I) shows a similar distribution, forming clusters on dendritic shafts and spines, which colocalize with synaptophysin labeling (H, arrows) and PSD-95/SAP90 labeling (J). Bar = 10 µm.
Domains regulating clustering and synaptic targeting
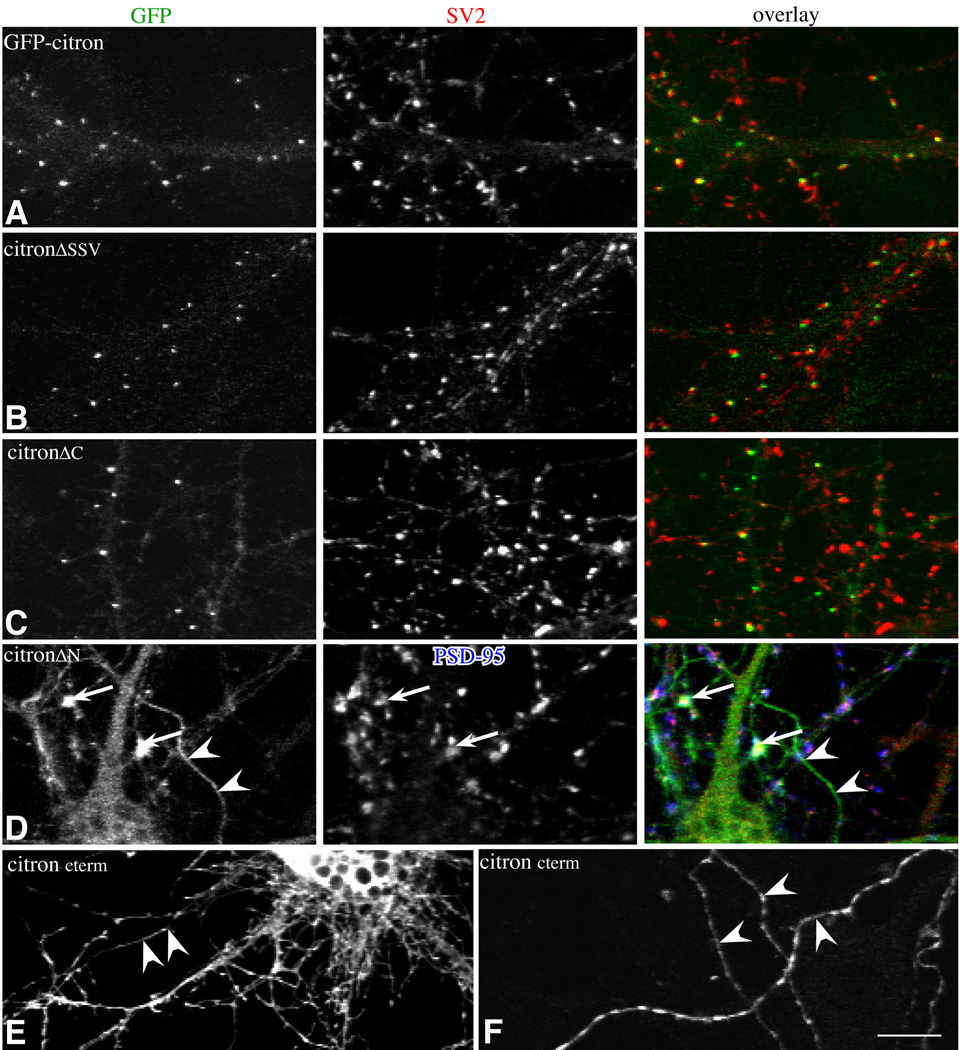
Citron contains several domains likely to be sites for protein-protein interactions (Fig. 3). These include a coiled-coil domain encompassing the Rho binding site, a cysteine rich zinc binding domain, and pleckstrin homology, CMG (citron/MRCK/Gek) and proline rich domains, in addition to the PDZ binding domain (SSV) that is known to interact with PSD-95/SAP90 (Furuyashiki et al., 1999, Zhang et al., 1999). We created a series of GFP-tagged citron mutants lacking particular domains and assessed the ability of the mutants to be targeted to the somatodendritic domain and clustered and restricted to postsynaptic sites. Full length citron and citron lacking its PDZ binding motif (citronΔSSV) form puncta in neurons and codistribute with PSD-95/SAP90 or SV2 in a manner that is indistinguishable (Table 1; Fig. 4A, B; Fig. 5) indicating that citron can form clusters and target to synapses independently of PSD-95/SAP90.
Figure 3.
Diagram shows full length, tagged citron, its domain structure, and truncation mutants used in this study. C1: cysteine rich zinc binding domain, PH: pleckstrin homology domain, PR: proline rich domain, RB: Rho binding domain.
Table 1.
Summary of distribution and crosslinking data using citron mutants.
| Mutant Construct |
Puncta in HEK293 cells |
Oligomerization by crosslinking |
Monomers following crosslinking |
Puncta in neurons |
Synaptic | Polarized to somatodendritic domain |
|---|---|---|---|---|---|---|
| GFP-CT | Yes | Yes | No | Yes | Yes | Yes |
| GFP-CTΔssv | Yes | Yes | No | Yes | Yes | Yes |
| GFP-CTΔN | No | No | Yes | Rare, large |
Yes | No |
| GFP-CTΔC | Yes | Yes | No | Yes | Partial | Yes |
| GFP-CTΔRB | Yes | Yes | No | Yes | Yes | Yes |
| GFP-CT1–704 | Yes | Yes | Yes | Yes | Yes | No |
| GFP-CT1–472 | Mixed | Yes | Yes | No | No | not tested |
| GFP-CT472–704 | Mixed | Yes | Yes | No | No | No |
| GFP-CT472–920 | Mixed | Yes | No | Yes | not tested | not tested |
| GFP-CT cterm | No | No | No | No | No | No |
Figure 4.
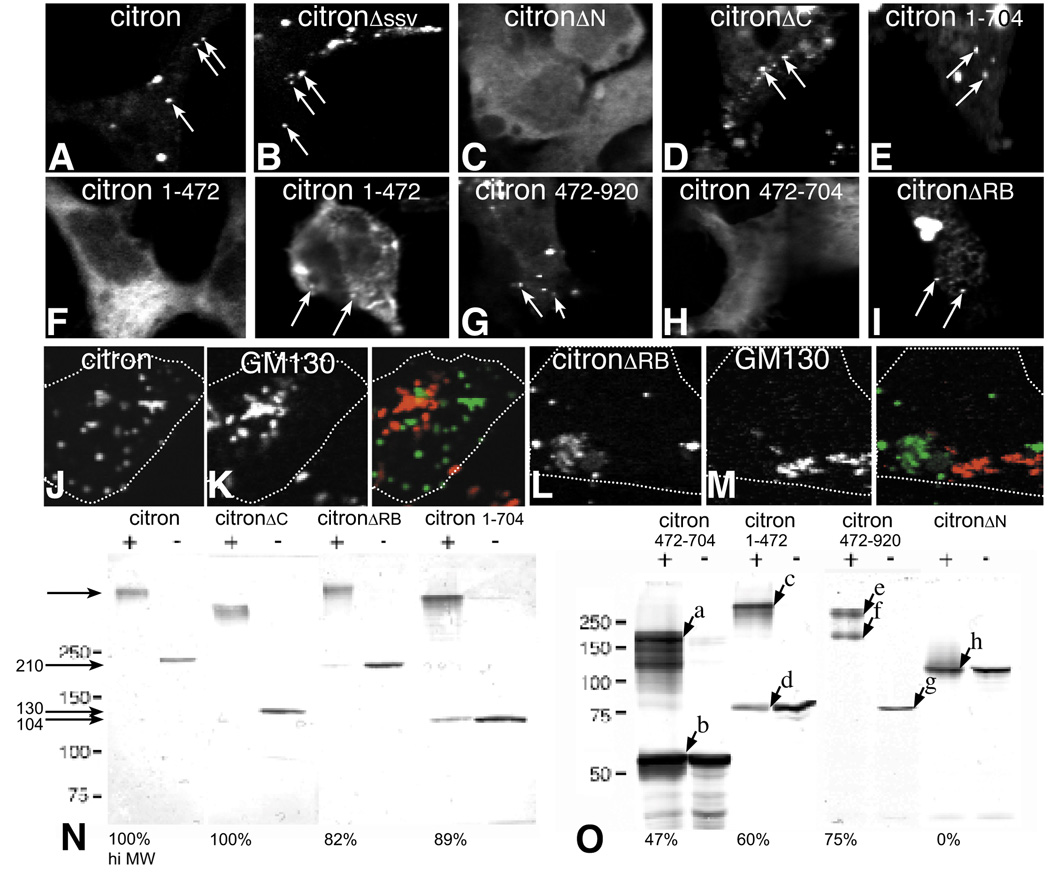
Fluorescence Images show cultured hippocampal neurons transfected with GFP-tagged full length and mutant citron and labeled for the markers indicated. Overlays of the images shown in the first (GFP-green) and second (SV2-red) columns (A–C) or (D) PSD-95 (blue) and SV2 (red) shown in the right hand column. All of the mutants show some degree of clustering, but the size and distribution of puncta in mutants containing the N-terminal CC domain (A–C) are most similar to wild type citron (A). Synaptic localization is decreased in mutants lacking the C-terminal half (C). Mutants lacking the N-terminal half are not restricted to the somatodendritic domain (D,E). A few poorly formed puncta show some synaptic localization (D; white in overlay). Citron cterm is most broadly distributed in axons and dendrites (E) and shows some clustering in axons labeled with GAP43 (F). Bar = 10 µm.
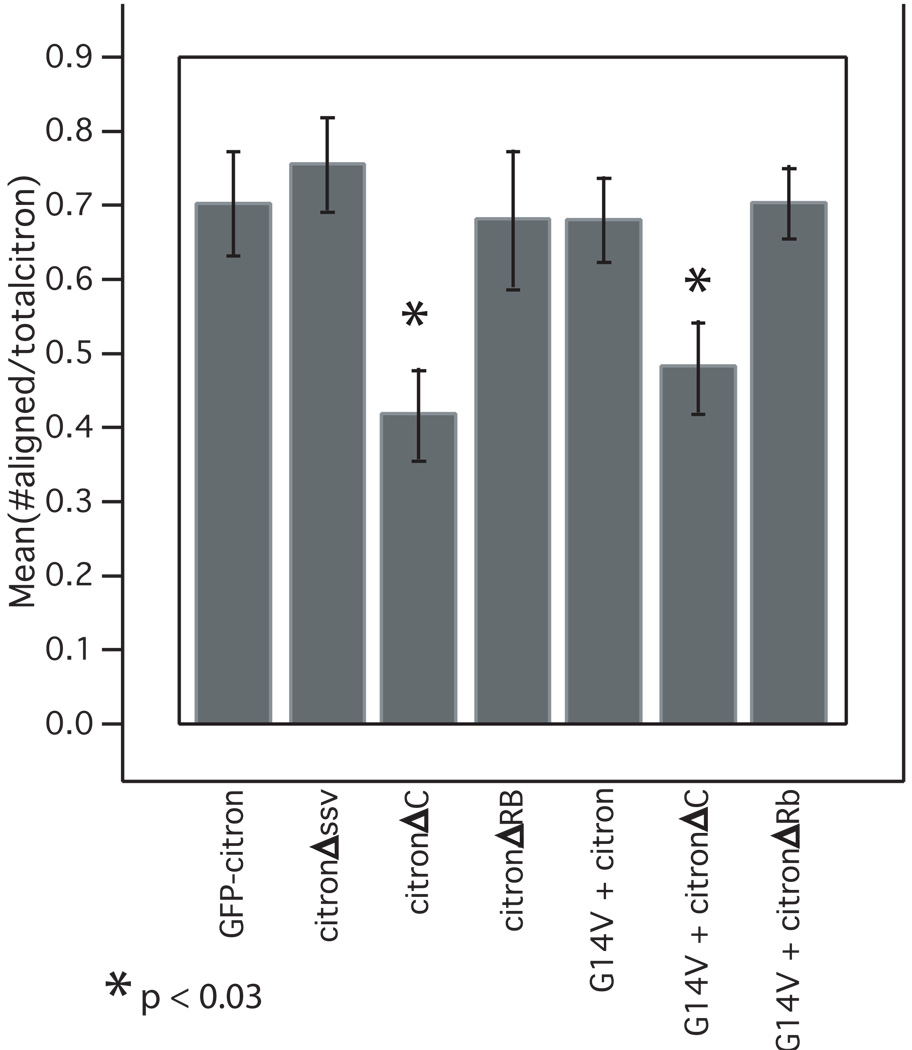
Figure 5.
The C-terminal domain mediates synaptic alignment. Bar graph in displays the ratio of the number of citron puncta aligned with presynaptic, synaptophysin-labeled puncta to the total number of citron puncta. Citron mutants lacking the C terminal domain show a significantly decreased association with synapses in the absence and presence of constitutively active RhoA (G14V). Means were compared using ANOVA and bars show SEM. Measurements were taken from an average of 8 neurons (range 5 – 12) taken from 2 – 3 separate cultures.
Mutants lacking the entire C-terminal half including cysteine rich zinc binding, pleckstrin homology, CMG and proline rich motifs (citronΔC) also form puncta in neurons. The density is similar to full length citron, but 42% fewer are associated with presynaptic vesicle clusters (Fig. 4, 5). The slight offset between synaptophysin and citron labeling suggests that the clusters that do localize to synapses are concentrated postsynaptically and excluded from presynaptic terminals (Fig. 4C, overlay). Thus, the CC domain is sufficient to cluster citron and could partially target these clusters to postsynaptic sites. The decrease in colocalization observed in mutants lacking the C-terminal half suggests that C-terminal motifs contain information important for a final stage of targeting or retention at synapses.
Citron lacking its coiled-coil domain (citronΔN) is distributed more diffusely throughout the cytoplasm of neurons. Additionally, it appears to be uniformly expressed in axons and dendrites indicating that its polarized distribution is lost (Fig. 4D). Together the data indicate that the CC domain is important for clustering citron at postsynaptic sites and for targeting citron to the somatodendritic domain. Surprisingly, citronΔN forms a very small number of unusually large clusters that coincide with synaptic markers (arrows, Fig 4D). Additional deletion of cysteine rich zinc binding and PH domains eliminates all synaptic clusters (e.g. citroncterm, Fig. 4E, F). Citroncterm concentrates in regularly distributed clusters in axons (Fig. 4G), but this is likely to be an artifact of axonal expression such that the mutant binds to axonal proteins (via proline rich or PDZ binding domains, for example) that would not normally be exposed to citron. Together with the results obtained using citronΔC, the data suggest that the C-terminal half contains information important for the recruitment of citron to postsynaptic sites. However, the mechanism that underlies this recruitment is clearly inefficient since the majority of citronΔN distributes diffusely in the cytoplasm.
Domains required for citron clustering
To more directly assess the citron domains responsible for clustering, the same series of GFP-citron mutants was introduced into HEK or L cells, neither of which express endogenous citron. In single optical sections taken through the center of the cell, full length citron can be seen to form widely distributed clusters (Fig. 6A). Similar to neurons, citron lacking its PDZ binding domain (citronΔSSV) shows a punctate distribution indistinguishable from full length citron (Fig 6B vs. 6A; Table 1), supporting that citron clustering is likely to be independent of any interaction with PDZ binding proteins. In fact, citronΔC formed puncta as well as native citron (Fig. 6D, E). Deleting the Rho binding domain alone also has no effect on clustering. In stark contrast, mutants lacking the entire coiled-coil domain (citroncterm and citronΔN) are diffusely distributed throughout the cytoplasm with no evidence of puncta (Fig. 6C and data not shown). To further define the contribution of the CC domain to the generation of puncta, further deletions were made. Mixed patterns of diffuse and punctate localization are observed with mutants containing only the first (citron1–472) or second (citron472–920) halves of the CC domain, but cells expressing the second half form puncta more consistently (Fig. 6F,G). Mutants expressing shorter pieces of the CC domain (e.g. citron472–704) produce no puncta at all (Fig. 6H). Together these data indicate that sequences contained within the coiled-coil domain of citron are both necessary and sufficient to generate citron puncta in heterologous cells, as well as in neurons (Fig. 4; table 1). Additionally, puncta formation appears to require a particular CC domain length rather than sequence in that some clustering is observed with either the N- or C-terminal halves (473 aa and 449 aa, respectively), but none is observed when the CC domain is reduced to 232 aa.
Figure 6.
The relationship between a punctate distribution (arrows) in heterologous cells and the formation of oligomers. Fluorescence images show that GFP-tagged full length citron (A) or citron lacking its C-terminal PDZ binding domain (B) are exclusively localized in puncta when expressed in HEK293 cells. Citron lacking most or all of the C-terminus (D, E) or the Rho binding domain (I) also form puncta, while citron lacking its N-terminal CC domain is diffusely localized (C). Subdividing the CC domain into either 1–472 amino acid (F) or 472–920 amino acid (G) halves greatly reduces the ability to form puncta with both diffuse and punctate labeling evident. Further truncation of the CC domain eliminates all puncta (H). Neither full length citron or citron lacking its Rho binding domain associated predictably with GM130 labeling (J–M). Dotted lines show cell outlines. To determine whether citron can form homo-oligomers, full length or mutant citron was expressed in HEK 293 cells and cross linked (+) (see Methods) or not (−) (N, O). All, but citronΔN migrate to a much larger size consistent with the formation of homotetramers. The size differences are most accurately determined for the smaller constructs where the sizes are estimated to be (in KDa): a = 200, b = 55; c = 320, d = 80, e = 310, f = 160, g = 78, h = 115. Bands a,c appear to be tetramer forms of band b,d, and bands e and f appear to be tetramer and dimer forms of g, respectively. The relative proportion of the crosslinked products migrating as a tetramer relative to total protein is indicated at the bottom of each cross-linked lane as a percent. Tetra-oligomer formation correlates with puncta formation, and the presence of monomers correlates with diffuse staining. Bar = 10 µm.
The broad distribution of clusters we see supports they are cytoplasmic. However, recent work suggests that citron can partially colocalize with Golgi (Camera et al, 2005). While we observe only very low levels of native citron in neuronal cell bodies (Fig. 1C, D), we asked whether overexpressed citron might colocalize with Golgi markers in HEK cells. We observe no consistent association between citron and GM130 labeling, a marker for Golgi membranes (Fig. 6J, K). Neither does GFP citron colocalize with a lysosomal marker (data not shown). Overexpressing citron absent the Rho binding domain can disrupt Golgi structure in neurons (Camera et al, 2005), but citronΔRB produced no detectable change in GM130 distribution in HEK cells (Fig. 6L, M). Together these data support that the citron clusters we observe in HEK are likely to be cytoplasmic.
Citron can self-multimerize
In many molecules coiled-coil domains form homodimeric strands. Since citron requires its CC domain to form puncta, it seemed a strong possibility that the mechanism for citron clustering would be self-multimerization. To address this, we expressed GFP-citron in HEK293 cells, crosslinked the cell extracts using BS3 and immunoblotted using an anti-GFP antibody. Crosslinked GFP-citron produces a very large molecular weight product that is outside the range delimited by the highest molecular weight standard (250 kDa, Fig. 6N). While it is very difficult to estimate accurately the size of this product, the size shifts observed with the smaller citron constructs suggests that GFP-citron forms a tetramer. For example, GFP-citron472–704 (55 kDa, Fig. 6Ob), GFP-citron1–472 (80 kDa, Fig. 6Od), GFP-citron472–490 (78 kDa, Fig. 6Og) all show crosslinked bands that migrate at sizes approximately four times the monomeric molecular weight (Fig. 6Oa, c, and e, respectively). The crosslinked bands are most likely homomers as apposed to complexes with other proteins since crosslinked complexes are about 4 times the monomeric protein. Additionally, fainter crosslinked bands at sizes corresponding to homodimers appear in many of the lanes, particularly following longer exposures (Fig. 6Of and data not shown). Citron mutants lacking the entire C-terminal half or those lacking the Rho binding domain alone form tetramers as well as full length citron (Fig. 3, Fig. 6N). Mutants lacking the coiled-coil domain (citronΔN) do not multimerize at all (Fig. 3, Fig. 6O), and any truncation of the coiled-coil region reduces the ability to form multimers (Fig. 6O, first 6 lanes). Some multimers can be detected with citron 1–472 and citron 472–704, both mutants showing a mixed punctate and diffuse cellular distribution pattern in heterologous cells, but in these cases the degree of multimerization appears to be insufficient to drive a consistent punctate pattern in cells (Fig. 6F, H, O). Notably, all of the mutants that form puncta in cells appear to be able to self-assemble into tetramers, and the degree of diffuse staining correlates with the appearance of monomers (Fig. 6 and Table 1). Together the data support that self-multimerization is likely to be the principal mechanism driving citron clustering (Table 1).
Rho activity and citron distribution
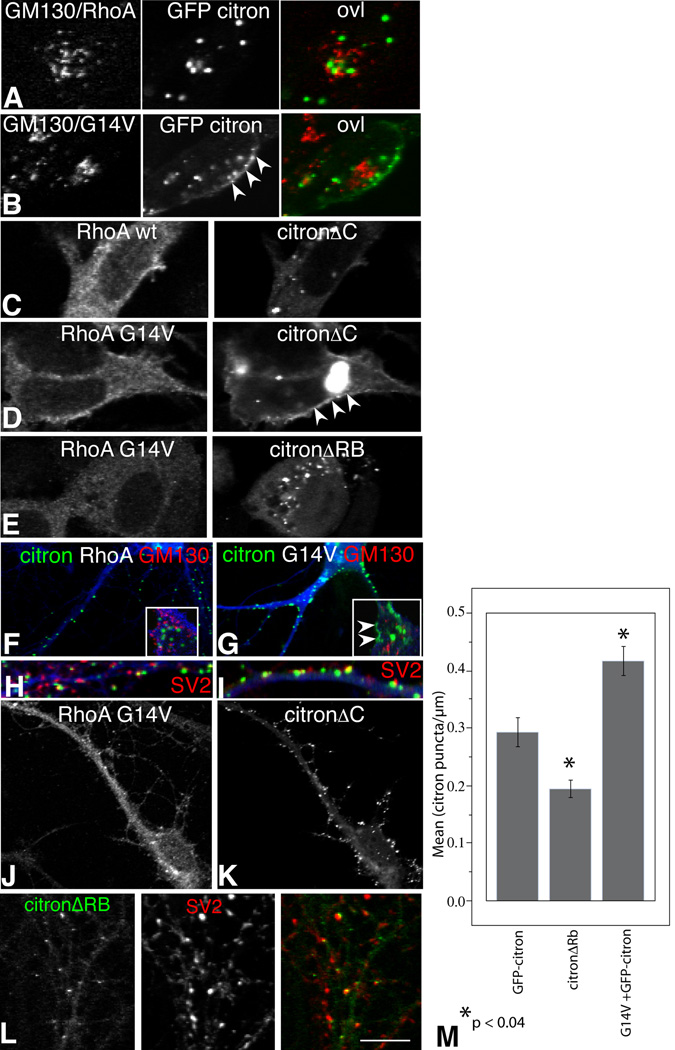
To determine whether Rho activation can alter citron distribution, we compared the localization of citron in HEK293 cells co-transfected with GFP-citron and RhoA or a mutant RhoA that is constitutively active (RhoAG14V). In cells expressing RhoA and citron, RhoA is diffusely distributed throughout the entire cell (similar to Fig. 7C) and citron forms widely distributed clusters as it does normally (Fig. 7A vs. Fig. 6A). Cells expressing RhoAG14V and citron direct a portion of citron to the cell periphery, such that citron spreads beneath the membrane where it forms either fewer or smaller clusters (Fig. 7B). The Rho-dependent change in citron localization is not paralleled by a similar redistribution of Golgi supporting that this effect is separable from any potential citron-Golgi interactions (Fig. 7A vs. 7B). CitronΔC is similarly redistributed (Fig. 7C, D), but selective deletion of the Rho binding site abolishes the submembranous distribution pattern (Fig. 7E, F). These data suggest that Rho activation promotes the recruitment or stabilization of citron beneath the membrane, similar to what has been observed for citron-K in HeLa cells (Eda et al, 2001). The increased recruitment could occur by changes in oligomerization, but we observe no detectable differences in the amount of crosslinked products when citron is co-expressed with RhoA or RhoAG14V (data not shown).
Figure 7.
Single confocal optical sections taken through the center of HEK293 cells (A–E) or cultured hippocampal neurons (F–L) expressing full length GFP-citron or the citron mutants indicated along with myc-tagged wild type RhoA (RhoA) or constitutively active RhoA (RhoAG14V) as indicated (see Fig. 3). Both RhoA wt and RhoAG14V are broadly distributed (A–D), but citron puncta that form in GFP-citron (A, B) or in GFP-citronΔC (C, D) become redistributed to the plasma membrane in cells expressing constitutively active RhoA (B and D, arrowheads). GM130 labeling for Golgi (A and B) shows little in the way of overlap with citron. Cells expressing citron lacking the RhoA binding domain along with active RhoA show no such clustering at the plasma membrane, supporting that this action on citron is direct (E). Citron puncta forming in neurons co-expressing citron and RhoA are indistinguishable from those forming in the absence of RhoA (compare G, and 4A). Coexpression with RhoA G14V increases puncta number (F, G, M), but the puncta show a similar degree of synaptic association as neurons expressing citron and RhoA H, I and Fig 5). Cell bodies shown in insets show labeling for GM130 (red), citron (green) and myc (blue). Colocalization between citron and Golgi is low, but Rho activation leads to some redistribution of citron beneath the plasma membrane (arrowheads in G) similar to HEK cells (B). In figure H, I, color codes are citron:green, Rho:blue, and SV2:red. Neurons coexpressing RhoA G14V and citronΔC show similar elevations in puncta number as full length citron, but the puncta show decreased association with synapses as citronΔC alone (J, K and Fig. 5). Mutants lacking the Rho binding domain show decreased cluster area and numbers, but puncta remain synaptically localized (L, M and Fig. 5). Bar graph in (M) shows that deleting the RhoA binding domain decreases citron puncta number while activating RhoA increases it. Means were compared using ANOVA and bars show SEM. Measurements were taken from an average of 20 neurons (range 16 – 23) taken from 2 – 3 separate cultures. Magnification Bar = 10 µm A–I and L; 20 µm J and K.
To determine whether neurons might also show Rho-mediated changes in citron localization, citron-GFP was cotransfected in hippocampal neurons along with Rho or RhoG14V. While RhoAG14V occasionally co-clusters with GFP-citron, it is more commonly distributed diffusely throughout the neuron (Fig. 7G). Constitutively active Rho produces an obvious increase in citron puncta density (Fig. 7F–I, M), and citron can also become clustered beneath the plasma membrane in cell bodies, similar to what we observed in HEK cells (compare insets, Fig. 7F, 7G). In some cases citron cluster size also appears increased (Fig. 7G), but this difference was not statistically significant. Most puncta appear on dendritic shafts (Fig. 7G, I), but are also observed on dendritic protrusions. Similar results are observed when RhoAG14V is co-expressed with the coiled-coil domain alone (Fig. 7J, K; 7K compared to 4C). Conversely, neurons expressing a citron mutant lacking the Rho binding region show a decreased density of significantly smaller clusters relative to GFP-citron controls (0.235 µm2 ± 0.01 for citronΔRB cluster area vs. 0.337 µm2 ± 0.01 for GFP-citron) (Fig. 7L, M). Importantly, in citron-GFP/RhoAG14V co-transfectants as well as in neurons expressing citronΔRB, the proportion of citron puncta associated with presynaptic terminals is similar to control neurons (Fig. 5, 7H, I). These results suggest that Rho activation distributes citron to a greater proportion of synapses.
Discussion
Excitatory synapses that form along the smooth dendrites of GABAergic interneurons in hippocampus contain high levels of citron postsynaptically. Such high levels appear to be distinctly characteristic of hippocampal interneurons, but here we find that citron is also clustered at excitatory synapses on dendritic spines of a subpopulation of glutamatergic pyramidal neurons taken from hippocampus and neocortex. Additionally, we find that citron is targeted to the somatodendritic domain and forms clusters using its N-terminal coiled-coil (CC) domain, most likely by forming homo-oligomers. The CC domain is also sufficient to direct more than half of the citron clusters to synases. Rho binding and activation regulate membrane targeting and cluster number, while interactions with citron’s C-terminal domains help to increase the fidelity of citron’s synaptic localization. Together our findings indicate that citron becomes restricted to a particular set of synapses according to a pattern that is dictated by cell type, protein-protein interactions and the activation state of RhoA. Thus, citron itself is downstream of RhoA signaling and is strategically positioned to orchestrate RhoA-mediated function at a heterogeneous population of synapses.
Citron localizes to spine and shaft synapses without PSD95/SAP90
Citron concentrates selectively at excitatory postsynaptic sites on GABAergic interneurons in a pattern that is nothing short of striking (Zhang et al., 1999). However, we also find that lower levels of citron cluster at glutamatergic synapses forming on spines and shafts in a subpopulation of a variety of glutamatergic neurons taken from hippocampus and neocortex. Citron’s restricted localization pattern seems to be governed principally by the high selectivity of protein expression in particular neurons--citron is expressed in most GABAergic neurons and a subpopulation of excitatory neurons in cortex and hippocampus. Consistent with this native localization pattern, exogenously introduced citron localizes to glutamatergic postsynaptic sites independent of cell type. These data indicate that citron’s localization to glutamatergic synapses is not unique to shaft synapses and suggest that citron’s actions can be exerted at different kinds of glutamatergic synapses. At the same time, citron is expressed in a discrete population of neurons, and within individual cells, it is not localized to every synapse, even when overexpressed. As a Rho target, citron’s likely role as an effector is restricted to a heterogenenous but circumscribed set of synapses. It will thus be important in future experiments to determine the functional consequences that result from the selective localization pattern.
Since PSD-95/SAP90 and citron show similar intraneuronal distribution patterns in hippocampal GABAergic neurons and become clustered in neurons on a similar developmental time scale (Zhang and Benson, unpublished), we hypothesized that citron might become localized to synapses via its association with PSD-95/SAP90 similar to the synaptic targeting that results from the interaction between a PDZ domain of Discs large (DLG) and the C terminus of Shaker K+ channels at the neuromuscular junction in Drosophila (Tejedor et al., 1997, Zito et al., 1997). However, citron clusters at synapses independent of any interaction with its PDZ binding domain. While synaptic targeting independent of PSD-95/SAP90 binding could conceivably occur by citronΔSSV forming tetramers with native citron, it is unlikely since synaptic targeting occurs in spiny neurons where native citron levels are exceedingly low or absent as well as in smooth dendrites where native citron levels are high. It is also clear that PSD-95/SAP90 does not require citron for its synaptic localization as citron is clustered at far fewer excitatory synapses (see also (Craven and Bredt, 1998, El-Husseini Ael et al., 2001). Citron binds principally to the third PDZ domain of PSD95/SAP90 as does neuroligin, a synaptic cell adhesion molecule that also appears to target synapses independent of it its interaction with PSD-95/SAP90 (Dresbach et al., 2004).
Molecular determinants responsible for clustering and targeting citron
In addition to citron’s tSXV tail, we find that the entire C-terminal half of citron is dispensable for citron clustering. Citron constructs containing the N- terminal CC domain alone form puncta in both heterologous cells and neurons. The ability of particular citron mutants to form puncta strongly correlates with the ability of the same mutants to form tetramers suggesting that citron self-oligomerization is the principal means by which citron clusters in neurons. The C1 and PH domains may negatively regulate the clustering ability of citron’s CC domain in that a recent report showed constructs containing these two domains in addition to the CC domain were diffusely localized (Camera et al, 2005). However, data presented here show that full length citron forms clusters as well as mutants lacking these domains suggesting that such an effect is likely to be minimal. Additionally, it seems that the neuronal clusters are also likely to contain additional protein(s) that can participate in citron’s synaptic localization. This idea gains support from our data showing that the deletion of the Rho binding domain reduces, but does not eliminate citron’s synaptic localization.
Constructs containing the CC domain alone also show partial synaptic localization suggesting that this domain encodes synapse targeting information or alternatively, this domain may interact with another dendritically targeted protein similar to what has been described for the CC domains in Homer which have been proposed to form complexes that crosslink mGluR at postsynaptic sites (Xiao et al., 1998). However, citron also requires its C-terminal half to attain the high degree of synaptic localization achieved by native citron. While there are a variety of candidate protein interaction domains such as C1, PH and citron homology domains in the C terminal half, it is worth noting that the region extending from the CMG domain to C-terminus can interact with profilin IIa (Camera et al., 2003) an actin binding and regulatory protein, which can be found enriched at synapses (Giesemann et al., 2003).
Recent work has shown that citron may also be important for the appropriate organization of Golgi in cell bodies of cultured hippocampal neurons (Camera et al., 2003). It is possible then that synaptic citron may serve as a mechanism to localize and organize Golgi membranes near synapses—an intriguing idea since synaptically localized Golgi elements could participate in the local synthesis and processing of membrane proteins (Horton and Ehlers, 2003). However, we were unable to find consistent patterns of interaction between citron and a Golgi marker suggesting this interaction may be weak or state dependent. At the very least, our data support that citron has a function outside that of Golgi organization.
Regulating citron at synapses
Members of the Rho family can regulate dendritic branching patterns and spine morphology (Luo et al., 1996, Li et al., 2000, Nakayama et al., 2000, Tashiro et al., 2000, Luo, 2002), but how they might influence the structure of the synaptic junction is not understood. In general, Rho GTPases are activated or inactivated locally, and the functional outcome is determined by the local composition of effector proteins. Activated Rho binds and activates citron-K (Di Cunto et al., 1998, Madaule et al., 1998, D'Avino et al., 2004), suggesting that citron which lacks the kinase domain, but is otherwise identical may be a key downstream signaling component driving changes in actin organization at the synapse. Here we show that citron itself is downstream of Rho activation, and that the interaction between citron and RhoA redistributes citron clusters peripherally where it appears less punctate and more smoothly distributed beneath the plasma membrane in HEK293 cells and in neuronal cell bodies. This is similar to what is observed in HeLa cells where activated Rho redistributes citron-K to the cleavage furrow in a process that is important for normal cytokinesis (Eda et al., 2001). In neurons, constitutively active Rho also causes an increase in the density of synaptic citron clusters. In dendrites it would be difficult to detect a shift toward the membrane, because in contrast to HEK293 cells, citron clusters associate with synaptic junctions placing them near the membrane with or without Rho activation. However, the postsynaptic density is a loosely layered, but precisely organized structure (Valtschanoff et al 2001; Naisbitt et al, 1999), and small changes in subsynaptic distribution would be predicted to alter the landscape of potential binding partners, thus, producing distinct functional outcomes.
Rho activation is important for regulating the number of synapses having citron clusters in neurons. It is not the sole requirement, since deleting the Rho binding domain diminishes, but does not eliminate synaptic citron. The consequences of changing the number of synapses having citron are not known, but recent work outlines some intriguing possibilities. Increases in profilin II decrease dendritic spine motility in an activity-dependent manner (Ackermann and Matus, 2003). Citron’s interaction with profilin II (Camera et al., 2003) may modulate this or serve to focus profilin action at synapses.
Acknowledgements
We thank Bettina Winckler, Stephen Salton, Tonya Anderson and the members of the Benson lab for their comments on the manuscript. This work was supported by NIH NS37731 and an Irma T. Hirschl Award.
References
- Ackermann M, Matus A. Activity-induced targeting of profilin and stabilization of dendritic spine morphology. Nat Neurosci. 2003;6:1194–1200. doi: 10.1038/nn1135. [DOI] [PubMed] [Google Scholar]
- Benson DL, Colman DR, Huntley GW. Molecules, maps and synapse specificity. Nat Rev Neurosci. 2001;2:899–909. doi: 10.1038/35104078. [DOI] [PubMed] [Google Scholar]
- Camera P, da Silva JS, Griffiths G, Giuffrida MG, Ferrara L, Schubert V, Imarisio S, Silengo L, Dotti CG, Di Cunto F. Citron-N is a neuronal Rho-associated protein involved in Golgi organization through actin cytoskeleton regulation. Nat Cell Biol. 2003;5:1071–1078. doi: 10.1038/ncb1064. [DOI] [PubMed] [Google Scholar]
- Craven SE, Bredt DS. PDZ proteins organize synaptic signaling pathways. Cell. 1998;93:495–498. doi: 10.1016/s0092-8674(00)81179-4. [DOI] [PubMed] [Google Scholar]
- D'Avino PP, Savoian MS, Glover DM. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J Cell Biol. 2004;166:61–71. doi: 10.1083/jcb.200402157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto F, Calautti E, Hsiao J, Ong L, Topley G, Turco E, Dotto GP. Citron rho-interacting kinase, a novel tissue-specific ser/thr kinase encompassing the Rho-Rac-binding protein Citron. J Biol Chem. 1998;273:29706–29711. doi: 10.1074/jbc.273.45.29706. [DOI] [PubMed] [Google Scholar]
- Dresbach T, Neeb A, Meyer G, Gundelfinger ED, Brose N. Synaptic targeting of neuroligin is independent of neurexin and SAP90/PSD95 binding. Mol Cell Neurosci. 2004;27:227–235. doi: 10.1016/j.mcn.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Eda M, Yonemura S, Kato T, Watanabe N, Ishizaki T, Madaule P, Narumiya S. Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J Cell Sci. 2001;114:3273–3284. doi: 10.1242/jcs.114.18.3273. [DOI] [PubMed] [Google Scholar]
- El-Husseini Ael D, Craven SE, Brock SC, Bredt DS. Polarized targeting of peripheral membrane proteins in neurons. J Biol Chem. 2001;276:44984–44992. doi: 10.1074/jbc.M103049200. [DOI] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20:847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- Furuyashiki T, Fujisawa K, Fujita A, Madaule P, Uchino S, Mishina M, Bito H, Narumiya S. Citron, a Rho-target, interacts with PSD-95/SAP-90 at glutamatergic synapses in the thalamus. J Neurosci. 1999;19:109–118. doi: 10.1523/JNEUROSCI.19-01-00109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesemann T, Schwarz G, Nawrotzki R, Berhorster K, Rothkegel M, Schluter K, Schrader N, Schindelin H, Mendel RR, Kirsch J, Jockusch BM. Complex formation between the postsynaptic scaffolding protein gephyrin, profilin, and Mena: a possible link to the microfilament system. J Neurosci. 2003;23:8330–8339. doi: 10.1523/JNEUROSCI.23-23-08330.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Aelst L, Cline HT. Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat Neurosci. 2000;3:217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- Madaule P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- Madaule P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. A novel partner for the GTP-bound forms of rho and rac. FEBS Lett. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P. Synaptic clustering of AMPA receptors by the extracellular immediate- early gene product Narp. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- Rao A, Kim E, Sheng M, Craig AM. Heterogeneity in the molecular composition of excitatory postsynaptic sites during development of hippocampal neurons in culture. J Neurosci. 1998;18:1217–1229. doi: 10.1523/JNEUROSCI.18-04-01217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. Essential role for dlg in synaptic clustering of Shaker K+ channels in vivo. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo H, Kandel ER. Using adenovirus-mediated gene transfer to study the role of the Rho GTPase in cultured hippocampal neurons. Proceedings of the Society for Neuroscience Annual Meeting. 1996;26:694. 619. [Google Scholar]
- Walikonis RS, Jensen ON, Mann M, Provance DW, Jr, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Zhang W, Vazquez L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito K, Fetter RD, Goodman CS, Isacoff EY. Synaptic clustering of Fascilin II and Shaker: essential targeting sequences and role of Dlg. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Garner CC. Cellular and molecular mechanisms of presynaptic assembly. Nat Rev Neurosci. 2004;5:385–399. doi: 10.1038/nrn1370. [DOI] [PubMed] [Google Scholar]