Abstract
The human microbiome refers to the community of microorganisms including prokaryotes, viruses and microbial eukaryotes that populate the human body. The National Institutes of Health launched an initiative that focuses describing the diversity of microbial species associated with health and disease. The first phase of this initiative includes the sequencing of hundreds of microbial reference genomes, coupled to metagenomic sequencing from multiple body sites. Here we present results from an initial reference genome sequencing of 178 microbial genomes. From 547,968 predicted polypeptides that correspond to the gene complement of these strains “novel” polypeptides that had both unmasked sequence length > 100 amino acids and no BLASTP match to any non-reference entry in the nr subset were defined. This analysis resulted in a set of 30,867 polypeptides, of which 29,987 (~97%) were unique. In addition, this set of microbial genomes allows for ~ 40% of random sequences from the microbiome of the gastrointestinal tract to be associated with organisms based on the match criteria used. Insights into pan-genome analysis suggest that we are still far from saturating microbial species genetic datasets. In addition, the associated metrics and standards used by the group for quality assurance are presented.
The human microbiome is the enormous community of microorganisms occupying the habitats of the human body. Different microbial communities are found in each of the varied environments of human anatomy. The aggregate microbial gene tally surpasses that of the human genome by orders of magnitude. The relationship of the microbial content to health and disease is one of the primary goals of human microbiome studies. The structure and function of any microbial community requires a detailed definition of the genomes that it encompasses and predicting and annotating their genes.
In 2007, the National Institutes of Health (NIH) initiated the Human Microbiome Project (HMP) as one of its Roadmap initiatives (1) to provide resources and build the research infrastructure. One component of the HMP is production of reference genome sequences for at least 900 bacteria from the human microbiome to catalog the microbial genome sequences from the human body and aid researchers conducting human metagenomic sequencing to assign species to sequences in their metagenomic data sets.
The HMP catalog of reference sequences is being produced by the NIH HMP Jumpstart Consortium of four Genome Centers: the Baylor College of Medicine Human Genome Sequencing Center; the Broad Institute; the J. Craig Venter Institute; and the Genome Center at Washington University. The challenges for the Jumpstart Consortium include selecting strains to sequence and identifying sources, creating standards for sequencing and annotation to ensure consistency and quality, and rapid release of information to the community.
Reference genome progress
To date, 239 genomes including 61 genomes at various stages of upgrading have been produced by the Jumpstart Consortium and released into public databases. At the time of manuscript preparation, 178 had been completely annotated and are presented in the analysis here. The process for selection of these strains is described in the supporting online material. The strains sequenced to date are distributed among body sites as follows: gastrointestinal tract (151), oral cavity (28), urogenital/vaginal tract (33), skin (18), respiratory tract (8), and also include one isolate from blood (see (2)). These are the five major body sites targeted by the HMP.
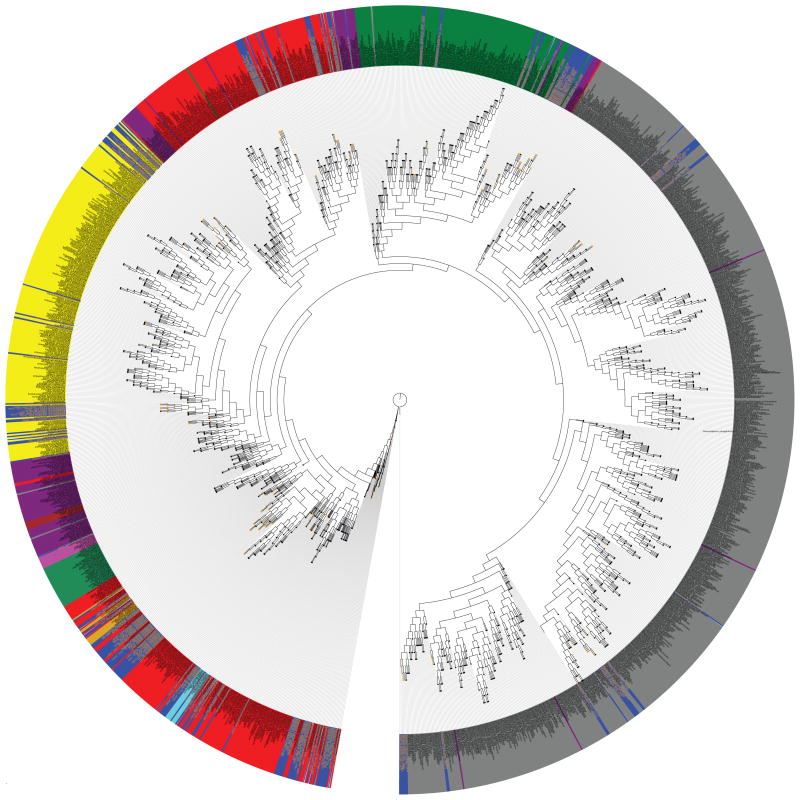
The broad phylogenetic distribution of the sequenced strains (Figure 1) represents a 16S rRNA overlay of HMP sequenced genomes on 16S rRNA sequences from cultured organisms with sequenced genomes (3). HMP-sequenced genomes represent two kingdoms (Bacteria and Archaea), 9 phyla, 18 classes, and 24 orders. Additional rRNA overlay figures broken down by individual body sites are available (4).
Figure 1. Phylogenetic tree of 16S rDNA sequences.
The tree was created using ~1500 16S rDNA representing single species. Organisms sequenced as part of the HMP project are highlighted in blue. Additional coloring indicates separation by phylum: yellow, Actinobacteria; dark green, Bacteroidetes; light green, Cyanobacteria; red, Firmicutes; cyan, Fusobacteria; dark red Planctomycetes; grey, Proteobacteria; magenta, Spirochaetes; light pink, TM7; tan, Tenericutes. The purpose of this analysis is not the details of the branching structure (which include minor known artifacts), but the overall distribution of the HMP strains (in blue) around the tree of life.
To obtain high quality draft genomes and a meaningful gene list, minimum standards were defined for assembly and annotation of draft genomes. Three reference bacterial genome assemblies were evaluated for efficacy of gene predictions and genome completeness. Based on the analysis, metrics for assembly characteristics and annotation characteristics were defined (for more details see supporting online materials). The quality of HMP genome assemblies are summarized in Table 1 and exceed the Jumpstart Consortium standards described in the supporting online materials with the exception of some genomes produced before standards were in place. More stringent metrics (N75 and N90 for contig and scaffold continuity) are presented, and nearly all genomes satisfy these higher standards.
Table 1.
Draft assembly metrics, organized by finishing status1
| Draft | Improved | ||||||
|---|---|---|---|---|---|---|---|
| Number of strains | 133 | 45 | |||||
| Metric | Passing standard | Pass % | Mean | Range | Pass % | Mean | Range |
| % of the genome included in contigs2 | >90% | 100% | 98.23% | 95.1–99.9% | 100% | 99.91% | 98.6–100% |
| % of the bases greater then 5x read coverage3 | >90% | 99% | 98.90% | 80.8–100% | 100% | 99.35% | 98.8–99.6% |
| Contig N50 | >5 KB | 100% | 102.61 kb | 11.12–861.67 kb | 100% | 517.92 kb | 58.03–3472.99 kb |
| Contig N75 | N/a | 99% | 54.82 kb | 4.97–556.76 kb | 100% | 340.20 kb | 30.56–2635.77 kb |
| Contig N90 | N/a | 90% | 25.54 kb | 2.01–240.69 kb | 100% | 211.51 kb | 14.96–2635.77 kb |
| Scaffold N502 | >20 KB | 100% | 883.93 kb | 50.56–3356.77 kb | 100% | 606.77 kb | 91.71–2898.42 kb |
| Scaffold N752 | N/a | 100% | 511.35 kb | 24.31–3237.97 kb | 100% | 378.22 kb | 52.32–2391.23 kb |
| Scaffold N902 | N/a | 99% | 282.14 kb | 11.74–2490.47 kb | 100% | 226.24 kb | 28.67–2391.23 kb |
| Average contig length | >5 KB | 100% | 31.52 kb | 5.62–180.70 kb | 100% | 174.70 kb | 23.26–1321.04 kb |
| % of core genes present in gene list | >90% | 99% | 99.63% | 86.4–100% | 100% | 99.90% | 98.5–100% |
Based on current assignments. Draft corresponds to Standard or High Quality Draft sequences, with no additional automated or manual attempts to improve assembly, beyond ensuring exclusion of contaminating sequence. Improved columns correspond to Improved-High-Quality-Draft submission, as defined in section 1.3. None of the reference genomes have been improved beyond this grade at this point.
Calculated only for strains with scaffold assemblies submitted to NCBI. The number of strains with scaffold assemblies, by grade: Draft (74), Improved (37).
Per base coverage not available for all reads, for example, those with some draft level of sequencing prior to the Jumpstart initiative or strains where a combination of technologies was used. The number of strains with per base read coverage: Draft (121), Improved (4).
Genome improvement
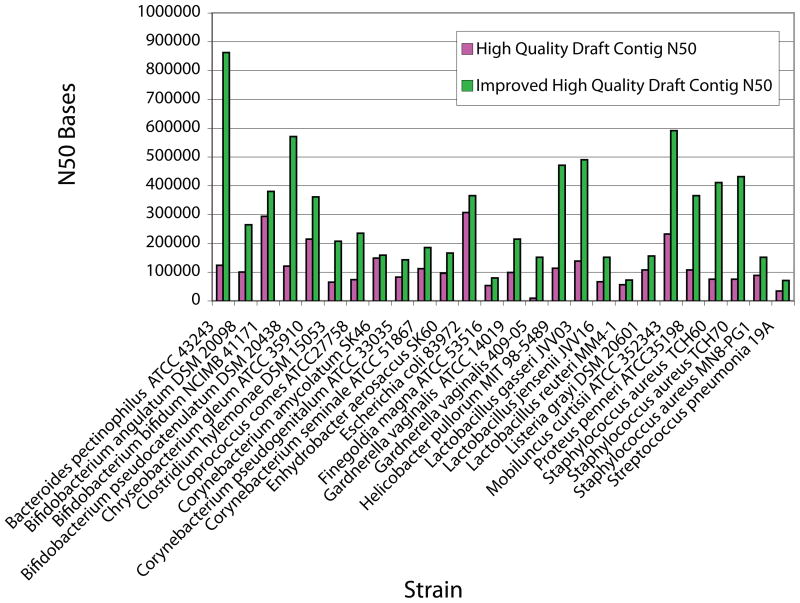
As described in supporting online materials, there are justifications for upgrading these High Quality Draft assemblies. The Jumpstart Consortium has completed initial improvement work on 26 bacterial genomes that differed significantly with respect to GC content and assembly metrics to explore the effort required and resulting benefits (Figure 2). The average contig N50 increased 3.63-fold, from 109 kb at draft to 396 kb after improvement. Bacteroides pectinophilus displays substantial improvement in N50, from 163 kb in the draft sequence to 862 kb after improvement. Lactobacillus reuteri illustrates the opposite extreme with improvement leading to a smaller contig N50 change, 56 kb to 72 kb. As more genomes improve and some graduate to higher levels of improvement, the assembly state or group of states most useful to the HMP scientific goals will be evaluated.
Figure 2. Contig N50 comparison for twenty-six draft and improved genomes.
High quality draft contig N50 bases are shown in magenta and improved high quality draft sequences are shown in green. These data represent the variety of approaches from the four data generation centers. The majority of shotgun data were produced on the Roche-454 platform, though some assemblies include paired Sanger reads to improve contiguity. All draft assemblies are based on the Roche-Newbler assembler, though some of the improved assemblies are based on PCAP (11) and the Celera assembler due to existing integration with finishing and improvement pipelines. Additional variation comes from the improvement approach. Directed Sanger reads from gap spanning PCR amplicons serves as the primary approach while some assemblies have been subjected only to manipulation of the shotgun data, making unrealized joins, removing poor quality data and placing unincorporated shotgun reads.
Pan-genome analysis
A bacterial species’ pan-genome can be described as the sum of the core genes shared among all sequenced members of the species, and the dispensable genes, or those genes unique to one or more strains studied. To start addressing questions about pan-genomes, we identified all species within our sequenced reference genome catalogue for which there was more than one sequenced and annotated genome. Of the nine species identified, four have five or more annotated genomes, generated either by the HMP or external projects publicly available at NCBI; five genomes being the minimum number for which a curve can reliably be fit to pan-genome data. These are Lactobacillus reuteri, Bifidobacterium longum, Enterococcus faecalis and Staphylococcus aureus. The genomic data used for the analysis consisted of both complete and draft genomes, the only requirement being that >90% of the genome is represented in the available annotated contigs or scaffolds.
Pan-genome curves (5) of the GIT isolates L. reuteri, B. longum, and E. faecalis (Figures S3-5) are consistent with an open pan-genome model, suggesting that more genome sequencing needs to be undertaken in order to characterize the actual make up of the species as a whole. Preliminary results suggest core genome sizes of approximately 1430 genes, 1800 genes and 1600 genes for B. longum, E. faecalis and L. reuteri, respectively. Based on the current core gene plots, L. reuteri (Figure S3) appears to be approaching a closed pan-genome model, with newly sequenced strains contributing very small numbers of new genes to the pan-genome; however we see an interesting community substructure within this species. Our current L. reuteri pan-genome analysis of seven isolates suggests four of the seven currently sequenced isolates are very similar to one another, contributing zero to two new genes to the pan-genome. Two further strains are also similar to one another, each contributing an intermediate number of new genes (~15–30), whereas one outlier strain contributes a distinct set of genes (~330). These findings are consistent with the comparison of average nucleotide identity with gene content discussed below for this species. It will be interesting to see whether additional sequencing of this species identifies other subgroups in addition to the three identified here, or whether this sample set is in fact largely representative of the species.
Similar findings for B. longum (Figure S4) suggest that four of the five currently sequenced genomes contribute approximately equally to the pan-genome (~50–150), with one outlier strain (ATCC 15697) contributing a much higher number of novel genes (~640). These data are consistent with differences in gene count across these genomes. Each of the five currently sequenced genomes of E. faecalis (Figure S5) contributes approximately equivalent numbers of new genes to the pan-genome. Our current datasets for these two species are still too small to determine whether we can realistically achieve a closed pan-genome, with newly sequenced isolates contributing on the order of 100 new genes each. It is unrealistic at this point to extrapolate how many additional genomes would need to be sequenced to see whether the number of new genes contributed by each new sequence continues to plateau around 100 new genes or approaches zero.
S. aureus pan-genome plots (Figure S6), representing isolates collected from the skin, urogenital tract and mucus membrane of mammals (human, bovine) are consistent with a closed pan-genome model, as previously suggested (6), with an estimated core size of 2295 genes and an estimated pan-genome size of ~3200 genes.
We performed a preliminary survey looking into the functions encoded by those genes unique to new gene datasets, and not found in the core dataset, based on gene product annotation and Enzyme Commission (EC) numbers, when available. It should be stressed that these genomes underwent automated annotation only, with no manual curation, so any trends seen should be considered putative only. Across all four species, the number of novel genes annotated only as hypothetical or conserved domain of unknown function ranged from 66% to 73%, comprising the bulk of the novel genes identified by the pan-genome analysis. Another predominant trend seen were unique family members corresponding to non-novel functions, e.g. functions also identified in the core dataset.
Potentially interesting categories of functions identified in novel gene sets unique to individual strains include accessory proteins involved in activation of urease, a virulence factor found in microorganisms associated with gastric ulceration, among other human health concerns(5) phage morphogenesis and regulation proteins; and small numbers of unique enzymes involved in metabolism of sugars and amino acids. Further work is needed to clean up annotations and to provide more consistent EC number assignments in order to confirm and build upon trends seen in this preliminary analysis. The HMP Data Analysis and Coordination Center (DACC) is mandated with adding value and updating annotations, which will allow for expansion of these analyses throughout this project.
Measuring diversity within genera
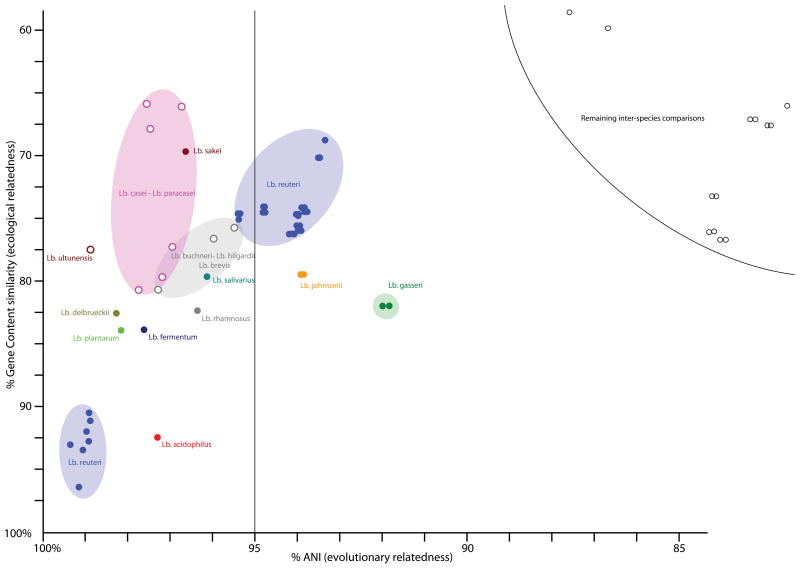
The genomic diversity among strains belonging to the same genus was explored by a measure for the evolutionary relatedness and gene content similarity in a pair wise fashion (Figure 3). The average nucleotide identity (ANI) is a measure for evolutionary relatedness based on sequence similarity between the set of shared genes (8). The measure of gene content similarity between two strains can provides a sense of functional or ecological relatedness, and one might predict that strains with a lower gene content similarity are more likely to be found in different habitats. The three genera selected for this comparison all contain at least 16 strains and include Lactobacillus (36 strains; Figure 3), Bifidobacterium (16 strains; Figure S7), and Bacteroides (21 strains; Figure S8). Genomes contributed by the HMP as well as those available in public databases were included in this analysis. High intra-species diversity was observed within genera in addition to inter-species diversity. Within Lactobacillus, several species showed significant diversity. For example, L. reuteri is represented by two main groups, one set (bottom left blue oval in Fig. 3) contains 7 different strains. Among the strains within that group, the %ANI and % gene content are above 98% and 90% respectively. In the second group (upper right blue oval in Fig. 3), the % ANI ranges between 96% and 93% with a gene content similarity lower than 78%. Previously, a value of 95%ANI was shown to correspond with the recommended cut-off of 70 % DNA–DNA reassociation for species delineation (9). This indicates that the L. reuteri strains obtained within the framework of the HMP significantly increased the known genomic diversity of this named species, as was also demonstrated by the pan-genome analysis. Other strains showing large intra-species diversity belong to L. johnsonii and L. gasseri.
Figure 3. Inter-strain diversity among Lactobacillus genomes.
Each point represents a whole-genome comparison between two Lactobacillus genomes and shows the percentage average nucleotide identity (ANI) on the x-axis as a measure of evolutionary distance, plotted against the percentage of gene content similarity on the y-axis. Only comparisons with ANI values above 85% are shown. The vertical line at 95% corresponds to a recommended cut-off of 70% DNA–DNA reassociation for species delineation. Different intra- and inter-species comparisons are color-coded, with full or open circles respectively, and labeled with given taxonomical name in corresponding color. Colored ovals assist in identifying related data points belonging to a single named species.
Among the strains of B. longum (Figure S7), four (two of which were contributed by the HMP) have pair-wise %ANI values at the higher end of the spectrum, ranging between 96% and 98%, but with relative low gene content similarity, i.e., below 82%, indicating a broad range in gene complements. One additional existing strain (ATCC 15697) has a %ANI below 95% and a gene content similarity below 65% and is therefore a clear evolutionary and ecological outlier.
The analysis of Bacteroides genomes has revealed several close common ancestries. Bacteroides sp. D4 and 9_1_42FAA are closely related to Bacteroides dorei (ANI > 95%), but still with a significant gene content difference, lower than 78% similarity. This suggests that the Bacteroides group may possess many closely related, yet ecologically distinct lineages
Novel genes
The 547,968 predicted polypeptides corresponding to the entire annotated gene complement of these strains (of which 516,631 (94%) were unique) were searched against the bacterial and viral divisions of NCBI’s non-redundant protein database (nr) using WU-BLASTP as described in the supporting information. Each polypeptide was also compared to a merged database of TIGRFAM and Pfam HMMs using version 2a of the HMMER3 package. A set of candidate “novel” polypeptides was defined by selecting those that had both (A) unmasked sequence length > 100 amino acids and (B) no BLASTP match to any non-reference entry in the nr subset. This analysis resulted in a set of 30,867 polypeptides, 5.6% of the total, of which 29,987 (~97%) were unique (10). Clustering this set with CD-HIT (11) resulted in 29,286 unique polypeptides at 98% sequence identity (~5% reduction), 28,857 polypeptides at 95% (~7% reduction), and 28,469 at 90% (~8% reduction). An alternate set of candidate novel polypeptides was also defined by modifying condition (A) above to filter on the number of bases not identified as low complexity sequences by SEG (12) (i.e., the sequence length after removing all seg-masked bases). This alternate initial set contains 28,693 polypeptides.
The above criteria were chosen by inspecting histograms of novel versus non-novel polypeptide counts at various E-value and sequence length thresholds and selecting cutoffs that seemed likely to minimize the number of false positives while not excluding too many true positives. The distribution of novel versus non-novel polypeptide counts overlaps at all E-value thresholds, making it impossible to pick a cutoff that does not exclude any true positives. Therefore a relatively high (100 aa) length threshold was selected in order to try to minimize noise or false positives, at the possible cost of losing some real novel polypeptides.
With ~1,300 completely sequenced bacterial genomes in GenBank(13) the observation that 5% of the genes annotated in the HMP genomes satisfy criteria for novelty underscores the remarkable diversity of bacterial proteins. In order to assess whether there is enriched novelty in the HMP-targeted genomes in relationship to previously sequenced prokaryotic genomes, we randomly selected 178 previously sequenced draft genomes from GenBank and ran the same analysis for comparison. This dataset resulted in 747,522 predicted polypeptides, of which 568,426 were unique. Of these, 14,269 polypeptides met our criteria for novelty, 1.9% of the total, of which 14,064 were unique, 2.5% of the unique total. Clustering resulted in a 2% reduction at 98% and a 3% reduction at 90%, indicating that this dataset does not contain as many highly similar protein predictions as the HMP novel set. This would suggest that there is enrichment in novelty in the HMP dataset of approximately 2:1 over the random dataset. While the human microbiome is generally thought to be less complex than soils, and certain other environmental microbiomes, it nevertheless clearly houses enormous microbial diversity yet to be described.
Analysis of metagenomic shotgun data
Because the HMP reference genomes that were sequenced had been selected primarily because they were isolates from human subjects, and had not been identified as strains seen in metagenomics studies, it was not known how much these genomes would help identify metagenomic sequences obtained from human microbial communities. The most useful reference genomes should expand our ability to interpret metagenomic data. We also used the stringent fragment recruitment technique (14) to compare metagenomic sequencing data to the reference genomes in nucleotide space(15). The stringency of this approach generally limits recruitment of metagenomic reads to organisms within the same genus but can resolve strain specific differences.
Publicly available metagenomic datasets from two human gastrointestinal studies were used in this analysis (16, 17), along with 454 reads from a Washington University dataset (which contributed the bulk of the 16.8 million reads that were tested). The reference genomes included 866 complete and 913 draft genomes available at NCBI including the HMP reference genomes with sequence reads available at the time of analysis. In total 62 HMP genomes showed significant levels of recruitment with 11.3 million metagenomic reads recruited (66% of all reads). Of these, a significant 6.9 million reads (41%) recruited best to the HMP reference genomes, based on the global percent identity (defined as the number of identities between read and reference divided by the length of the read). A read is considered to be a best hit to an HMP genome if the best global percent identity includes a match to an HMP genome. Many of these reads would not have been recruited at all if not for the availability of the HMP reference genomes: between 20% and 40% of the reads were recruited only because of the presence of the HMP genomes.
These results show that a significant number of the genomes sequenced as part of the HMP project are directly adding to our understanding the human microbiome. These results also show that specific genomes are useful references across a wide range of individuals despite the strain specific diversity noted above. Despite the large number of genomes available, a significant amount of the metagenome (33%) is still not well represented by any reference genome. It is likely that the 900 genomes target of the HMP will reduce this number of unidentified reads further without redundancy in genome selection. It should be noted that this analysis focused on the gastrointestinal tract and it is likely that additional genomes exist in other body sites, and thus the composition of the 900 genomes should address these organisms.
Data release, future plans and conclusions
The Jumpstart Centers have made significant progress with respect to the generation of a set of reference genomes that describe the human microbiome. We have made every effort to ensure that all strains are available in public repositories, and to release these genomes and their associated data, assemblies and annotations in accordance with NIH policy (18). In addition, all data and SOPs are available through the Data Analysis and Coordination Center (DACC; (19)) where we welcome community input and feedback.
Human microbiome research groups from around the world have launched an International Human Microbiome Consortium (IHMC), which together will sequence more than 1000 human microbial bacterial reference genomes. This includes the 900 reference strains that are being sequenced by the HMP Jumpstart Centers, 100 genomes sequenced as part of the EU-funded MetaHIT project(20), and additional genomes produced by international efforts. Other strains are being sequenced as part of the DOE Genomic Encyclopedia of Bacteria and Archaea (GEBA; (21, 22)) project. All of these strains appear on the DACC.
Nevertheless, the human microbiome is much more complex than this set of genomes, and is likely to exceed it by orders of magnitude. In addition to the significant number of cultured strains, many unculturable strains remain to be defined, and significant intraspecies diversity still needs to be described. Thus, this initial effort is only a beginning, is valuable, and not only contributes to the catalog of reference strains but also builds infrastructure for strain selection and acquisition, developing methods for sequencing unculturables, defining standards for the various deliverables, providing online access to the large new data set, and for addressing many other issues.
Of particular note is the development of standards that will be applied to the 900 genomes that are being sequenced. This will provide a new and higher level of uniformity to microbial genome data. The Jumpstart Consortium members are also in discussion with other consortia interested in standards to extend this uniformity beyond the HMP.
While this report and the initial stage of the HMP focus on bacteria, this effort is currently being expanded to produce reference genomes for eukaryotic microbes and viruses. These other components of the human microbiome have not been forgotten, but the initial focus on bacteria has allowed necessary infrastructure to be developed for the large task ahead. This can now be readily deployed for other organisms. It is our ultimate goal to sample the human microbiome as completely as possible.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge James Warren, Jingkun Zhang, R Gerald Fowler, Peter Pham, Dan Haft, Jeremy Selengut, Tanja Davidsen, Phil Goetz, Derek Harkins, Susmita Shrivastava, Sergey Koren, Brian Walenz, Les Foster, Indresh Singh, Yu-hui Rogers and the JCVI Joint Technology Center; Jian Xu, Shiaw-Pyng Yang and Seth Schobel for bioinformatics support; the Broad Genome Sequencing Platform, Yi Han, Viktoriya Korchina, Mark Scheel, Rebecca Thornton and the BCM-HGSC production team, Laura Courntey, Catrina Fronick, Otis Hall, Michelle O’Laughlin, Mark Cunningham, David O’Brien, Brenda Theising and the GCWU production team for sequencing; Jeff Gordon, Floyd Dewhirst, Brenda Wilson, Bryan White, Robert Mandrell, Martin Blaser, Roy H. Stevens, Sharon Hillier, Yang Liu, Zeli Shen, David Schauer, James Fox, Milton Allison, Chris D. Sibley, Delphine M. Saulnier, and Glenn R. Gibson for providing strains; and Maria Y. Giovanni, Carl L. Baker, Vivien Bonazzi, Carolyn D. Deal, Susan Garges, Robert W. Karp, R. Wayne Lunsford, Jane Peterson, Michael Wright, Tsegahiwot T. Belachew, and Christopher R. Wellington for funding agency management. We would like to acknowledge the National Institutes of Health for funding this project to The J. Craig Venter Institute (N01 AI 30071; U54-AI084844), Washington University (U54-HG003079; U54-HG004968), Baylor College of Medicine (U54-HG003273; U54-HG004973) and Broad Institute (HHSN272200900017C; U54-HG004969). Finally, funding for Emma Allen-Vercoe was from the Crohn’s and Colitis Foundation of Canada; Dirk Gevers had secondary affiliation at the Laboratory of Microbiology (WE 10), Department of Biochemistry and Microbiology, Faculty of Sciences, Ghent University, KL Ledeganckstraat 35, 9000 Ghent, Belgium and is indebted to the Fund for Scientific Research, Flanders (Belgium), for a postdoctoral fellowship and research funding for the duration of this project; Michael Surette would like to acknowledge the Canadian Cystic Fibrosis Foundation and the Canadian Institutes of Health Research for funding of his research for this project.
The Human Microbiome Jumpstart Reference Strains Consortium
Manuscript Preparation
Karen E. Nelson 1
George M. Weinstock 2
Sarah K. Highlander 3,4
Kim C. Worley 3,5
Heather Huot Creasy 6
Jennifer Russo Wortman 7,6
Douglas B. Rusch 8
Makedonka Mitreva 9
Erica Sodergren 2
Asif T. Chinwalla 2
Michael Feldgarden 9
Dirk Gevers 9
Brian J. Haas 9
Ramana Madupu 8
Doyle V. Ward 9
Principal Investigator
Bruce W. Birren 9
Richard A. Gibbs 3,5
Sarah K. Highlander 3,4
Barbara Methe 1
Karen E. Nelson 1
Joseph F. Petrosino 3,4
Robert L. Strausberg 1
Granger G. Sutton 8
George M. Weinstock 2
Owen R. White 10,6
Richard K. Wilson 2
Annotation
Asif T. Chinwalla 2
Heather Huot Creasy 6
Scott Durkin 8
Michelle Gwinn Giglio 6
Sharvari Gujja 9
Brian J. Haas 9
Sarah K. Highlander 3,4
Clint Howarth 9
Chinnappa D. Kodira 11
Nikos Kyrpides 12
Ramana Madupu 8
Teena Mehta 9
Makedonka Mitreva 9
Donna M. Muzny 3,5
Matthew Pearson 9
Kymberlie Pepin 2
Amrita Pati 12
Xiang Qin 3,5
Kim C. Worley 3,5
Jennifer Russo Wortman 7,6
Chandri Yandava 9
Qiandong Zeng 9
Lan Zhang 3,5
Assembly
Aaron M. Berlin 9
Lei Chen 2
Theresa A. Hepburn 9
Justin Johnson 8
Jamison McCorrison 8
Jason Miller 8
Pat Minx 2
Donna M. Muzny 3,5
Chad Nusbaum 9
Xiang Qin 3,5
Carsten Russ 9
Granger G. Sutton 8
Sean M. Sykes 9
Chad M. Tomlinson 2
Sarah Young 9
Wesley C. Warren 2
Kim C. Worley 3,5
Data Analysis
Jonathan Badger 13
Jonathan Crabtree 6
Heather Huot Creasy 6
Michael Feldgarden 9
Dirk Gevers 9
Sarah K. Highlander 3,4
Ramana Madupu 8
Victor M. Markowitz 14
Makedonka Mitreva 2
Donna M. Muzny 3,5
Joshua Orvis 6
Joseph F. Petrosino 3,4
Douglas B. Rusch 8
Granger G. Sutton 8
Doyle V. Ward 9
Kim C. Worley 3,5
Jennifer Russo Wortman 7,6
DNA Sequence Production
Andrew Cree 3,5
Steve Ferriera 15
Lucinda L. Fulton 2
Robert S. Fulton 2
Marcus Gillis 1
Lisa D. Hemphill 3,5
Vandita Joshi 3,5
Christie Kovar 3,5
Donna M. Muzny 3,5
Manolito Torralba 1
Xiang Qin 3,5
Funding Agency Management
Kris A. Wetterstrand 16
Genome Improvement
Amr Abouellleil 9
Aye M. Wollam 2
Christian J. Buhay 3,5
Yan Ding 3,5
Shannon Dugan 3,5
Michael G. FitzGerald 9
Lucinda L. Fulton 2
Robert S. Fulton 2
Mike Holder 3,5
Jessica Hostetler 1
Ramana Madupu 8
Donna M. Muzny 3,5
Xiang Qin 3,5
Granger G. Sutton 8
Project Leadership
Bruce W. Birren 9
Sandra W. Clifton 2
Sarah K. Highlander 3,4
Karen E. Nelson 1
Joseph F. Petrosino 3,4
Erica Sodergren 2
Robert L. Strausberg 1
Granger G. Sutton 8
George M. Weinstock 2
Owen R. White 10,6
Strain Management
Emma Allen-Vercoe 17
Jonathan Badger 13
Sandra W. Clifton 2
Heather Huot Creasy 6
Ashlee M. Earl 9
Candace N. Farmer 2
Michelle Gwinn Giglio 6
Marcus Gillis 1
Sarah K. Highlander 3,4
Konstantinos Liolios 12
Karen E. Nelson 1
Erica Sodergren 2
Michael G. Surette 18
Granger G. Sutton 8
Manolito Torralba 1
Doyle V. Ward 9
George M. Weinstock 2
Jennifer Russo Wortman 7,6
Qiang Xu 19
Submissions
Asif T. Chinwalla 2
Craig Pohl 2
Scott Durkin 8
Granger G. Sutton 8
Katarzyna Wilczek-Boney 3,5
Dianhui Zhu 3,5
 Corresponding author.
Corresponding author.
1. Human Genomic Medicine, J. Craig Venter Institute, 9704 Medical Center Drive, Rockville, Maryland, 20850, USA
2. The Genome Center, Washington University School of Medicine, 4444 Forest Park Ave, St. Louis, Missouri, 63108, USA
3. Human Genome Sequencing Center, Baylor College of Medicine, BCM226, One Baylor Plaza, Houston, Texas, 77030, USA
4. Department of Molecular Virology and Microbiology, BCM280, Baylor College of Medicine, One Baylor Plaza, Houston, Texas, 77030, USA
5. Department of Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, Houston, Texas, 77030, USA
6. Institute for Genome Sciences, University of Maryland School of Medicine, 801 W. Baltimore St. Baltimore, Maryland, 21201, USA
7. Department of Medicine, University of Maryland School of Medicine, Department of Genetics, 801 W. Baltimore St. Baltimore, Maryland, 21201, USA
8. Bioinformatics, J. Craig Venter Institute, 9704 Medical Center Drive, Rockville, Maryland, 20850, USA
9. Genome Sequencing and Analysis Program, Broad Institute, 7 Cambridge Center, Cambridge, Massachusetts, 02142, USA
10. Department of Epidemiology and Preventive Medicine, University of Maryland School of Medicine, 801 W. Baltimore St., Baltimore, Maryland, 21201, USA
11. Genome Sequencing and Analysis Program, 454 Sequencing, 15 Commercial Street, Branford, Connecticut, 06405, USA
12. DOE-Joint Genome Institute, 2800 Mitchell Drive, Walnut Creek, California, 94598, USA
13. Microbial and environmental genomics, J. Craig Venter Institute, 10355 Science Center Drive, La Jolla, California, 92121, USA
14. Biological Data Management and Technology Center, Lawrence Berkeley National Laboratory, Berkeley, California, 94720, USA
15. Sequencing, J. Craig Venter Institute, 9704 Medical Center Drive, Rockville, Maryland, 20850, USA
16. NHGRI, 5635 Fishers Lane, Bethesda, Maryland, 20892, USA
17. Molecular and Cellular Biology, University of Guelph, 50 Stone Road, Guelph, Ontario, N1G 2W1, Canada
18. Microbiology & Infectious Diseases, University of Calgary, 3330 Hospital Drive, Calgary T2N4N1 Canada, Alberta, T2N4N1, Canada
19. Osel Inc., 4008 Burton Drive, Santa Clara, California, 95054, USA
References and Notes
- 1.The NIH Common Fund Human Microbiome Project. Division of Program Coordination, Planning and Strategic Initiatives, National Institutes of Health, U.S. Department of Health and Human Services; ( http://nihroadmap.nih.gov/hmp/) [Google Scholar]
- 2.HMP Project Catalog. Human Microbiome Project Data Analysis Coordinating Center; ( http://www.hmpdacc.org/project_catalog.html) [Google Scholar]
- 3.16S rDNA for cultured bacteria. ( http://bioinfo.unice.fr/blast/documentation/alphabetical_list.html)
- 4.Reference Genomes of the Human Microbiome Project. Human Microbiome Project Data Analysis Coordinating Center; ( http://hmpdacc.org/reference_genomes.php) [Google Scholar]
- 5.Mobley HL, Island MD, Hausinger RP. Microbiol Rev. 1995 Sep;59:451. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tettelin H, et al. Proc Natl Acad Sci U S A. 2005 Sep 27;102:13950. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tettelin H, Riley D, Cattuto C, Medini D. Curr Opin Microbiol. 2008 Oct;11:472. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Konstantinidis KT, Ramette A, Tiedje JM. Appl Environ Microbiol. 2006 Nov;72:7286. doi: 10.1128/AEM.01398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goris J, et al. Int J Syst Evol Microbiol. 2007 Jan;57:81. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 10.Materials and methods are available as supporting material on Science Online.
- 11.Li W, Wooley JC, Godzik A. PLoS One. 2008;3:e3375. doi: 10.1371/journal.pone.0003375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wootton JC, Federhen S. Computers & Chemistry. 1993;17:149. [Google Scholar]
- 13.Welcome to the National Center for Biotechnology Information. National Center for Biotechnology Information, U.S. National Library of Medicine; ( http://www.ncbi.nlm.nih.gov/) [Google Scholar]
- 14.Rusch DB, et al. PLoS Biol. 2007 Mar;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genome Selection Page Organized by Coverage. J. Craig Venter Institute; ( http://gos.jcvi.org/users/hmpGenomes/genomes.html) [Google Scholar]
- 16.Turnbaugh PJ, et al. Nature. 2009 Jan 22;457:480. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill SR, et al. Science. 2006 Jun 2;312:1355. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovanni MY. Genome Sequencing Centers NIAID Data and Reagent Sharing and Release Guidelines. National Institute of Allergy and Infectious Diseases, National Institues of Health, U.S. Department of Health and Human Services; ( http://www.niaid.nih.gov/dmid/genomes/mscs/data_release.htm) [Google Scholar]
- 19.Documentation and SOPs. Human Microbiome Project Data Analysis Coordinating Center; ( http://www.hmpdacc.org/sops.php) [Google Scholar]
- 20.Qin J, et al. Nature. 2010 Mar 4;464:59. [Google Scholar]
- 21.A Genomic Encyclopedia of Bacteria and Archaea (GEBA) Joint Genome Institute, U.S. Department of Energy Office of Science; ( http://www.jgi.doe.gov/programs/GEBA/) [Google Scholar]
- 22.Wu D, et al. Nature. 2009 Dec 24;462:1056. [Google Scholar]
- 23.Huang X, Wang J, Aluru S, Yang SP, Hillier L. Genome Res. 2003 Sep;13:2164. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon D, Abajian C, Green P. Genome Res. 1998 Mar;8:195. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 25.Gordon D, Desmarais C, Green P. Genome Res. 2001 Apr;11:614. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.