Abstract
Recently identified genetic determinants for enhanced susceptibility to Crohn’s disease (CD) included polymorphisms in the ATG16L1 and IRGM1 loci suggesting that the autophagy pathway plays a role in the pathogenesis of this disease. We have generated and analyzed three mouse models with diminished expression of autophagy proteins and show how the loss of function of various autophagy components contributes to CD pathogenesis. In the mouse small intestine, the common cellular target of Atg16L1, Atg5, and Atg7 is the Paneth cell, a specialized epithelial cell whose main function is the delivery of antimicrobial factors into the intestinal lumen by production and secretion of its characteristic cytoplasmic granules. Autophagy-deficient Paneth cells exhibited a striking loss of function in this granule exocytosis pathway. Transcriptional analysis revealed a gain of function whereby the gene expression associated with inflammatory responses was increased in autophagy-deficient Paneth cells. Importantly, we validated these findings by analyzing intestinal tissues from CD patients. Similar Paneth cell abnormalities were observed in CD patients homozygous for the ATG16L1 risk allele. Thus, one role for the autophagy pathway in CD pathogenesis is through selective effects on the biology and specialized properties of Paneth cells.
Keywords: ATG16L1, ATG5, ATG7, Paneth cell, Crohn’s disease, inflammatory bowel disease, intestine, exocytosis, adipocytokine, PPAR
Atg16L1 mutant mice display reduced autophagy and Paneth cell abnormalities
Over 30 genetic loci are now associated with increased susceptibility to CD.1 This major type of inflammatory bowel disease is most commonly characterized by uncontrolled, transmural inflammation in the distal ileum (small intestine), though the colon can be involved as well. One CD susceptibility allele is in the predicted autophagy gene ATG16L1.2–5 To determine the role of Atg16L1 in the intestine, we generated two mouse lines with gene trap-mediated disruptions of the Atg16L1 locus, Atg16L1HM1 and Atg16L1HM2 (collectively referred to as Atg16L1HM).6 These mice displayed reduced (hypomorphic, HM) expression of Atg16L1 protein, and both fibroblasts and intestinal tissue from these mice were compromised in autophagy activity indicating that Atg16L1, like yeast Atg16, is a bona fide autophagy protein.
The intestinal epithelium is constantly exposed to a wide and diverse population of commensal microbes (microbiota)7 and is sporadically exposed to enteric pathogens. Mice deficient in Nod2, another CD susceptibility gene, displayed increased sensitivity to oral infection by Listeria Monocytogenes.8 Thus, one potential mechanism by which autophagy contributes to intestinal homeostasis is through clearance of intracellular pathogens, a well-documented function of autophagy.9–11 However, we did not detect increased susceptibility to oral L. monocytogenes infection in Atg16L1HM mice, suggesting that there may be differences in the roles of Nod2 and Atg16L1 in the small intestine. Instead, in the absence of exogenous bacterial infection, we found striking morphological abnormalities in Paneth cells that are a highly specialized epithelial cell located at the base of epithelial invaginations called crypts of Lieberkuhn. Paneth cells likely shape the intestinal microbiota via secretion of granule contents including antimicrobial peptides and lysozyme.12 By light and ultrastructural analysis, Paneth cells in Atg16L1HM mice displayed a reduction in the number of granules, dramatically increased cytoplasmic vesicles, and abnormal mitochondria. Furthermore, lysozyme, which is normally packaged efficiently into the granules, displayed striking defects in its distribution including diffuse cytoplasmic expression.
To understand the nature and potential etiology of these morphological abnormalities, we examined the transcriptional profile of Atg16L1-deficient Paneth cells. Microarray analysis comparing ileal crypt base cells (which are primarily Paneth cells)13 procured by laser capture microdissection from Atg16L1HM mice and controls revealed an increase in transcripts associated with PPAR signaling, acute phase reactants, adipocytokine signaling, and lipid metabolism in the Atg16L1HM mice. Many of these genes are directly implicated in inflammation, and two of these transcripts, leptin and adiponectin, are known to be increased in CD patients.14, 15 Thus, in addition to its role in maintaining the granule exocytosis pathway, Atg16L1 is an important brake for the expression of pro-inflammatory genes in Paneth cells.
Identification of a novel pathological hallmark of CD associated with ATG16L1
Since Atg16L1 is important for the biology of Paneth cells in mice, we examined the role of the human ATG16L1 CD risk allele. Compared to control ileal samples from CD patients that did not carry the risk alleles, ileal specimens from CD patients homozygous for the ATG16L1 risk allele displayed Paneth cell abnormalities remarkably similar to the Atg16L1HM mice. In addition to the morphological abnormalities, in comparison with controls, human Paneth cells with the CD risk polymorphism of ATG16L1 were more likely to overexpress leptin protein, an observation that was prompted by the transcriptional analysis in mice. Since the role of NOD2 is controversial in both secretion as well as production of anti-microbial peptides by Paneth cells,16–21 our analysis only included samples from patients that did not carry the NOD2 CD risk alleles. In the future, it will be interesting to examine the effect of polymorphisms in NOD2 and other CD susceptibility genes using the same assays.
The role of the autophagy pathway in Paneth cell abnormalities
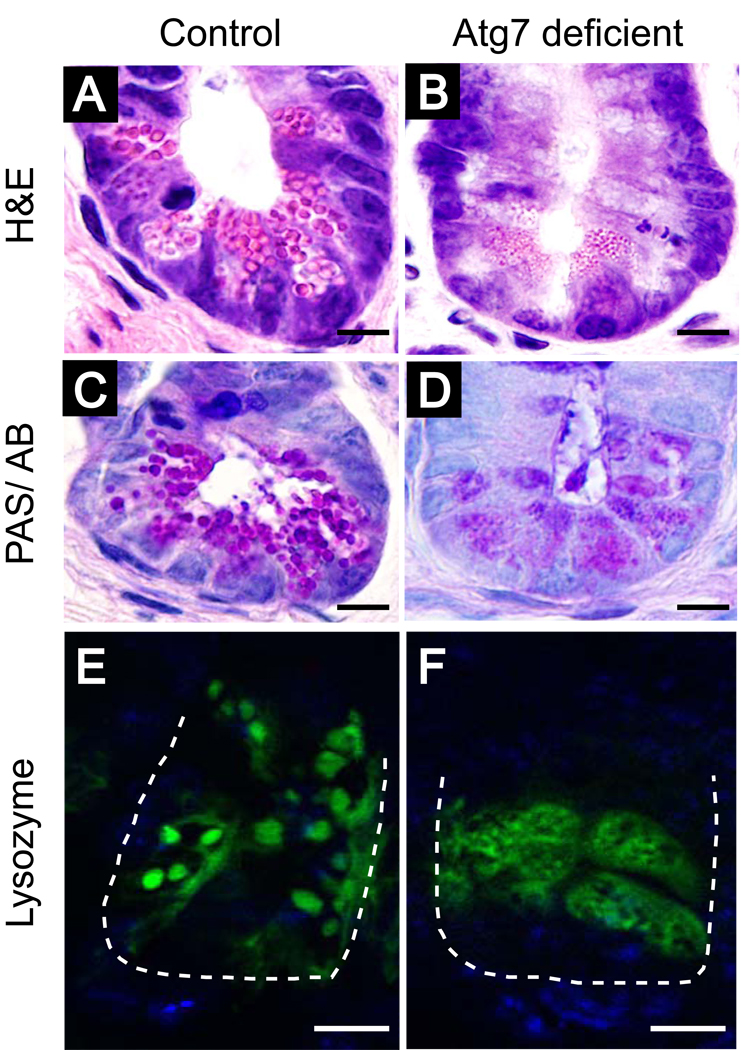
The ATG16L1 variant associated with CD encodes a threonine to alanine substitution that may result in reduced protein expression,22 but Atg16L1 may have roles outside of autophagy. To further examine the potential role of the autophagy pathway in Paneth cell and CD, we generated mice that were deficient in Atg5 in the intestinal epithelium by breeding Atg5flox/flox mice23 with villin-Cre mice that express the Cre recombinase specifically in the intestinal epithelium including Paneth cells24 (Atg5flox/floxvillin-Cre). Remarkably, these mice display morphological abnormalities in their Paneth cells that are indistinguishable from the Atg16L1-deficient mice. Moreover, like the Atg16L1HM mice, Atg5flox/floxvillin-Cre mice do not display obvious morphological abnormalities outside the Paneth cells within the ileal epithelium. Subsequently, we have generated mice with a conditional deletion of Atg7 in the intestinal epithelium (Atg7flox/floxvillin-Cre). By histological and immunohistochemical characterization, the ileal pathology in these mice is indistinguishable from that observed in Atg16L1HM and Atg5flox/floxvillin-Cre mice (Fig. 1). Thus, these results suggest that a defect in the autophagy pathway in the intestinal epithelium is responsible for the Paneth cell pathology.
Figure 1.
Loss of Atg7 in the intestinal epithelium leads to Paneth cell abnormalities similar to those associated with loss of function for both Atg16L1 and Atg5. Atg7flox/flox mice30 were bred with mice expressing the Cre recombinase from the villin promoter (villin-Cre)24 to generate mice with conditional deletion of Atg7 in the intestinal epithelium (Atg7flox/floxvillin-Cre). (A–D) Histological examination of the distal (ileal) small intestine reveals morphologically aberrant Paneth granules in mice with loss of Atg7. Ileal sections from Atg7flox/flox control (A,C,E) or Atg7flox/floxvillin-Cre mice (B,D,F) were stained with hematoxylin and eosin (A,B), PAS/alcian blue (C,D), or goat-anti lysozyme antisera and Alexaflour 488-labled donkey anti-goat Ig (E,F). Mice with loss of Atg7 in the intestinal epithelium contain Paneth cell granules that are abnormal in size, morphology, and number. Instead of labeling granules as in Atg7flox/flox control Paneth cells, lysozyme staining displayed abnormal diffuse cytoplasmic distribution in Atg7flox/floxvillin-Cre samples. Green represents lysozyme, blue represents nuclei, and the dashed line denotes the crypt unit. Bars=10 µm.
How does an autophagy defect lead to such specific effects on Paneth cells, while other small intestinal epithelial lineages and stem cells do not exhibit obvious morphological abnormalities? Paneth cells, in contrast to other intestinal epithelial cells, contain abundant endoplasmic reticulum (ER). Loss of key autophagy proteins may disrupt autophagy-mediated organelle turnover thus disrupting ER homeostasis. Studies examining the intestinal role of Xbp1, a transcription factor important in ER homeostasis, provides evidence that Paneth cells are particularly sensitive to ER stress.25 Indeed, electron microscopy of Atg16L1-deficient Paneth cells reveals degeneration of mitochondria and replacement of ER with excess vesicular structures. However, loss of Xbp1 does not completely mirror autophagy deficiency, as Paneth cells die in the former model but not in the latter. Another possible explanation for the vesicle formation is that there are autophagosome-independent roles for autophagy proteins as has been previously reported.11, 26–29
Conclusion
Using mice with mutations in Atg16L1 and Atg5, and now reported here Atg7, we have described a novel role for the autophagy pathway in Paneth cells that we confirmed in human CD samples. One consequence of this lineage-specific role of autophagy is that future studies will have to investigate the function of Atg16L1 in relevant cell-types in vivo. For instance, to examine the precise effect of the ATG16L1 polymorphism, a knock-in mouse model in which the endogenous Atg16L1 is replaced by the CD risk allele of ATG16L1 would be particularly useful. It may also be informative to cross Atg16L1HM mice to other models for CD susceptibility such as Nod2 mutant mice. Moreover, in addition to a clear genetic component, there is also an ill-defined environmental component of CD. Therefore, it will be critical to determine what factors contribute to the observed Paneth cell defect and if environmental damage such as enteric pathogens leads to pathological changes in Atg16L1-deficient mice. Rarely are individual pathological hallmarks of a complicated disease associated with a specific gene, and thus, Atg16L1HM mice provide a unique opportunity to advance our understanding of a complex and debilitating disease.
Acknowledgement
This work was supported by grant U54 AI057160 Project 6 and the Broad Foundation (K.C. and H.W.V.), and the Pew Foundation (K.P. and T.S.S.). KC is the Lallage Feazel Wall Fellow of the Damon Runyon Cancer Research Foundation (DRG-1972-08).
References
- 1.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van GA, Zelenika D, Franchimont D, Hugot JP, de VM, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 4.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De LV, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 6.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, Takada H, Goldman WE, Fukase K, Silverman N, Yoshimori T, Kurata S. Autophagic control of listeria through intracellular innate immune recognition in drosophila 1. Nat. Immunol. 2008;9:908–916. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter EM, Bevins CL, Ghosh D, Ganz T. The multifaceted Paneth cell. Cell Mol. Life Sci. 2002;59:156–170. doi: 10.1007/s00018-002-8412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stappenbeck TS, Mills JC, Gordon JI. Molecular features of adult mouse small intestinal epithelial progenitors. Proc Natl Acad Sci U. S. A. 2003;100:1004–1009. doi: 10.1073/pnas.242735899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbier M, Vidal H, Desreumaux P, Dubuquoy L, Bourreille A, Colombel JF, Cherbut C, Galmiche JP. Overexpression of leptin mRNA in mesenteric adipose tissue in inflammatory bowel diseases. Gastroenterol. Clin. Biol. 2003;27:987–991. [PubMed] [Google Scholar]
- 15.Yamamoto K, Kiyohara T, Murayama Y, Kihara S, Okamoto Y, Funahashi T, Ito T, Nezu R, Tsutsui S, Miyagawa JI, Tamura S, Matsuzawa Y, Shimomura I, Shinomura Y. Production of adiponectin, an anti-inflammatory protein, in mesenteric adipose tissue in Crohn's disease. Gut. 2005;54:789–796. doi: 10.1136/gut.2004.046516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, Schroder JM, Bevins CL, Fellermann K, Stange EF. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal α-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, Shen B, Schaeffeler E, Schwab M, Linzmeier R, Feathers RW, Chu H, Lima H, Jr., Fellermann K, Ganz T, Stange EF, Bevins CL. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc Natl Acad Sci U. S. A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simms LA, Doecke JD, Walsh MD, Huang N, Fowler EV, Radford-Smith GL. Reduced α-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn's disease. Gut. 2008;57:903–910. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 19.Voss E, Wehkamp J, Wehkamp K, Stange EF, Schröder JM, Harder J. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J Biol. Chem. 2006;281:2005–2011. doi: 10.1074/jbc.M511044200. [DOI] [PubMed] [Google Scholar]
- 20.Lala S, Ogura Y, Osborne C, Hor SY, Bromfield A, Davies S, Ogunbiyi O, Nunez G, Keshav S. Crohn's disease and the NOD2 gene: a role for paneth cells. Gastro. 2003;125:47–57. doi: 10.1016/s0016-5085(03)00661-9. [DOI] [PubMed] [Google Scholar]
- 21.Ogura Y, Lala S, Xin W, Smith E, Dowds TA, Chen FF, Zimmermann E, Tretiakova M, Cho JH, Hart J, Greenson JK, Keshav S, Nunez G. Expression of NOD2 in Paneth cells: a possible link to Crohn's ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 24.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 25.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 27.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U. S. A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, Mizushima N, Oshumi Y, Jung YK. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 29.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 30.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]