Abstract
This work investigates the interaction of N-terminal gp41 fusion peptide (FP) of human immunodeficiency virus type 1 (HIV-1) with model membranes in order to elucidate how FP leads to fusion of HIV and T-cell membranes. FP constructs were (i) wild-type FP23 (23 N-terminal amino acids of gp41), (ii) water-soluble monomeric FP that adds six lysines on the C-terminus of FP23 (FPwsm), and (iii) the C-terminus covalently linked trimeric version (FPtri) of FPwsm. Model membranes were (i) LM3 (a T-cell mimic), (ii) 1,2-dioleoyl-sn-glycero-3-phosphocholine, (iii) 1,2-dioleoyl-sn-glycero-3-phosphocholine/30 mol% cholesterol, (iv) 1,2-dierucoyl-sn-glycero-3-phosphocholine, and (v) 1,2-dierucoyl-sn-glycero-3-phosphocholine/30 mol% cholesterol. Diffuse synchrotron low-angle x-ray scattering from fully hydrated samples, supplemented by volumetric data, showed that FP23 and FPtri penetrate into the hydrocarbon region and cause membranes to thin. Depth of penetration appears to depend upon a complex combination of factors including bilayer thickness, presence of cholesterol, and electrostatics. X-ray data showed an increase in curvature in hexagonal phase 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, which further indicates that FP23 penetrates into the hydrocarbon region rather than residing in the interfacial headgroup region. Low-angle x-ray scattering data also yielded the bending modulus KC, a measure of membrane stiffness, and wide-angle x-ray scattering yielded the Sxray orientational order parameter. Both FP23 and FPtri decreased KC and Sxray considerably, while the weak effect of FPwsm suggests that it did not partition strongly into LM3 model membranes. Our results are consistent with the HIV FP disordering and softening the T-cell membrane, thereby lowering the activation energy for viral membrane fusion.
Keywords: fusion peptide, trimer, T-cell mimic, structure, bending modulus
Introduction
Infection by enveloped viruses, such as human immunodeficiency virus type 1 (HIV-1), requires fusion of the virion membrane with the target cell membrane in order to transfer viral RNA into the target cell.1 Enveloped viruses use ectodomain glycoproteins to first dock with receptors on the T-cell membrane and then perturb the target membrane to form a pore.2 On the HIV-1 ectodomain, the glycoproteins gp160 are typically assembled as homotrimers3 and proteolytically cleaved to gp120 and gp41, which remain non-covalently associated. gp120 binds to the CD4 glycoprotein on the T cell4 and a chemokine coreceptor, primarily CCR5 and CXCR4.5 This binding causes a conformational change in gp41 that exposes the fusion peptide (FP23) that contains a highly hydrophobic sequence of 23 amino acids at the N-terminus of gp41.3,6,7 FP interacts in a nonspecific way with the target membrane and perhaps also with the virion membrane;8 this interaction provides the perturbation that allows a pore to form, thus allowing release of the HIV RNA into the target cell. The importance of FP23 is indicated by its ability to fuse and/or lyse liposomes and erythrocytes,9,10 and mutations with a polar residue in this sequence drastically reduce fusogenic activity.11,12 Although some investigations have studied longer13 and shorter14 N-terminal FP constructs from HIV (the 12-mer being the minimal FP15), FP23 is the most common choice.9,10,13 It may also be noted that a recent work presents evidence to show that the main pathway is endocytosis of the HIV virion, rather than fusion with the plasma membrane, and then the virion membrane fuses with the endosomal membrane near the nuclear membrane in lymphoid CEMss cells expressing CD4 and CXCR4.16 Even in this scenario, the mechanism of fusion between the virion and endosomal membranes would likely be similar to that at the plasma membrane, since the same HIV ectodomain is exposed at the virion membrane surface.17
Atomic-resolution structure is often achievable and valuable. The crystal structure of gp41 of simian immunodeficiency virus has been obtained18 as has the crystal structure of HIV-1 gp41 ectodomain.3 These structures share a similar coiled-coil core, which is thought to stabilize the interaction of the viral and T-cell membranes. In contrast, the crystal structure of the N-terminal FP has not been determined, since it is normally removed prior to crystallization due to its extreme hydrophobicity.3,18 Even a 3D solution structure of ectodomain of simian immunodeficiency virus gp41 did not contain the 26 N-terminal amino acids.19 Furthermore, the membrane with which FP interacts is in a fluid state; hence, it should not be expected that physiological concentrations of FP will induce a crystalline state. Even if a method could be found to obtain a crystalline state of FP with lipids, it is unlikely that such a state would be biologically relevant because it would not be fully hydrated and the disorder due to the fluid nature of the membranes would not be present. Instead of atomic-level crystallography that utilizes sharp diffraction peaks, we use the diffuse x-ray scattering that is produced by fully hydrated, fluctuating bilayers to determine disordered structure at the nanoscale.20
Since gp41 acts as a trimer during HIV infection, a trimer construct of FP (FPtri) could be an important tool for studying its membrane interactions. A water-soluble monomer (FPwsm) had been synthesized, by attaching six lysines to the native FP, in order to ease synthesis steps.21 A similar FPwsm produced by genetic expression was found useful for solution NMR studies.22 FPtri enhanced lipid mixing in LM3 model fusion studies by 40 times compared to FPwsm.21 Solid-state NMR has shown that there is a strong correlation between membrane insertion depth and fusogenicity in a model membrane system of PC:PG (~4:1) with 30 mol% cholesterol.23 In the present work, we study FP23 as well as the FPwsm and FPtri constructs in LM3. LM3 is a T-cell lipid membrane mimic consisting of a mixture of six lipids [1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE):1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS):soy phosphatidylinositol (PI):egg sphingomyelin (ESM):cholesterol (Chol)], in a 10:5:2:1:2:10 mole ratio,24,25 which our work shows is a liquid ordered bilayer that becomes more disordered when FP is added. As controls, we also compare results of adding FP23 to simpler lipid bilayers, starting with the favorite fully fluid 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) (diC18:1PC) bilayer, and we also add cholesterol. However, the DOPC bilayer is considerably thinner than LM3; thus, we have also studied 1,2-dierucoyl-sn-glycero-3-phosphocholine (diC22:1PC; with and without 30 mol% cholesterol), which is 7 Å thicker than DOPC.26 In addition to structure, our synchrotron low-angle x-ray scattering (LAXS) data determine the bending modulus, KC, which is a measure of membrane stiffness. Using our wide-angle x-ray scattering (WAXS) analysis,27 we also determine the orientational Sxray order parameters. In a third type of diffraction experiment, we show that FP23 increases the curvature of the hexagonal II phase forming lipid 1,2-dioleoyl-sn-glycero-3-phos-phoethanolamine (DOPE), thus confirming that FP23 penetrates into the hydrocarbon interior. Finally, our volumetric data are fundamental for quantitative structural analysis.
Results
Volumes
The volume shown for LM3 is the mole fraction weighted average of lipid (including cholesterol) in the LM3 mixture. Upon addition of FP23 mole fraction, xFP23 =[FP23]/([FP23]+[lipids]), of FP23 to any of the three bilayers in Table 1, the measured volume is VM = xFP23VFP +(1 −xFP23)VLipid. Using VFP23 =2664 Å3 calculated from volumes of amino acids in crystals†, we show in the VL column in Table 1 that the volume of the average lipid molecule increased negligibly for LM3 (1218 Å3 versus 1211 Å3) but decreased significantly for the di-monounsaturated lipids DOPC and diC22:1PC, suggesting that FP23 had a strong condensing effect on some lipids.
Table 1.
Volume results
Hydration issues and MLV data
Our best scattering data are obtained from oriented stacks of bilayers. For biological relevance, it is important that those samples be well hydrated to provide enough water between neighboring bilayers in the stack so that there is ample water space in which hydrophilic parts of added peptide could reside and so that interactions between bilayers do not significantly perturb the structure they would have as single bilayers. Preliminary to obtaining oriented x-ray data that required hydrating from water vapor, it was valuable to obtain the repeat D spacings from multilamellar vesicle (MLV) samples in excess water. Compared to MLVs of zwitterionic lipids that have well-defined finite D spacings, lipids with net charge have electrostatic forces that may repel neighboring membranes so strongly that they are “unbound” with essentially infinite, nonobservable values of D. Table 2 reports that the addition of x=0.091 positively charged FP23 unbound neutral DOPC and diC22:1PC bilayers. Negatively charged LM3 bilayers were also unbound; they became bound when enough positively charged FP23 was added to reduce their net charge. FP23 at x=0.048 did not induce binding of LM3 membranes, but FPtri did induce binding at x=0.016. Although these two concentrations of FP23 and FPtri have the same number of native amino acids, FPtri was synthesized with additional lysines to make it water soluble and the extra charge accounts for binding at lower concentrations than FP23. FPwsm at x=0.048 has the same number of native amino acids and essentially the same number of added lysines as FPtri at x=0.016. We suggest that FPwsm does not cause unbound LM3 bilayers to bind to each other because a larger fraction of FPwsm partitions into the water than FPtri, thereby leaving a larger net negative charge on LM3.
Table 2.
Summary of fully hydrated, MLV D spacings
| Peptide/lipid | x | D (±0.5 Å) |
|---|---|---|
| LM3 | 0 | Unbound |
| FP23/LM3 | 0.048 | Unbound |
| FP23/LM3 | 0.091 | 64.8 |
| FPwsm/LM3 | 0.048 | Unbound |
| FPtri/LM3 | 0.016 | 65.4 |
| DOPC | 0 | 64.0 |
| FP23/DOPC | 0.048 | 67.0 |
| FP23/DOPC | 0.091 | Unbound |
| diC22:1PC | 0 | 70.0 |
| FP23/diC22:1PC | 0.048 | 70.7 |
| FP23/diC22:1PC | 0.091 | Unbound |
Diffuse x-ray scattering from oriented samples
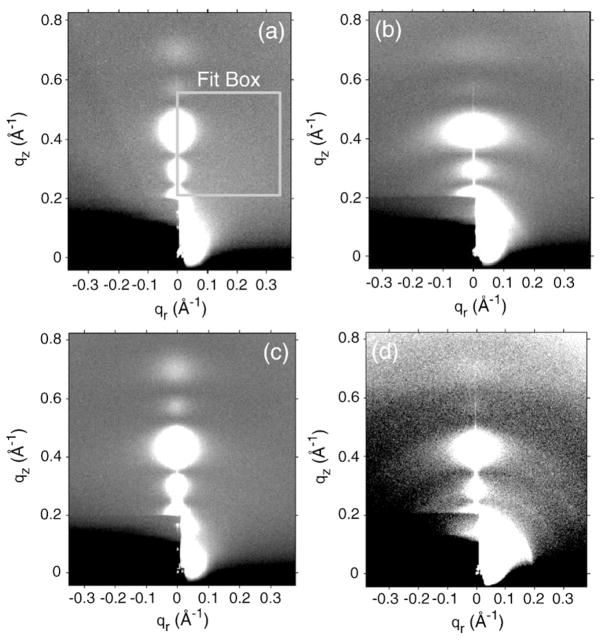
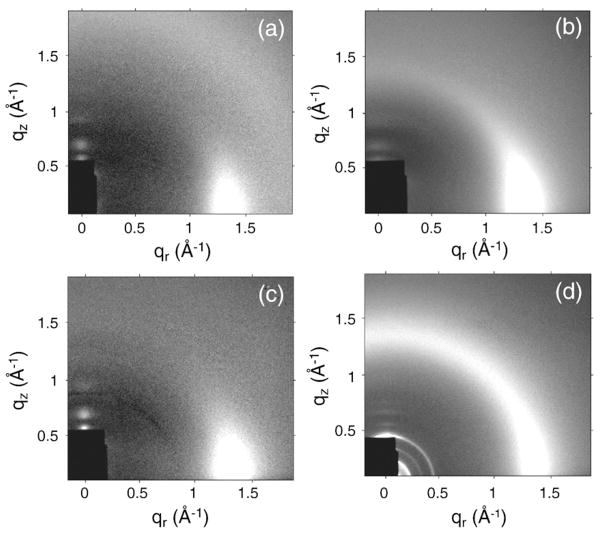
Figure 1 shows some diffuse LAXS data for oriented samples based on LM3 bilayers. All these samples were hydrated far enough to have quite large D spacings with an ample water cushion between adjacent bilayers. Such extensive hydration causes the oriented stacks to fluctuate and produce diffuse lobes of scattering,20 shown as circular or ellipsoidal white lobes in Fig. 1. Compared to the LM3 data in Fig. 1a, the addition of FP23 in Fig. 1b and the addition of FPtri in Fig. 1d caused the diffuse lobes to become broader in the qr direction due to increased fluctuations. In contrast, the water-soluble monomeric form of FP23, 4.8 mol% FPwsm/LM3 sample in Fig. 1c, had a LAXS pattern quite similar to that of the control, LM3 in Fig. 1a. This indicates that FPwsm had the least perturbing effect on the LM3 structure, perhaps remaining in the solvent. The broadening of diffuse lobes observed in Fig. 1b and d is not due to a significant increase in mosaic spread, since the mosaic spread in these samples was measured to be <0.3°, only slightly greater than our best oriented samples with a mosaic spread <0.1°. CCD images of diC22:1PC and DOPC with 4.8 mol% FP23 have been reported previously.28
Fig. 1.
CCD images (high intensity is white) of (a) LM3 (D=120 Å), (b) 4.8 mol% FP23/LM3 (D=143.7 Å), (c) 4.8 mol% FPwsm/LM3 (D=74.7 Å), and (d) 1.6 mol% FPtri/LM3 (D=65.4 Å). The beam and first two Bragg orders are visible through the rectangular, molybdenum semitransparent beam stop in the lower left-hand corner. Data were analyzed for KC in fit boxes such as the one shown in (a). Grayscale was optimized for comparison.
Structure
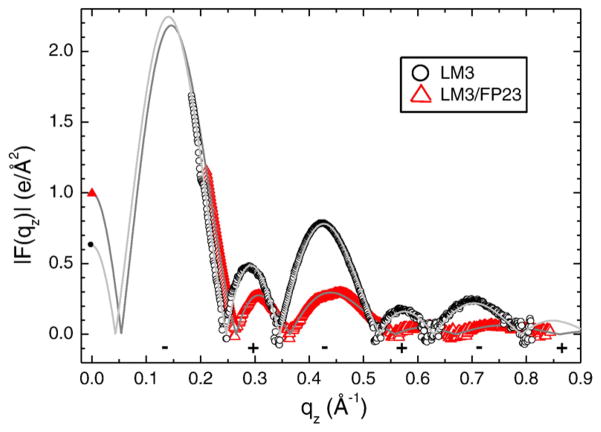
Figure 2 shows the form factors obtained for LM3 with and without 4.8 mol% FP23 as examples of data used for the structure determination. The open symbols show the relative form factors |F(qz)| obtained from the LAXS data in Fig. 1a and b. The continuous lines come from electron density model fits to these |F(qz)| data and provide the absolute scaling factor.
Fig. 2.
Form factor data for LM3 and 4.8 mol% FP23/LM3. Open symbols are from LAXS intensity data and continuous lines are best fits to the SDP model. The F(0) points (filled symbols) come from volume measurements [Eq. (1)]. Phases (±) are shown beneath the lobes.
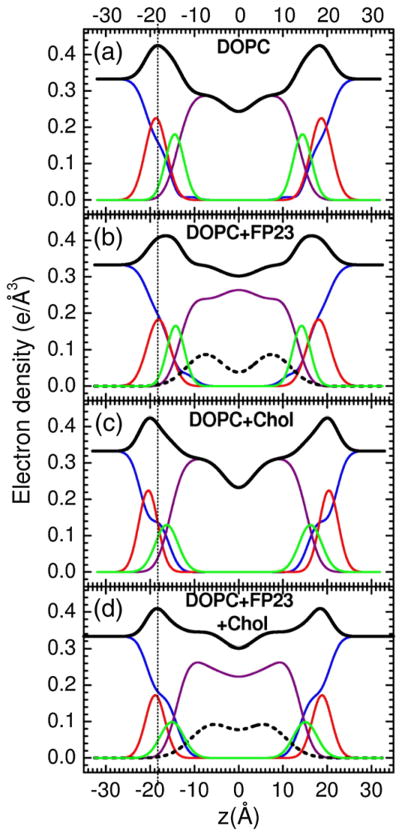
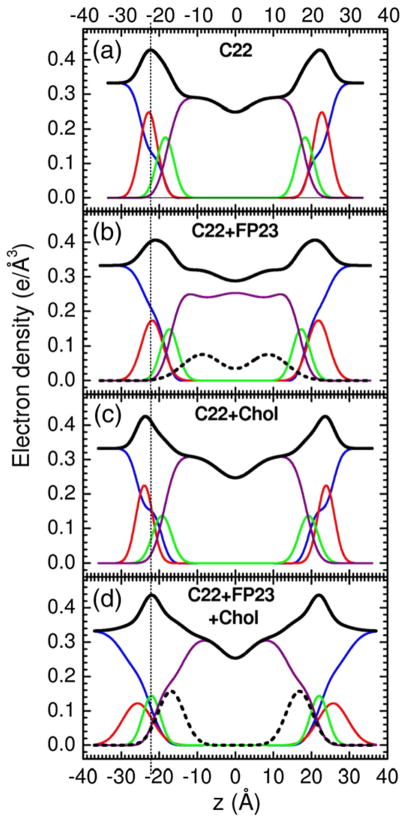
Figures 3–5 show electron density profiles obtained from LAXS data. The most robustly determined quantity is the head-to-head thickness, DHH, which is defined as the distance between the maxima in the total electron density profiles. Figures 3–5 show that all FP constructs decreased DHH. With no FP, the distance DPP, which is defined as the distance between the phosphate groups, was close to DHH. When peptide was added, DHH generally became smaller than DPP because the location of the total maximum involves both the peptide (0.43 e/Å3) and the electron-dense phosphatidylcholine group (0.54 e/Å3). For both FP23 and FPtri in LM3, the mean position of the peptide was inside the hydrocarbon region, but closer to the glycerol–carbonyl headgroup Gaussian than to the center of the bilayer. The control membrane with nearly the same thickness as LM3 is diC22:1PC. Similarly to LM3, Fig. 4 shows that FP23 caused ~3 Å thinning in diC22:1PC, while 30 mol% cholesterol caused a 3-Å thickening. As shown in Fig. 4, the position of FP23 in diC22:1PC changed dramatically with cholesterol, from ~9 to ~17 Å from the bilayer center. In the latter position, FP23 is close to the glycerol–carbonyl headgroup, as in LM3. Since LM3 contains 33 mol% cholesterol, this suggests that cholesterol has the effect of displacing FP23 from deep within the hydrocarbon interior to nearer the carbonyl/glycerol moiety in these fairly thick bilayers. Similarly to the thicker lipids, FP23 caused ~3.5 Å thinning in DOPC (see Fig. 5) while cholesterol caused ~4 Å thickening, and these two effects cancelled each other in the sample with both components. However, unlike the thicker lipids, cholesterol did not have the effect of moving FP23 closer to the headgroup region.
Fig. 3.
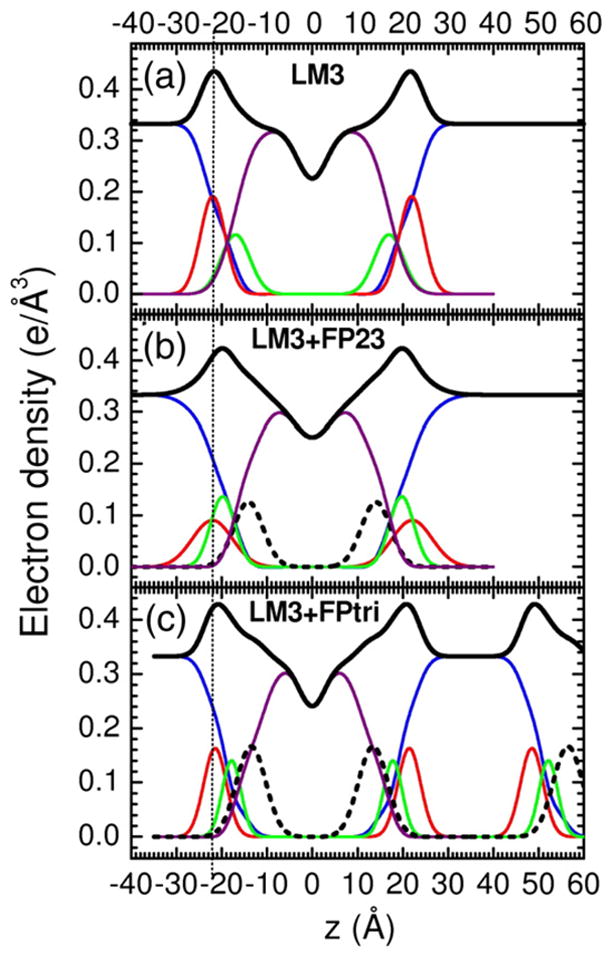
Electron density versus distance from the bilayer center z from samples of (a) LM3, (b) 4.8 mol% FP23/LM3, and (c) 1.6 mol% FPtri/LM3. Line colors: Black is total electron density, blue is water, red is phosphate plus choline or other average headgroup moiety, green is glycerol–carbonyl, purple is hydrocarbon region including cholesterol when present, and black dotted line is peptide. (c) shows part of a second adjacent bilayer at large values of z, and the presence of a large water spacing between adjacent bilayers is emphasized in all panels.
Fig. 5.
Electron density profiles of (a) DOPC, (b) 6.3 mol% FP23/DOPC, (c) 30 mol% cholesterol/DOPC, and (d) 6.3 mol% FP23/DOPC/30 mol% cholesterol. Line colors and D/2 values are as in Fig. 4.
Fig. 4.
Electron density profiles of (a) diC22:1PC, (b) 6.3 mol% FP23/diC22:1PC, (c) 30 mol% cholesterol/diC22:1PC, and (d) 6.3 mol% FP23/diC22:1PC/30 mol% cholesterol. Only one central bilayer is shown with the profiles cut off at ±D/2 and therefore only half the water space between bilayers is shown. Line colors are as in Fig. 3.
Secondary structure
It is of interest to know if the peptide position or Gaussian width is correlated with its secondary structure. Circular dichroism (CD) data (not shown) indicated well-defined α-helix in dry 9.1 mol% FP23/LM3, but that changed upon hydration to a poorly determined mixture of β-sheet and random coil. Primarily β-sheet structure was indicated for hydrated 9.1 mol% FP23/diC22:1PC, whereas 9.1 mol% FP23/DOPC appeared to be a mixture of α-helix, β-sheet, and random coil. In our hands, there was no clear correlation between CD secondary structure and the peptide position or its Gaussian width obtained from LAXS.
Bending modulus KC
The bending modulus KC was obtained from LAXS data of oriented samples with results summarized in Table 3. Consistent with earlier results for DOPC and diC22:1PC bilayers,28 addition of FP generally decreased KC dramatically. The new data for LM3 bilayers further show that FPtri is almost as effective per native amino acid (comparing three times the mole fraction) in lowering KC as FP23. FPwsm is not as effective as either, consistent with the LAXS result shown in Fig. 1, suggesting that it may not partition as completely into the LM3 bilayer as FP23 and FPtri. Table 3 also includes previous results that cholesterol has little effect on KC of lipids with two monounsaturated chains.29,30
Table 3.
Bending modulus KC
| Lipid | xChol | Peptide | xFP | KC/kT |
|---|---|---|---|---|
| LM3 | 0.33 | 0 | 32±2 | |
| LM3 | 0.33 | FP23 | 0.048 | 5.3±0.3 |
| LM3 | 0.33 | FP23 | 0.091 | 4.4±0.3 |
| LM3 | 0.33 | FPwsm | 0.048 | 25±2 |
| LM3 | 0.33 | FPtri | 0.016 | 6.6±1.3 |
| DOPC | 0 | 19±0.9 | ||
| DOPC | FP23 | 0.063 | 6.2±0.428 | |
| DOPC | 0.3 | 0 | 18±0.4 | |
| DOPC | 0.3 | FP23 | 0.063 | 5.8±0.7 |
| diC22:1PC | 0 | 31±3 | ||
| diC22:1PC | FP23 | 0.063 | 2.4±0.828 | |
| diC22:1PC | 0.3 | 0 | 30±3 | |
| diC22:1PCl | 0.3 | FP23 | 0.063 | 2.0±0.8 |
Order parameter
Figure 6 shows WAXS obtained from oriented samples at 30 °C. The WAXS intensity for LM3 in Fig. 6a is concentrated near the equator, typical of liquid-ordered (Lo) phases with high orientational order27. Upon addition of FP23, the WAXS intensity in Fig. 6b extends further off the equator due to greater orientational disorder. By contrast, WAXS scattering upon addition of monomeric FPwsm in Fig. 6c is closer to control LM3 in Fig. 6a, which again suggests weaker partitioning into the LM3 membrane. The maximal disordering effect compared to the LM3 control was obtained with trimeric FP23/LM3 as shown in Fig. 6d.
Fig. 6.
Grayscale CCD images of 2D WAXS of (a) LM3, (b) 4.8 mol% FP23/LM3, (c) 4.8 mol% FPwsm/LM3, and (d) 1.6 mol% FPtri/LM3. White is high intensity; lighter background intensity in (a) is due to condensed water on the silicon wafer.
Figure 7 shows the x-ray orientational order parameter Sxray. As suggested by the raw data in Fig. 6, FPtri decreased Sxray most in LM3 bilayers, even when plotted at the same concentration of native amino acids. By contrast, FPwsm had little effect on chain order. Figure 7 also shows that FP23 decreased Sxray in DOPC and diC22:1PC with and without cholesterol. As previously reported, cholesterol increased Sxray in pure lipid bilayers.29,30 The larger value of Sxray in LM3 than in diC22:1PC or DOPC, even with cholesterol, suggests that LM3 should be thought of as a liquid-ordered bilayer.
Fig. 7.
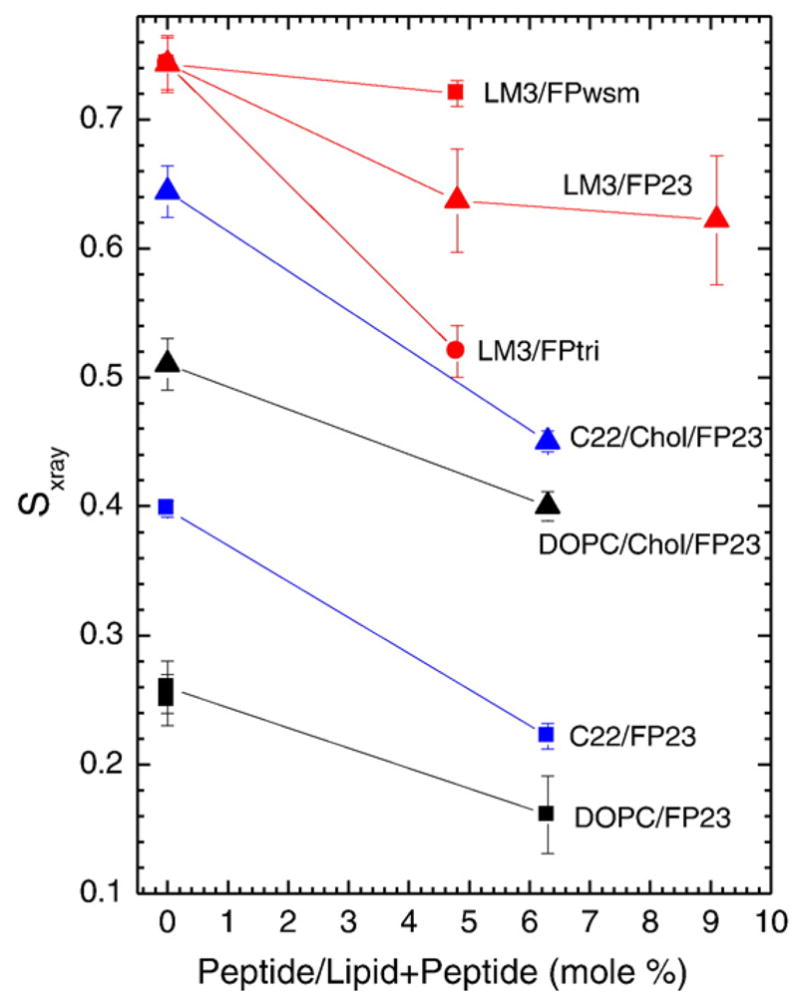
Sxray order parameters as a function of increasing peptide concentration; 1.6 mol% FPtri/LM3 is plotted at the equivalent native amino acid concentration, 4.8 mol% FPtri/LM3, for appropriate comparison. Lines are to guide the eye.
Spontaneous curvature
A standard method to study spontaneous curvature employs lipids such as DOPE that form hexagonal II phases. Our initial goal was to see how FP23 changes the spontaneous curvature of DOPE, although our result is perhaps more illuminating regarding the location of FP23. When FP23 was added to pure DOPE in the mole ratios shown in Fig. 8, there was a decrease in the Dhex spacing up to 2 mol% FP23. The same trend was observed in the presence of 16% tetradecane (TD), which facilitates the formation of HII phase by providing hydrocarbon to fill in the interstices between cylinders to relieve packing stress.31,32 At higher concentration, FP23 caused an increase in the Dhex spacing. This could be due to aggregation of FP23 in the highly constrained, tubular HII phase, as has been suggested previously in a study of feline leukemia virus FP in DOPE.33 Since a substantial decrease of KC was previously observed at 2 mol%,28 and since the D spacing was still smaller than that of the control DOPE, this concentration was chosen for the hydration curve in Fig. 9.
Fig. 8.
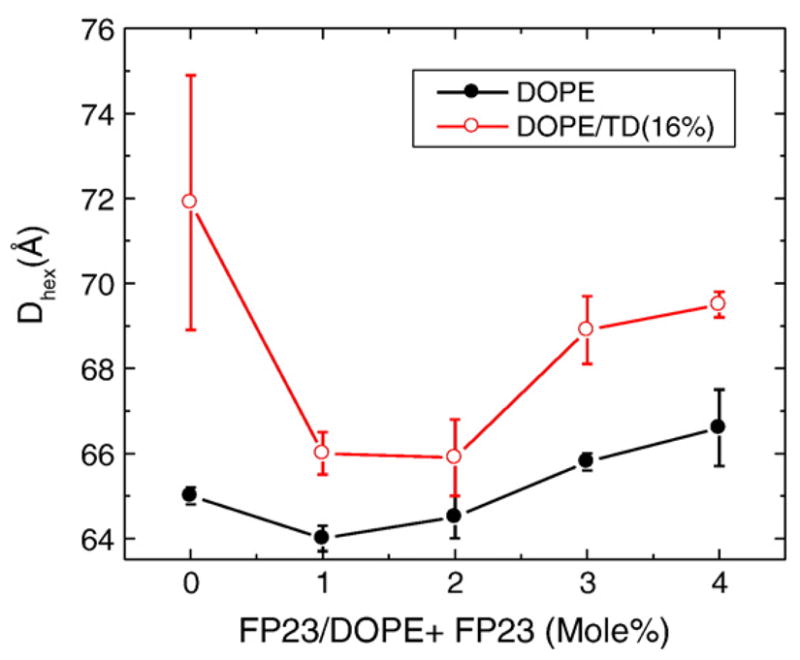
Effect of increasing mole percent of FP23 on Dhex of DOPE. Black filled circles have no added TD, and red open circles were prepared with 16% TD. All samples are in excess water.
Fig. 9.
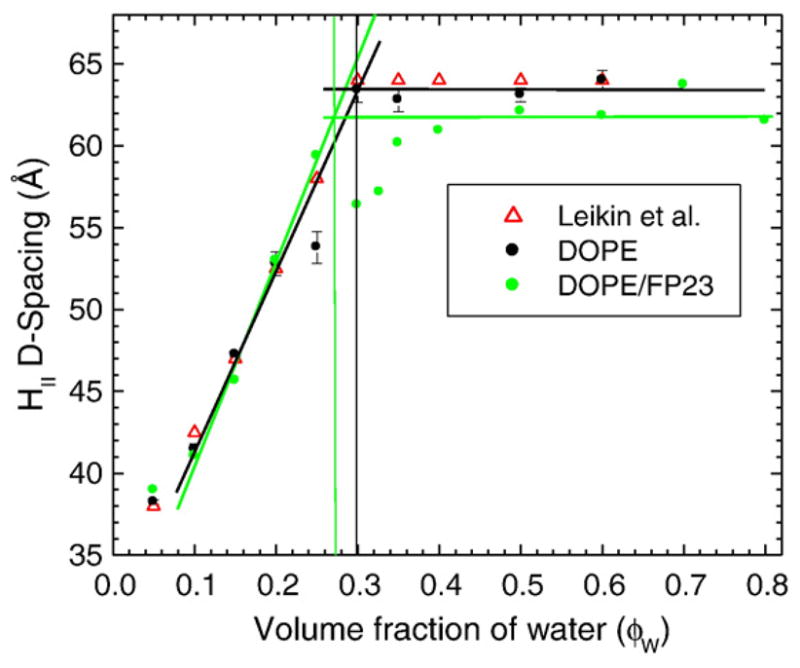
Hydration curves at T=24.5±0.5 °C of DOPE (black filled circles) and DOPE with 2 mol% FP23 (green filled circles). The data of Leikin et al. for pure DOPE are plotted for comparison as open red triangles.34
Figure 9 shows the hydration curve for pure DOPE and 2 mol% FP23/DOPE. The data for pure DOPE of Leikin et al. are plotted for comparison.34 Figure 9 shows that the initial slope of the hydration curve of DOPE is nearly the same with and without FP23, but the maximum, fully hydrated value of Dhex is smaller when FP23 is added to DOPE, even when the scatter in the data was considered. At all hydration levels, all samples were in the HII phase as evidenced by Dhex Bragg orders related as 1, 1/√3, 1/2; the three lowest Bragg orders were used to determine Dhex, even though six or more HII Bragg orders were observed in several of the drier samples.
Following the methods of Rand et al.,32,34–36 we quantified curvature as the inverse of the radius of the pivotal plane, defined as that radius at which the area does not change as the monolayer bends upon hydration. Figure 10 shows the location of the pivotal plane as well as other locations of interest. These include RW, the radius of the HII water cylinder. We have supplemented these standard quantities with RS, the radius from the center of the HII tube to the lipid steric thickness, which bisects the headgroup region with associated water, RC, the radius to the start (Gibbs dividing surface) of the hydrocarbon region, and R1, the effective lipid radius, including void regions. Using these radii and the equations of Leikin et al.,34 we calculate three areas: AW, the area per lipid molecule at the Luzzati plane (RW), AP, the area per unit cell at the pivotal plane (R0P), and A1, the area corresponding to R1. Table 4 gives numerical values for these quantities. Agreement with Leikin et al. for DOPE is satisfactory.34 The main result is that FP23 decreases R0P, thereby increasing the spontaneous curvature of the mixture beyond that of the already highly curved HII DOPE lipid; this result is suggested by the directly measured decrease in Dhex.
Fig. 10.
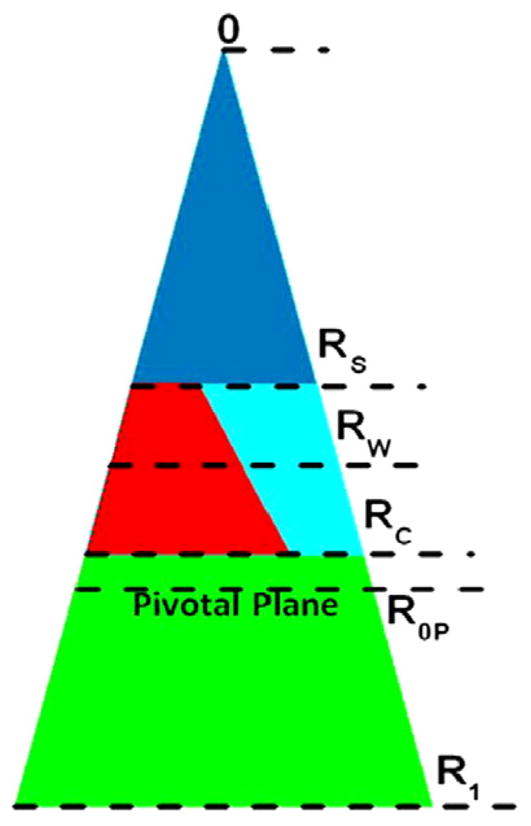
Schematic for the packing of lipid and water in the hexagonal II phase. The cylindrical water tubes extend in the direction perpendicular to the page. The center of each tube is at 0. The triangle extending from 0 to R1 represents the piece-of-pie space occupied by a monolayer of lipid and peptide and its associated water. The small dark blue (color online) triangle between 0 and RS (the steric radius) is occupied by water. The lower green trapezoid between R1 and RC (the Gibbs dividing surface for the hydrocarbon chains) represents the hydrophobic volume. The interfacial region between RS and RC represents the volume of the interfacial region; this is occupied by interfacial water (the right-hand light blue trapezoid) and the lipid headgroup and interfacial part of the peptide (the left-hand red trapezoid). The pivotal plane is at R0P. Radii are roughly to the scale of values for pure DOPE in Table 5.
Table 4.
Radial and areal results for DOPE and 2 mol% FP23/DOPE
| Sample | ϕW | Dhex | RW | R0P | RS | RC | R1 | AW | AP | A1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Leikin et al.34 | 0.30 | 63.8 | 21.1 | 28.5 | — | — | — | 49.6 | 64.2 | — |
| DOPE | 0.30 | 63.3 | 21.0 | 27.3 | 16.9 | 25.6 | 38.5 | 50.0 | 65.0 | 91.6 |
| DOPE/FP23 | 0.28 | 62.3 | 20.0 | 25.6 | 15.8 | 24.5 | 37.8 | 50.1 | 64.2 | 94.5 |
Units are in the appropriate powers of angstroms.
Table 5 summarizes the measured volumes and calculated volumes. V1 is the volume/unit cell where V1 = VL for pure DOPE and V1 = VDOPE + xVDOPE/FP23 (definition of unit cell as in Ref. 34) for DOPE with FP23, where x=0.02. Volumes were obtained from the measured densities as described in Materials and Methods. VP, the volume of the non-aqueous unit cell contents between 0 and R0P, was obtained by plotting (AW/AP)2 versus [AW/(VLRW)], where the negative slope=2VP/VL.34 VH is the volume/headgroup, determined previously as 252 Å3 for the lipid DLPE37 and assumed to be the same for DOPE; of course, this volume fills only part of the volume in the interfacial region between RC and RS, as shown in Fig. 10. VC is the volume/hydrocarbon region=VL − VH. VW is the volume of water between 0 and RW, and VW′ is the volume of water closely associated with the headgroup. From VW′, we calculate nW′ = 6.1 for DOPE, which compares well with that of DLPE, which has nW′ =6.2 (incorrectly reported as 4.7 in Ref. 37). The results shown in Tables 3 and 4 are for fully hydrated samples.
Table 5.
Volume results for DOPE and 2 mol% FP23/DOPE
| Sample | ϕW | V1 | VP | VH | VC | VW | VW′ | nW′ |
|---|---|---|---|---|---|---|---|---|
| Leikin et al.34 | 0.30 | 1235 | 375 | — | — | 524 | — | — |
| DOPE | 0.30 | 1226 | 364 | 252 | 974 | 525 | 183 | 6.1 |
| DOPE/FP23 | 0.28 | 1287 | 321 | 252 | 1035 | 501 | 189 | 6.3 |
Volumes are expressed in cubic angstroms.
Discussion
Although the most appropriate comparison was expected to be FPtri with the monomeric FPwsm from which FPtri was constructed, our LAXS (Fig. 1), KC (Table 3), WAXS (Fig. 6), and Sxray (Fig. 7) results show that monomeric FPwsm perturbs the LM3 bilayer far less than either monomeric FP23 or trimeric FPtri. We suggest that FPwsm did not partition into the LM3 membrane. This is at first surprising since both FPwsm and FPtri have the same proportion of additional lysines chemically attached to the C-terminus of wild-type FP23 in order to increase their solubility. We suggest that chemical attachment of three FPwsm peptides, each containing five or six lysines, reduces the average positive charge per lysine compared to FPwsm, due to lowering of the collective lysine pK values by adjacent positively charged lysines. This could shift a net hydrophilicity for FPwsm to a net hydrophobicity for FPtri. Comparing FPtri to FP23, while FP23 is clearly more hydrophobic per native amino acid, the factor of three could make FPtri even more net hydrophobic than FP23. Another factor in the comparison of the solubilities of FPtri and FP23 is the interaction of the more positively charged FPtri with the negatively charged LM3. It may also be that highly charged FPwsm may bind to the negatively charged headgroups in LM3, but it is clear that both FPtri and FP23 bind to and strongly perturb our model LM3 membrane and we will henceforth exclude FPwsm from a comparison of trimer versus monomer.
As emphasized in the Introduction, the structure of a peptide in fully hydrated, fluid-phase membranes is more amorphous than the atomic-level structure one obtains from crystallography. This perspective is reinforced by our CD results that there is no single secondary structure of FP23 in our bilayers, and this in turn is consistent with the literature that reports many different secondary structures.38–40 Although there is general agreement among liquid-state NMR studies that FP23 in detergent is largely helical,41–43 for membrane-associated FP23, populations of both α-helical and β-sheet conformations have been observed with β-sheet favored at higher peptide–lipid ratios and in membranes containing cholesterol.44–46 It has even been suggested that conversion from α-helical to β-structure is a step in membrane fusion.47 Also, simulations,48,49 experiments,50,51 and calculations52,53 have estimated a tilt angle of a helical FP, and FTIR experiments have shown that a β-sheet FP can also be tilted.54,55 Since we modeled peptides roughly as Gaussians, tilt angles cannot be calculated from our work. Secondary structure may not be so relevant, since both α-helix and β-sheet have been observed during fusion.39,56–60 The structure that we report focuses on different kinds of quantities, such as the average location of the peptide in our different model membranes.
For HIV studies, our most relevant model membrane is the LM3 mixture that we have characterized for the first time. Figures 3–5 show that LM3 is nearly as thick (DHH ~ 42 Å) as the long-chain diC22:1PC bilayer (DHH ~ 44 Å) and considerably thicker than the DOPC bilayer (DHH ~ 37 Å). LM3 is also as stiff as the diC22:1PC bilayer (Table 3) and LM3 has an even higher order parameter Sxray than diC22:1PC with similar cholesterol mole fraction (Fig. 7). This characterizes LM3 as an ordered fluid (Lo) phase. A better comparison of cholesterol-containing LM3 might be to DOPC/chol and diC22:1PC/chol. Then, LM3 DHH thickness is closer to that of DOPC/chol, but Sxray and KC remain closer to those of diC22:1PC/chol. A major difference is that LM3 has the negatively charged POPS and PI lipids. This makes a considerable difference in whether the repeat D spacing remains finite (Table 2) and it could make a difference for the location of positively charged FP.
Figures 4 and 5 show that FP23 penetrates well into the hydrocarbon region in DOPC and diC22:1PC without cholesterol. This is consistent with our result that FP23 increases the negative spontaneous curvature of DOPE monolayers in hexagonal II phase because, if FP23 packed into the interfacial headgroup region, it would counteract the negative curvature that DOPE has due to its relatively small headgroup area compared to its chain area.61 We should emphasize that the methodology for extracting structural information from x-ray scattering of peptides in bilayers is important for this conclusion. We were able to obtain excellent fits to our scattering data with the peptide Gaussian in the interfacial region when we did not impose volume conservation in our older H2 electron density model.62 Although the H2 model only involved small (<5%) violations of volume conservation for pure lipid bilayers, those violations grew (~25%) when FP was added because there was nothing to prevent artifactually packing FP into an already crowded interfacial region. In contrast, no volume conservation violations were allowed in the SDP method and our attempts to force FP into the interfacial region failed conspicuously to obtain sensible distributions of the component groups and their volumes.
When cholesterol was added to DOPC, Fig. 5 shows that FP23 remained deep in the hydrocarbon region, in contrast to our result for LM3 (Fig. 3) where both FP23 and FPtri were located closer to the carbonyl groups but still well within the hydrocarbon region. One might try to account for this difference by the negatively charged PS and PI headgroups in LM3 attracting the positively charged FP towards the interfacial region. While electrostatics undoubtedly play a role, they are not likely to be the sole factor for three reasons. First, our results for uncharged, thicker diC22:1PC/chol bilayers (Fig. 4) also have FP23 closer to the interface. Second, recent work23 reported that FPtri inserted closer to the bilayer center in the charged lipid system DTPC:DTPG (~4:1) and 30 mol% cholesterol, which had lipid chains with 14 carbons, and presumably had comparable thickness to LM3 and DOPC/chol in the present study. These first two reasons suggest that a larger thickness may be a factor in drawing the peptide closer to the headgroup region. Third, electrostatics alone would not lead to the result that the location of FP23 with only one positively charged arginine is so similar in Fig. 3 to the location of FPtri with 16 lysines in addition to 3 arginines. We therefore suggest that the position of FP in cholesterol-containing membranes depends upon other bilayer properties such as the thickness, the order parameter, and the bending modulus.
Any plausible pathway for fusion requires considerable bending and restructuring of the membranes. We showed previously that KC, the bending modulus, is dramatically lowered when FP23 was added to DOPC and diC22:1PC, which are both in the Lα (Ld) fluid state.28 Table 3 now shows that KC is similarly lowered when FP23 and FPtri are added to LM3, an Lo model lipid bilayer for the target T-cell membrane. Reduction of the bending modulus KC lowers the free energy F =1/2 KC (C − C0)2 required to induce curvature C in flat membranes that have spontaneous curvature C0 =0; thus, the activation free-energy barrier Fact for the curvature Ctrans of a transition state would generally be smaller for smaller KC. In addition, our membrane curvature results in DOPE HII phase show that C0 is increased, which might also act to lower Fact, depending upon details of the fusion pathway. This idea has been discussed by Siegel.63 We also find that FP mimics disorder all our lipid membranes (Figs. 6 and 7), which could help to prepare for a mixing of lipids from the two fusing membranes. These results are consistent with our recent finding that FP23 uncouples monolayers in a DOPC bilayer.64
A macroscopically appealing view of the role of the FP, inspired perhaps by science fiction movies showing three-legged pods attacking a mother ship, is that of a nail or battering ram that pokes a hole in the host T-cell membrane. The lack of definitive secondary peptide structure, as well as the flexibility of membranes, suggests more microscopic views. Our finding that FP penetrates into the hydrocarbon region is consistent with the hypothesis that FP binds sufficiently strongly that it could provide an anchor that could then be raised (or lowered) by conformational changes in gp41, which could then drag a substantial part of the host cell membrane with it, thereby initiating a pore. However, our favorite hypothesis is more subtle, namely, the penetration of FP disorders (Fig. 7) and softens the T-cell membrane (Table 3), thereby lowering the activation free-energy barrier for fusion of the virion membrane with the cell membrane, thereby allowing viral RNA to pass into the cell and infect it. This view focuses on fundamental energetics rather than details of the kinetic reaction pathway that would likely include many variations.
Materials and Methods
Materials
Lipids
LM3 is a mixture of six lipids (POPC:POPE:POPS:PI: ESM:Chol, 10:5:2:1:2:10) that mimics the T-cell lipid composition.24,25 Lyophilized lipids were purchased from Avanti, Alabaster, AL: POPC (Lot 160–181PC-174), POPE (Lot 160–181PE-109), POPS (Lot 160–181PS-203), PI (Lot PPI-150), ESM (Lot ESM-101), and cholesterol (Lot CH-55). DOPC (Lot 181–228), diC22:1PC (Lot 221PC-33), and DOPE (Lots 181PE-304 and 243) were also purchased from Avanti Polar Lipids in the lyophilized form and used without further purification.
For LM3, the lipids were mixed by dissolving each lipid in HPLC chloroform (Aldrich, St. Louis, MO) and then combining the stock solutions in the mole ratios shown above. The LM3 mixture has an average molecular weight of 751.1. Mixtures of DOPC and diC22:1PC with 30 mol% cholesterol were made by mixing stock solutions in chloroform. Thin-layer chromatography using chloroform:methanol:7 N NH4OH (46:18:3, v/v) and a molybdic acid stain performed on samples after x-ray at the Cornell High Energy Synchrotron Source (CHESS) revealed 0.1–0.5% lysolecithin formation on all samples.
Peptides
FP23 (AVGIGALFLGFLGAAGSTMGARS) was synthesized by the Peptide Synthesis Facility at the Pittsburgh Biotechnology Center (>95% purity). Both a water-soluble monomer form of FP23 (FPwsm) (AVGIGALFLGFL-GAAGSTMGARSWKKKKKKA) and a water-soluble trimer form of FPwsm (FPtri) were synthesized as previously described.65 The lyophilized peptides were weighed using a Mettler AE163 analytical balance and then dissolved in hexafluoroisopropanol (Aldrich). Lyophilized lipids were similarly weighed and dissolved in chloroform. Evaporation of organic solvent from the stock solutions was prevented by the use of Agilent glass vials (Agilent Technologies, Santa Clara, CA) with a Teflon septum that was punctured by a Hamilton syringe. These stock solutions were combined and quantitatively pipetted using a repeating dispenser on two Hamilton syringes (1 ml and 25 μl). The mole percent of FPtri was chosen to be one-third that of FPwsm and FP23 to have the same native amino acid fraction relative to the lipid. The solvent was allowed to evaporate in a fume hood over 1–2 days.
Methods
Differential scanning calorimetry
It is important that LM3, the mixture of lipids designed to mimic the T-cell lipid membrane composition, was in a fluid phase at 30 °C, where the experiments were performed. Differential scanning calorimetry at 76 °C/h was carried out using a Microcal MC-II (Northampton, MA) on 0. 5-mg/ml samples hydrated by temperature cycling as described for MLV samples. It appeared that a subgel phase66 could form in LM3 after incubating near 0 °C; however, the highest transition temperature of LM3 was less than 15 °C. For comparison, the calculated Tm for LM3, based on a weighted average of Tm values of the component lipids, is 16 °C. Additional differential scanning calorimetry of the other lipid/peptide mixtures had no transitions above 13 °C, indicating that all of the membranes studied were in a fluid phase at 30 °C.
X-ray sample preparation, oriented stacks
Four milligrams of lipid or lipid/peptide dried mixture (in duplicate) was dissolved in 200 μl chloroform: hexafluoroisopropanol:methanol (1:1:1 v/v) mixture and this was plated onto a 30 mm×15 mm×1 mm silicon wafer, using the rock-and-roll technique.67 After drying overnight in a glove box, residual solvent was removed by 1–2 h in a vacuum oven. The samples were trimmed to a 5 mm×30 mm strip in the center of the wafer. Hydration of oriented samples from water vapor was carried out in a thick-walled hydration chamber.68
Multilamellar vesicle
Unoriented MLVs in excess water for volumetric measurements, calorimetry, and fully hydrated D spacing measurements were prepared by pipetting the stock solutions described above into Nalgene vials and removing solvent in a fume hood and vacuum oven. The dried samples were mixed with milli-Q water and then thermally cycled three times with vortexing between −20 °C and 50 °C before loading into 1-mm-diameter glass capillaries for x-ray or 3- ml Nalgene vials for calorimetry or 1-ml glass vials for volume measurements. For the DOPE hydration curves, water was added to the dried mixtures quantitatively using a Mettler AE163 analytical balance and samples were allowed to equilibrate for 4–5 days in the refrigerator at 1–4 °C before loading into x-ray capillaries. TD was purchased from Aldrich Chemical Co. and added to DOPE in the mole percent 16.8±1.4% TD.
X-ray scattering experiments, oriented stacks
X-ray data of oriented fluid-phase lipids and lipid/peptide samples at 30 °C were obtained at CHESS on two separate trips using the G1 station where the wavelength was set with a WB4/C multilayer monochromator to 1.1797 or 1.1803 Å with a full width at half maximum of ±0.012 Å and the total beam intensity was ~1011 photons/s. Beam size was 0.26 mm H and 1 mm V. The sample was ~10 μm thick along the normal to the ~2000 bilayers. Its dimension along the direction of the beam for LAXS and WAXS was narrow (5 mm). The flat samples were rotated from −3° to 7° in θ relative to the beam during the 10- to 20-s LAXS exposures and they were x-rayed at fixed θ for the 10- to 20-s WAXS exposures. For WAXS, θ=0.2° was used to first collect lipid scattering and then θ=−0.2° was used to collect background chamber scattering that was subtracted from the lipid data. Data were collected using a Flicam CCD (Finger Lakes Instrumentation, Lima, NY) with a 1024×1024 pixel array and a pixel size of 69.78 μm/pixel. The sample-to-CCD distance was 399.7 or 370.9 mm for LAXS and 154.7 or 149 mm for WAXS. Temperature was controlled with a Neslab Controller (Portsmouth, NH) and monitored using a Cole-Parmer thermistor thermometer (Vernon Hills, IL).
MLV samples
MLV samples were x-rayed at 25 °C (DOPE) and 30 °C (all others) using a Rigaku RUH3R microfocus rotating anode (Woodlands, TX) equipped with a Xenocs FOX2D focusing collimation optic. Beam size was 1 mm×1 mm, and 5-min scans were collected using a Rigaku Mercury CCD detector with a pixel size of 68.0 μm/pixel; silver behenate (D=58.367 Å) was used for calibration. Temperature was controlled with a Julabo F25-ED (Allentown, PA) monitored with a Cole-Parmer thermistor thermometer.
X-ray data analysis, LAXS
The analysis of LAXS diffuse data has been described in previous publications26,68 and will be reviewed here only briefly. The scattering intensity for a stack of oriented bilayers is the product: I(q)=S(q)|F(qz)|2/qz, where q=(qr,qz), S(q) is the structure interference factor, F(qz) is the bilayer form factor, and is the usual low-angle approximation to the Lorentz factor for narrow oriented samples and a tall beam for which all the sample remains in the beam for all relevant q. The first step of the analysis obtains the bilayer bending modulus (KC), the compression modulus (B), and |F(qz)|2/qz.
In the second step, x-ray data were fit to models using the SDP program.69 SDP has the advantage over previous programs62,70 in that volume is rigorously conserved at all z values along the bilayer normal. The experimental volume V per unit cell provides an important constraint between area A, the various thicknesses, and the zeroth order form factor F(0):71
| (1) |
where n is the number of electrons in the unit cell chosen to be one lipid (averaged for LM3) and the amounts of peptide and cholesterol corresponding to their mole fractions. Our models had Gaussians for the molecular components consisting of phosphatidylcholine (PC) or phosphatidylother (PO) for LM3, carbonyl/glycerol (CG), and the terminal methyls (M) on the hydrocarbon chains. The total hydrocarbon region, which also included cholesterol when present, was modeled by a symmetric pair of error functions from which the volumes of the methyl and peptide were subtracted. The headgroup volume was fixed to 295 Å3 for LM3 based on a weighted average of the volumes of LM3 component headgroups or 331 Å3 for PC lipids, and for all samples, the volumes of the three Gaussians were soft constrained with harmonic penalty terms for deviations from values obtained from simulations and previous analyses of pure lipids.69 For some samples, it was necessary to constrain the widths of some of the components that became either unrealistically wide or narrow.
WAXS
WAXS emanates primarily from chain scattering that is affected by interchain distances and chain orientational order. The orientational order is obtained from the angular dependence I(φ) (where tan φ=qz/qr) of WAXS data from oriented samples that are analyzed following Mills et al.27 An x-ray order parameter
| (2) |
provides a measure of the distribution of local tilt angles β.
Volume determination
Lipid molecular volume in fully hydrated MLV was determined by two methods: (1) neutral buoyancy and (2) vibrating tube densimetry. In method 1, weighed mixtures of D2O and H2O (1.5–3 ml) were used to hydrate ~3 mg of the dried lipid mixtures by temperature cycling, as for MLV. The hydrated samples were temperature equilibrated for 2 days. Visual inspection determined if the sample was floating, sinking, or neutrally buoyant and the latter samples provided the density of the sample. Conversion to molecular volume used the molecular weight of the lipid or lipid mixture and Avogadro’s number. In method 2, sample density ρS and water density ρW were measured at 30±0.001 °C using an Anton-Paar DMA4500 (Ashland, VA) vibrating tube densimeter and molecular volume was calculated for a sample with lipid or mixture mass mL and water mass mW using
| (3) |
where ML was the weighted average of the component molecular weights in LM3 or in mixtures of lipids and peptides.
CD spectroscopy
CD spectroscopy was carried out using a Jasco (Easton, MD) CD spectropolarimeter. Samples with lipid and lipid/peptide mixtures were oriented onto one inner side of a quartz cuvette using the same rock-and-roll procedure as for x-ray LAXS samples. CD spectra of the dried, oriented samples were collected first. Then, the cuvette was slowly filled with water so as not to perturb the lipid film and CD spectra were obtained within 10–30 min. During this procedure, the lipid/peptide films remained on the cuvette wall but became opaque due to absorption of water. This hydrated sample is most similar to our oriented samples, fully hydrated through the vapor. Individual scans were collected in 1 min over the range 250 to 180 nm; 5–10 scans were averaged for each sample and temperature was controlled to 30 °C. Secondary structures were assigned based on comparison to published data.72–74
Spontaneous curvature
Following established procedures,34–36 the location of the Gibbs dividing surface for water RW relative to the center of the water tubes (see Fig. 10) was calculated using the x-ray measured hexagonal spacing Dhex and the measured volume fraction of water φW
| (4) |
The corresponding area per unit cell (one lipid and mole fractional peptide) with volume V1 is given by
| (5) |
Following an established recipe,36 the radius of the pivotal plane R0P was obtained at the maximum level of hydration determined in the Dhex plot versus ϕ(Fig. 9). Areas and radii are related by Aj = fAW and Rj = fRW, where
| (6) |
and Vj is the volume between 0 and Rj. It is also convenient to define g=Aj/Rj = AW/RW. Then, the volume between R2 and R1 is given by
| (7) |
which allows one to calculate Ri from Rj, Vij, and g. The radius R1, which includes the measured volume of lipid plus peptide plus associated water at full hydration, is given by . [This value of R1 is the same as that obtained from RW and the measured volume VM and using Eq. (7).] For pure DOPE, we calculated RC from the volume of DOPE hydrocarbon chains using VC and Eq. (4). For added FP23, RC depends upon how the peptide volume is partitioned between hydrocarbon and interfacial regions. The steric radius RS was obtained by subtracting an estimated thickness 8.7 Å of DOPE headgroups37 from RC. The location of the line drawn in Fig. 10 between the headgroup volume and the interfacial water volume satisfies (non-uniquely) the Gibbs dividing surface constraint that the interfacial water volume between RC and RW must equal the headgroup volume between RW and RS.
Acknowledgments
This research was supported by National Institutes of Health (NIH) Grant GM 44976 (S.T.-N. and J.F.N.), NIH Grant AI 47153 (W.Q. and D.P.W.), and the Howard Hughes Medical Institute (R.C.). X-ray scattering data were taken at CHESS, which is supported by the National Science Foundation and the NIH/National Institute of General Medical Sciences under National Science Foundation award DMR-0225180. We especially thank Dr. Arthur Woll for obtaining our beam and for general support during our data collection at the G1 station. We would also like to thank Dr. Jianjun Pan, Dr. Thalia Mills, Deren Guler, Alex Greenwood, and Dipon Ghosh for help with x-ray data collection; David Lenkner for data analysis; and Drs. Gilman Toombes and David Siegel for useful discussions. We also thank the Center for Molecular Analysis at Carnegie Mellon University for use of its CD spectrometer.
Abbreviations
- FP
fusion peptide
- HIV-1
human immunodeficiency virus type 1
- LAXS
low-angle x-ray scattering
- WAXS
wide-angle x-ray scattering
- MLV
multilamellar vesicle
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine
- PI
soy phosphatidylinositol
- ESM
egg sphingomyelin
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- diC22:1PC
1,2-dierucoyl-sn-glycero-3-phosphocholine
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- TD
tetradecane
- CHESS
Cornell High Energy Synchrotron Source
Footnotes
References
- 1.Blumenthal R, Clague MJ, Durell SR, Epand RM. Membrane fusion. Chem Rev. 2003;103:53–69. doi: 10.1021/cr000036+. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez LD, Hoffman LR, Wolfsberg TG, White JM. Virus–cell and cell–cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 3.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 4.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 6.Weissenhorn W, Dessen A, Calder LJ, Harrison SC, Skehel JJ, Wiley DC. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 7.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 8.Jahn R, Lang T, Sudhof TC. Membrane fusion. Cell. 2003;112:519–533. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 9.Gordon LM, Curtain CC, Zhong YC, Kirkpatrick A, Mobley PW, Waring AJ. The amino-terminal peptide of HIV-1 glycoprotein 41 interacts with human erythrocyte membranes: peptide conformation, orientation and aggregation. Biochim Biophys Acta. 1992;1139:257–274. doi: 10.1016/0925-4439(92)90099-9. [DOI] [PubMed] [Google Scholar]
- 10.Slepushkin VA, Andreev SM, Sidorova MV, Melikyan GB, Grigoriev VB, Chumakov VM, et al. Investigation of human immunodeficiency virus fusion peptides. Analysis of interrelations between their structure and function. AIDS Res Hum Retroviruses. 1992;8:9–18. doi: 10.1089/aid.1992.8.9. [DOI] [PubMed] [Google Scholar]
- 11.Freed EO, Delwart EL, Buchschacher GL, Jr, Panganiban AT. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mobley PW, Waring AJ, Sherman MA, Gordon LM. Membrane interactions of the synthetic N-terminal peptide of HIV-1 gp41 and its structural analogs. Biochim Biophys Acta. 1999;1418:1–18. doi: 10.1016/s0005-2736(99)00014-0. [DOI] [PubMed] [Google Scholar]
- 13.Kliger Y, Aharoni A, Rapaport D, Jones P, Blumenthal R, Shai Y. Fusion peptides derived from the HIV type 1 glycoprotein 41 associate within phospholipid membranes and inhibit cell–cell fusion—structure–function study. J Biol Chem. 1997;272:13496–13505. doi: 10.1074/jbc.272.21.13496. [DOI] [PubMed] [Google Scholar]
- 14.Martin I, Schaal H, Scheid A, Ruysschaert JM. Lipid membrane fusion induced by the human immunodeficiency virus type 1 gp41 N-terminal extremity is determined by its orientation in the lipid bilayer. J Virol. 1996;70:298–304. doi: 10.1128/jvi.70.1.298-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charloteaux B, Lorin A, Crowet JM, Stroobant V, Lins L, Thomas A, Brasseur R. The N-terminal 12 residue long peptide of HIV gp41 is the minimal peptide sufficient to induce significant T-cell-like membrane destabilization in vitro. J Mol Biol. 2006;359:597–609. doi: 10.1016/j.jmb.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 16.Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang ZN, Mueser TC, Kaufman J, Stahl SJ, Wingfield PT, Hyde CC. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J Struct Biol. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- 19.Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, et al. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu YF, Nagle JF. Diffuse scattering provides material parameters and electron density profiles of biomembranes. Phys Rev E. 2004;69:040901–040904. doi: 10.1103/PhysRevE.69.040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang R, Yang J, Weliky DP. Synthesis, enhanced fusogenicity, and solid state NMR measurements of cross-linked HIV-1 fusion peptides. Biochemistry. 2003;42:3527–3535. doi: 10.1021/bi027052g. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Tamm LK. Structure and plasticity of the human immunodeficiency virus gp41 fusion domain in lipid micelles and bilayers. Biophys J. 2007;93:876–885. doi: 10.1529/biophysj.106.102335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiang W, Sun Y, Weliky DP. A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc Natl Acad Sci USA. 2009;106:15314–15319. doi: 10.1073/pnas.0907360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brugger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich HG. The HIV lipidome: a raft with an unusual composition. Proc Natl Acad Sci USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 27.Mills TT, Toombes GES, Tristram-Nagle S, Smilgies DM, Feigenson GW, Nagle JF. Order parameters and areas in fluid-phase oriented lipid membranes using wide angle x-ray scattering. Biophys J. 2008;95:669–681. doi: 10.1529/biophysj.107.127845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tristram-Nagle S, Nagle JF. HIV-1 fusion peptide decreases bending energy and promotes curved fusion intermediates. Biophys J. 2007;93:2048–2055. doi: 10.1529/biophysj.107.109181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan JJ, Tristram-Nagle S, Nagle JF. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys Rev E. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan JJ, Mills TT, Tristram-Nagle S, Nagle JF. Cholesterol perturbs lipid bilayers nonuniversally. Phys Rev Lett. 2008;100:198103. doi: 10.1103/PhysRevLett.100.198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirk GL, Gruner SM. Lyotropic effects of alkanes and headgroup composition on the L-alpha-Hii lipid liquid crystal phase transition—hydrocarbon packing versus intrinsic curvature. J Phys (Paris) 1985;46:761–769. [Google Scholar]
- 32.Rand RP, Fuller NL, Gruner SM, Parsegian VA. Membrane curvature, lipid segregation, and structural transitions for phospholipids under dual-solvent stress. Biochemistry. 1990;29:76–87. doi: 10.1021/bi00453a010. [DOI] [PubMed] [Google Scholar]
- 33.Davies SMA, Epand RF, Bradshaw JP, Epand RM. Modulation of lipid polymorphism by the feline leukemia virus fusion peptide: implications for the fusion mechanism. Biochemistry. 1998;37:5720–5729. doi: 10.1021/bi980227v. [DOI] [PubMed] [Google Scholar]
- 34.Leikin S, Kozlov MM, Fuller NL, Rand RP. Measured effects of diacylglycerol on structural and elastic properties of phospholipid membranes. Biophys J. 1996;71:2623–2632. doi: 10.1016/S0006-3495(96)79454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Rand RP. The influence of cholesterol on phospholipid membrane curvature and bending elasticity. Biophys J. 1997;73:267–276. doi: 10.1016/S0006-3495(97)78067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rand RP, Fuller NL. Structural dimensions and their changes in a reentrant hexagonal–lamellar transition of phospholipids. Biophys J. 1994;66:2127–2138. doi: 10.1016/S0006-3495(94)81008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durell SR, Martin I, Ruysschaert JM, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 39.Reichert J, Grasnick D, Afonin S, Buerck J, Wadhwani P, Ulrich AS. A critical evaluation of the conformational requirements of fusogenic peptides in membranes. Eur Biophys J. 2007;36:405–413. doi: 10.1007/s00249-006-0106-2. [DOI] [PubMed] [Google Scholar]
- 40.Haque ME, Koppaka V, Axelsen PH, Lentz BR. Properties and structures of the influenza and HIV fusion peptides on lipid membranes: implications for a role in fusion. Biophys J. 2005;89:3183–3194. doi: 10.1529/biophysj.105.063032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaroniec CP, Kaufman JD, Stahl SJ, Viard M, Blumenthal R, Wingfield PT, Bax A. Structure and dynamics of micelle-associated human immunodeficiency virus gp41 fusion domain. Biochemistry. 2005;44:16167–16180. doi: 10.1021/bi051672a. [DOI] [PubMed] [Google Scholar]
- 42.Tamm LK, Lai AL, Li Y. Combined NMR and EPR spectroscopy to determine structures of viral fusion domains in membranes. Biochim Biophys Acta. 2007;1768:3052–3060. doi: 10.1016/j.bbamem.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabrys CM, Weliky DP. Chemical shift assignment and structural plasticity of a HIV fusion peptide derivative in dodecylphosphocholine micelles. Biochim Biophys Acta. 2007;1768:3225–3234. doi: 10.1016/j.bbamem.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Parkanzky PD, Bodner ML, Duskin CA, Weliky DP. Application of REDOR subtraction for filtered MAS observation of labeled backbone carbons of membrane-bound fusion peptides. J Magn Reson. 2002;159:101–110. doi: 10.1016/s1090-7807(02)00033-2. [DOI] [PubMed] [Google Scholar]
- 45.Wasniewski CM, Parkanzky PD, Bodner ML, Weliky DP. Solid-state nuclear magnetic resonance studies of HIV and influenza fusion peptide orientations in membrane bilayers using stacked glass plate samples. Chem Phys Lipids. 2004;132:89–100. doi: 10.1016/j.chemphyslip.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Zheng Z, Yang R, Bodner ML, Weliky DP. Conformational flexibility and strand arrangements of the membrane-associated HIV fusion peptide trimer probed by solid-state NMR spectroscopy. Biochemistry. 2006;45:12960–12975. doi: 10.1021/bi0615902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Epand RM, Epand RF. Modulation of membrane curvature by peptides. Biopolymers. 2000;55:358–363. doi: 10.1002/1097-0282(2000)55:5<358::AID-BIP1009>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 48.Kamath S, Wong TC. Membrane structure of the human immunodeficiency virus gp41 fusion domain by molecular dynamics simulation. Biophys J. 2002;83:135–143. doi: 10.1016/S0006-3495(02)75155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maddox MW, Longo ML. Conformational partitioning of the fusion peptide of HIV-1 gp41 and its structural analogs in bilayer membranes. Biophys J. 2002;83:3088–3096. doi: 10.1016/S0006-3495(02)75313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradshaw JP, Darkes MJM, Harroun TA, Katsaras J, Epand RM. Oblique membrane insertion of viral fusion peptide probed by neutron diffraction. Biochemistry. 2000;39:6581–6585. doi: 10.1021/bi000224u. [DOI] [PubMed] [Google Scholar]
- 51.Brasseur R, Vandenbranden M, Cornet B, Burny A, Ruysschaert JM. Orientation into the lipid bilayer of an asymmetric amphipathic helical peptide located at the N-terminus of viral fusion proteins. Biochim Biophys Acta. 1990;1029:267–273. doi: 10.1016/0005-2736(90)90163-i. [DOI] [PubMed] [Google Scholar]
- 52.Horth M, Lambrecht B, Khim MCL, Bex F, Thiriart C, Ruysschaert JM, et al. Theoretical and functional-analysis of the SIV fusion peptide. EMBO J. 1991;10:2747–2755. doi: 10.1002/j.1460-2075.1991.tb07823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brasseur R. Differentiation of lipid-associating helices by use of 3-dimensional molecular hydrophobicity potential calculations. J Biol Chem. 1991;266:16120–16127. [PubMed] [Google Scholar]
- 54.Castano S, Desbat B. Structure and orientation study of fusion peptide FP23 of gp41 from HIV-1 alone or inserted into various lipid membrane models (mono-, bi- and multibi-layers) by FT-IR spectroscopies and Brewster angle microscopy. Biochim Biophys Acta. 2005;1715:81–95. doi: 10.1016/j.bbamem.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 55.Charloteaux B, Lorin A, Brasseur R, Lins L. The “Tilted Peptide Theory” links membrane insertion properties and fusogenicity of viral fusion peptides. Protein Pept Lett. 2009;16:718–725. doi: 10.2174/092986609788681724. [DOI] [PubMed] [Google Scholar]
- 56.Peisajovich SG, Epand RF, Pritsker M, Shai Y, Epand RM. The polar region consecutive to the HIV fusion peptide participates in membrane fusion. Biochemistry. 2000;39:1826–1833. doi: 10.1021/bi991887i. [DOI] [PubMed] [Google Scholar]
- 57.Nieva JL, Nir S, Muga A, Goni FM, Wilschut J. Interaction of the HIV-1 fusion peptide with phospholipid vesicles: different structural requirements for fusion and leakage. Biochemistry. 1994;33:3201–3209. doi: 10.1021/bi00177a009. [DOI] [PubMed] [Google Scholar]
- 58.Rafalski M, Lear JD, DeGrado WF. Phospholipid interactions of synthetic peptides representing the N-terminus of HIV gp41. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- 59.Del Angel VD, Dupuis F, Mornon JP, Callebaut I. Viral fusion peptides and identification of membrane-interacting segments. Biochem Biophys Res Commun. 2002;293:1153–1160. doi: 10.1016/S0006-291X(02)00353-4. [DOI] [PubMed] [Google Scholar]
- 60.Vaccaro L, Cross KJ, Kleinjung J, Straus SK, Thomas DJ, Wharton SA, et al. Plasticity of influenza haemagglutinin fusion peptides and their interaction with lipid bilayers. Biophys J. 2005;88:25–36. doi: 10.1529/biophysj.104.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Israelachvili JN, Mitchell DJ. Model for packing of lipids in bilayer membranes. Biochim Biophys Acta. 1975;389:13–19. doi: 10.1016/0005-2736(75)90381-8. [DOI] [PubMed] [Google Scholar]
- 62.Klauda JB, Kucerka N, Brooks BR, Pastor RW, Nagle JF. Simulation-based methods for interpreting x-ray data from lipid bilayers. Biophys J. 2006;90:2796–2807. doi: 10.1529/biophysj.105.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegel DP. Fourth-order curvature energy model for the stability of bicontinuous inverted cubic phases in amphiphile–water systems. Langmuir. 2010;26:8673–8683. doi: 10.1021/la904838z. [DOI] [PubMed] [Google Scholar]
- 64.Shchelokovskyy P, Tristram-Nagle S, Dimova R. Effect of the HIV-1 fusion peptide on the mechanical properties and leaflet coupling of lipid bilayers. New J Phys. 2010 doi: 10.1088/1367-2630/13/2/025004. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiang W, Weliky DP. HIV fusion peptide and its cross-linked oligomers: efficient syntheses, significance of the trimer in fusion activity, correlation of beta strand conformation with membrane cholesterol, and proximity to lipid headgroups. Biochemistry. 2009;48:289–301. doi: 10.1021/bi8015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tristram-Nagle S, Wiener MC, Yang CP, Nagle JF. Kinetics of the subtranstion in dipalmitoylphosphatidylcholine. Biochemistry. 1987;26:4288–4294. doi: 10.1021/bi00388a016. [DOI] [PubMed] [Google Scholar]
- 67.Tristram-Nagle SA. Preparation of oriented, fully hydrated lipid samples for structure determination using X-ray scattering. Methods Mol Biol. 2007;400:63–75. doi: 10.1007/978-1-59745-519-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kučerka N, Liu YF, Chu NJ, Petrache HI, Tristram-Nagle ST, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multi-lamellar arrays and from unilamellar vesicles. Biophys J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kučerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data. Biophys J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wiener MC, White SH. Fluid bilayer structure determination by the combined use of x-ray and neutron diffraction. 2 Composition-space refinement method. Biophys J. 1991;59:174–185. doi: 10.1016/S0006-3495(91)82209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagle JF, Wiener MC. Relations for lipid bilayers—connection of electron-density profiles to other structural quantities. Biophys J. 1989;55:309–313. doi: 10.1016/S0006-3495(89)82806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terzi E, Holzemann G, Seelig J. Self-association of beta-amyloid peptide (1–40) in solution and binding to lipid membranes. J Mol Biol. 1995;252:633–642. doi: 10.1006/jmbi.1995.0525. [DOI] [PubMed] [Google Scholar]
- 73.Mao D, Wallace BA. Differential light scattering and absorption flattening optical effects are minimal in the circular dichroism spectra of small unilamellar vesicles. Biochemistry. 1984;23:2667–2673. doi: 10.1021/bi00307a020. [DOI] [PubMed] [Google Scholar]
- 74.Chen FY, Lee MT, Huang HW. Evidence for membrane thinning effect as the mechanism for peptide-induced pore formation. Biophys J. 2003;84:3751–3758. doi: 10.1016/S0006-3495(03)75103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]