Abstract
RNA interference (RNAi) depends on the production of small RNA to regulate gene expression in eukaryotes. Two RNAi systems exist to control repetitive selfish elements in Neurospora crassa. Quelling targets transgenes during vegetative growth, whereas meiotic silencing by unpaired DNA (MSUD) silences unpaired genes during meiosis. The two mechanisms require common RNAi proteins, such as RNA-directed RNA polymerases, Dicers, and Argonaute slicers. We have previously demonstrated that, while Quelling depends on the redundant dicer activity of DCL-1 and DCL-2, only DCL-1 is required for MSUD. Here, we show that QDE-2-interacting protein (QIP), an exonuclease that is important for the production of single-stranded siRNA during Quelling, is also required for MSUD. QIP is crucial for sexual development and is shown to colocalize with other MSUD proteins in the perinuclear region.
NEUROSPORA crassa is a filamentous fungus that grows by hyphal tip extension and branching (Glass et al. 2000). Since septa (cross walls) between individual cells are normally incomplete, deleterious elements such as viruses or selfish DNA can easily infiltrate the entire network of hyphae (known as the mycelium). To combat these repetitive elements, several genome surveillance systems have evolved and are maintained in N. crassa. For example, repeat-induced point mutation, a premeiotic process operating during the haploid dikaryotic stage, introduces extensive GC-to-AT mutations to duplicated sequences (Cambareri et al. 1989). Quelling and meiotic silencing by unpaired DNA (MSUD), on the other hand, target transcripts generated from potential intruders during vegetative growth and meiosis, respectively (Romano and Macino 1992; Shiu et al. 2001). These surveillance mechanisms presumably allow N. crassa to be virtually free of active transposons and viruses (Catalanotto et al. 2006).
In the Quelling model, large tandem arrays of a transgene often alert the host defense mechanism, presumably during DNA replication (Nolan et al. 2008). QDE-3, a DNA helicase (Cogoni and Macino 1999b), may play a role in resolving secondary structures of tandem transgenes, enabling the transcription of single-stranded aberrant RNA and their conversion to double strands by QDE-1, a DNA- and RNA-directed RNA polymerase (DdRp and RdRP, respectively; Cogoni and Macino 1999a; Lee et al. 2009). The double-stranded RNA (dsRNA) molecules are further processed into 21- to 25-nucleotide small interfering RNA (siRNA) by the redundant dicer activity of DCL-1 and DCL-2 (Catalanotto et al. 2004). The siRNA duplexes are then loaded into the QDE-2 Argonaute, a component of the RNA-induced silencing complex (RISC) (Catalanotto et al. 2002). One of the siRNA strands (the passenger strand) is nicked by the slicer activity of QDE-2 and later degraded by the QDE-2-interacting protein (QIP) (Maiti et al. 2007). The remaining single-stranded siRNA (the guide strand) can subsequently recognize homologous mRNA by base complementarity and target them for QDE-2-dependent cleavage.
In addition to propagation through mycelial growth and dispersal of conidia (asexual spores), N. crassa can also enter a sexual cycle. After fertilization and nuclear proliferation, opposite mating-type nuclei, A and a, pair and migrate into a dikaryotic hypha (for review, see Shiu and Glass 2000). Karyogamy occurs between A and a nuclei in the ascus (spore sac) mother cell, with meiosis and ascospore development following immediately afterward. Meiosis represents a window of opportunity for the expansion of selfish elements, as the two genomes are aligned intimately during the homologous pairing stage. In N. crassa, the mechanism known as MSUD exists to silence unpaired and potentially harmful genes. In MSUD, a gene not paired with a homologous partner generates a signal that silences all copies of that gene during sexual development. The current MSUD model suggests that an unpaired gene is detected and transcribed into aberrant RNA. The aberrant RNA are converted into dsRNA by the SAD-1 RdRP (Shiu and Metzenberg 2002), whose localization is controlled by the SAD-2 protein (Shiu et al. 2006). The dsRNA in turn are diced into siRNA by DCL-1 (Alexander et al. 2008). SMS-2, an Argonaute protein, is responsible for the siRNA-guided destruction of mRNA (Lee et al. 2003). The involvement of DCL-1 in both Quelling and MSUD suggests that a crosstalk exists between the two RNA-silencing mechanisms. In this work, we have set out to determine whether QIP, a part of the Quelling machinery, is also required for MSUD.
MATERIALS AND METHODS
Strains, media, and growth:
The Neurospora strains used in this study are described in Table 1. Auxotrophic and other mutant strains were acquired from the Fungal Genetics Stock Center (FGSC; McCluskey 2003). The description of individual genes, including their mapping information, can be obtained from the Neurospora Compendium (Perkins et al. 2001) and the e-Compendium (http://bmbpcu36.leeds.ac.uk/∼gen6ar/newgenelist/genes/gene_list.htm). The qipΔ deletion strain contains the replacement of a sequence encompassing the qip open reading frame (positions −206 to 2070; supporting information, Figure S1) with a hygromycin-resistant gene (hph) (Colot et al. 2006). The qipfs (frameshift) allele was constructed from the wild-type gene by cleavage at the NsiI site (positions 149–154) followed by a Klenow fill-in reaction. Preparation of culturing and crossing media was as previously described (Westergaard and Mitchell 1947; Vogel 1964). Homokaryons were isolated using the method of Ebbole and Sachs (1990). Standard procedures for growth, crosses, and other Neurospora manipulations were followed throughout (Davis and de Serres 1970).
TABLE 1.
Neurospora strains used in this study
| Strain | Genotype |
|---|---|
| F1-05 | fl a |
| F2-01 | fl A |
| F2-06 | fl; qipΔ∷hph A |
| F2-29 | rid RΔ∷hph; fl A |
| F2-35 | his-3+∷act+; fl A |
| F2-36 | his-3+∷BmlR; fl A |
| F3-23 | rid his-3+∷asm-1+; fl; Asm-1Δ∷hph A |
| P3-07 | Oak Ridge wild-type A (FGSC 2489) |
| P3-08 | Oak Ridge wild-type a (FGSC 2490) |
| P3-25 | mep Sad-1Δ∷hph a |
| P5-52 | Sad-1Δ∷hph rid his-3 a |
| P6-07 | rid A |
| P6-08 | rid a |
| P6-62 | rid his-3+∷sad-2-rfp; inv Sad-2RIPa |
| P9-39 | qipΔ∷hph a (FGSC 12130) |
| P10-16 | rid his-3+∷hH1-gfp a |
| P10-18 | rid his-3+∷dcl-1-gfp; dcl-1Δ∷hph mus-52Δ∷bar A |
| P11-21 | rid his-3; qipΔ∷hph; mus-51Δ∷bar a |
| P11-36 | rid his-3+∷r+; mus-52Δ∷bar A |
| P11-38 | rid his-3+∷qipfs; mus-52Δ∷bar A |
| P11-41 | rid his-3+∷r+; qipΔ∷hph; mus-51Δ∷bar a |
| P12-13 | rid his-3+∷hH1-gfp; mus-51Δ∷bar; qipΔ∷hph a |
| P12-28 | rid his-3; qipΔ∷hph; mus-51Δ∷bar A |
| P13-15 | sad-1-gfp∷hph A |
| P14-05 | rid his-3+∷qip-gfp; qipΔ∷hph; mus-51Δ∷bar A |
| P15-02 | rid his-3+∷sms-2-rfp; mus-52Δ∷bar; qip-gfp∷hph A |
| P15-03 | rid his-3+∷sms-2-rfp; mus-52Δ∷bar; qip-gfp∷hph a |
FGSC, Fungal Genetics Stock Center.
Nucleic acid methods and transformation:
Standard molecular techniques were used according to Sambrook and Russell (2001). Fungal DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). Custom oligonucleotide primers, as listed in Table S1, were obtained from Integrated DNA Technologies (Coralville, IA). DNA amplification by polymerase chain reaction (PCR) was conducted in a PTC-100 Peltier Thermal Cycler (MJ Research, Waltham, MA), using either the AccuPrime Pfx system (Invitrogen, Carlsbad, CA) or the Expand Long Range dNTPack (Roche Applied Science, Indianapolis). PCR products, when necessary, were cloned into the pCRII-TOPO vector (Invitrogen). Bacterial plasmid DNA was purified with the HiSpeed Plasmid Midi Kit (Qiagen). DNA sequencing was performed by the University of Missouri DNA core (Columbia, MO). For integration at the his-3 locus, the qipfs allele and various green and red fluorescent protein (GFP and RFP, respectively) constructs were built using pBM61 (Margolin et al. 1997), pMF272 (Freitag et al. 2004), and pMF334 (Freitag and Selker 2005), respectively. For GFP integration at native loci, fusion PCR products from genomic DNA and a gfp-hph-containing plasmid (pTH1067.9) were obtained by double-joint PCR (Yang et al. 2004; Yu et al. 2004) and were used as the transforming DNA. DNA-mediated gene placement in Neurospora was performed according to Margolin et al. (1997).
Reverse-transcriptase PCR:
Total RNA extraction was performed as previously described (Shiu and Glass 1999). Poly(A+) mRNA was enriched using the Oligotex mRNA kit (Qiagen). Reverse transcription, using a first-strand cDNA synthesis kit (Amersham Biosciences, Piscataway, NJ), was conducted according to the manufacturer's specifications. Primers used in the PCR amplification of a region spanning four qip introns are listed in Table S1. The PCR product of the qip cDNA is 1414 bp in length, as compared to the 1666 bp of that of genomic DNA. The identities of reverse transcriptase PCR (RT–PCR) products, from vegetative (P3-07) and perithecial (F1-05 × P3-07) mRNA, were confirmed by DNA sequencing. Our intron 2 sequence (Figure S1), which is based on cDNA sequencing, is 96 nucleotides shorter than the sequence depicted in version 3 of the qip predicted open reading frame and is in agreement with the sequence found in version 4 (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
Sample preparation and cytological methods:
Perithecia fixation, mounting, and viewing using the Zeiss LSM510 were as described (Alexander et al. 2008). Some samples were imaged using a Zeiss LSM710 confocal laser scanning microscope, equipped with a PlanNeofluar ×40 (NA1.3) oil immersion objective and standard Zeiss software (ZEN). Multi-fluorophore images were scanned sequentially. Visualization of the GFP was achieved by use of a 488-nm Argon laser line for excitation with the detector set to collect emission bandwidth at 494–536 nm; RFP visualization was achieved by use of a 560-nm diode laser line for excitation with the detector set to collect emission bandwidth at 565–620 nm; and DAPI visualization was achieved by use of a 405-nm diode laser excitation with the detector set to collect emission bandwidth at 410–470 nm.
RESULTS
qip is expressed during both vegetative and sexual stages:
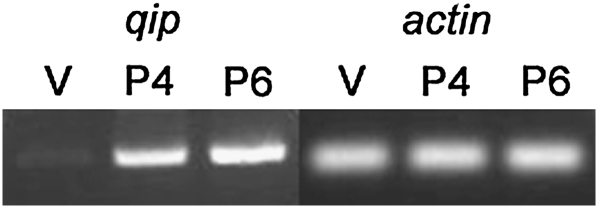
qip encodes a 600-amino-acid polypeptide and is located between mus-52 and tim14, on the right arm of linkage group III (Figure S1). To determine the expression pattern of qip, we obtained total RNA from mycelia (vegetative cells) and two perithecial (fruiting body) preparations (4 and 6 days after fertilization). cDNA products of qip, whose identities were confirmed by sequencing, could be detected from all conditions tested (Figure 1). The expression of qip in the perithecial tissue suggests that qip may play a role during sexual development.
Figure 1.—
qip is expressed in both asexual and sexual tissues. RT–PCR products for qip and actin (control) are shown (1414 and 227 bp, respectively). RNA from vegetative (V) and perithecial (P4 and P6: perithecial at 4 and 6 days) preparations were used for the amplification reactions.
A cross homozygous for qipΔ is barren:
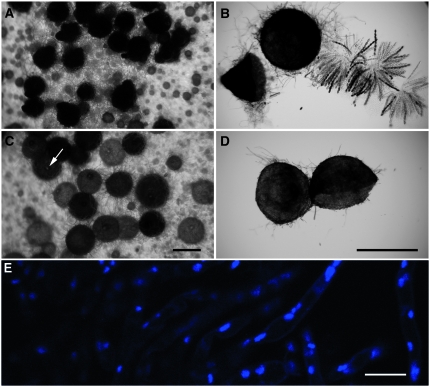
Maiti et al. (2007) did not report any vegetative defect other than a Quelling deficiency in a qip strain. To determine if qip is important for sexual development, we performed crosses heterozygous and homozygous for qipΔ. While ascospores are produced in a qip+ × qipΔ cross, a cross homozygous for qipΔ is completely barren. Although pigmented perithecia are produced in such a cross, they have just a hint of a beak (Figure 2C). Perithecial contents showed no asci, not even their rudiments (Figure 2D). DAPI staining showed only fluorescent nuclei in the background paraphysal tissue (Figure 2E). Perithecial development was apparently arrested very early. Crosses homozygous for dcl-1Δ have a similar phenotype (Alexander et al. 2008), suggesting that both of these genes are important for early sexual development.
Figure 2.—
Perithecial examination of various crosses demonstrates the requirement of qip in early sexual development. (A and B) qip+ × qip+ (F2-01 × P3-08). Normal perithecia and rosettes of eight-spored asci can be seen from the control cross. (C and D) qipΔ × qipΔ (F2-06 × P9-39). Undersized beaks (arrow in C) and the absence of asci (perithecial cross section in D) in a cross homozygous for qipΔ. Bars, 500 μm. (E) Only paraphysal tissue is found in perithecia from a qipΔ × qipΔ (F2-06 × P9-39) cross, suggesting a severe defect in perithecial development in a qip-null background. DAPI stain was used. Bar, 10 μm.
qipΔ does not act as a dominant suppressor of meiotic silencing:
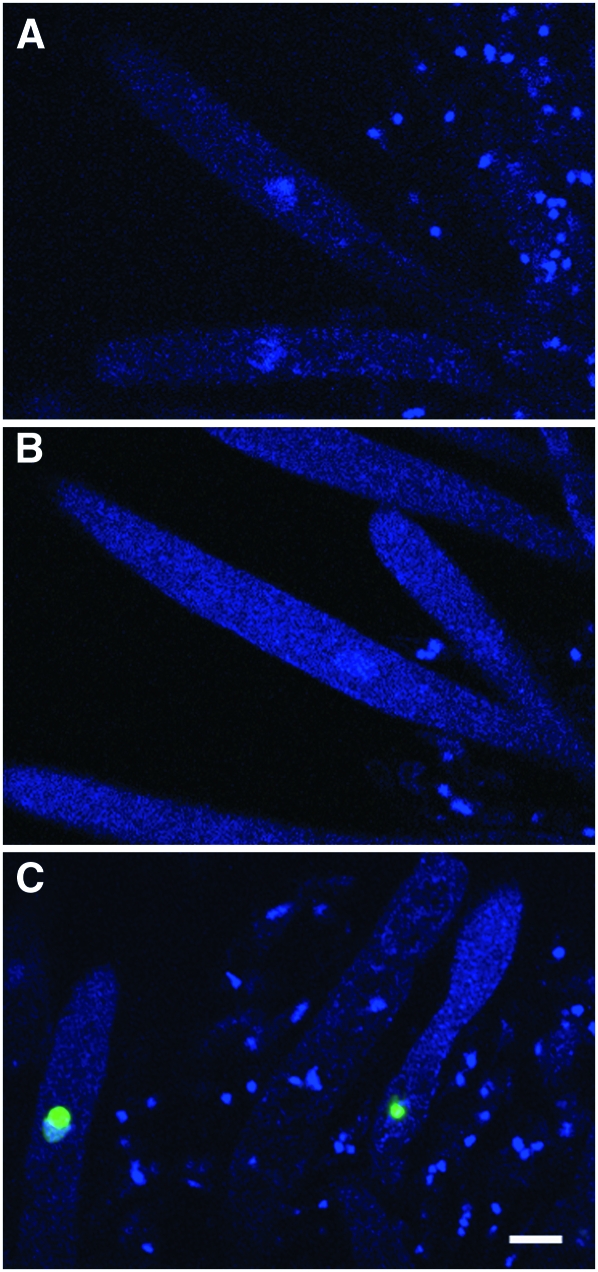
QIP is important for the degradation of the passenger siRNA strand in Quelling (Maiti et al. 2007). While there are two paralogs for both RdRP and Argonaute in the N. crassa genome (one set for Quelling and another for MSUD), only one qip gene is present. These observations suggest either that MSUD utilizes a different method of passenger strand removal or that QIP is important for both vegetative and meiotic silencing, as is the case for DCL-1 (Alexander et al. 2008). Many deletion mutants of genes encoding components of the MSUD machinery, such as Sad-1Δ and Sad-2Δ (Shiu et al. 2001, 2006), act as a dominant suppressor of meiotic silencing in a cross. For example, the unpaired r+ (Round spore) gene is silenced in a wild type × RΔ cross (which gives round spores) while it is expressed in a Sad-1Δ × RΔ cross (which gives wild-type spindle-shaped spores resembling an American football) (Shiu et al. 2001). The logic behind the dominant suppression in a Sad-1Δ × RΔ cross is that the sad-1+ gene itself is unpaired, allowing the silencer to silence itself and thereby defeating the silencing mechanism (Shiu and Metzenberg 2002). To determine whether qip suppresses MSUD in a dominant fashion, we introduced qipΔ to crosses containing various unpaired genes, including actin, ascospore maturation-1, β-tubulin, and Round spore. These unpaired genes, in an MSUD-proficient background, lead to various aberrant ascus/ascospore phenotypes (lollipop asci, white ascospores, elongated asci, and round ascospores, respectively). Our data indicate that the presence of a single qipΔ allele does not suppress the meiotic silencing of any unpaired gene tested (Table 2). We reached the same conclusion with a visual gfp expression assay using a histone hH1-gfp reporter gene (Raju et al. 2007; Alexander et al. 2008). In a qipΔ × ∷hH1-gfp cross, the unpaired hH1-gfp gene is silenced as usual and does not give rise to fluorescent nuclei in developing asci (Figure 3, A and B). Taken together, these results indicate that qipΔ, unlike Sad-1Δ or Sad-2Δ, does not act as a dominant MSUD suppressor in a cross.
TABLE 2.
qipΔ, unlike Sad-1Δ, does not act as a dominant suppressor of MSUD
| Unpaired gene | Parent 1 (mat A): ectopic insertion at his-3 (∷) and/or deletion (Δ) | Parent 2 (mata): mutation at sad-1 or qip | Ascospore count/phenotype | Predominant ascus phenotype |
|---|---|---|---|---|
| actin | ∷act+ | Wild type | 208 × 103 | Lollipop asci |
| ∷act+ | qipΔ | 320 × 103 | Lollipop asci | |
| ∷act+ | Sad-1Δ | 2408 × 103 | Normal | |
| Ascospore maturation-1 | ∷asm-1+; Asm-1Δ | Wild type | 1.6% black | White ascospores |
| ∷asm-1+; Asm-1Δ | qipΔ | 1.4% black | White ascospores | |
| ∷asm-1+; Asm-1Δ | Sad-1Δ | 86% black | Black ascospores | |
| β-tubulin | ∷BmlR | Wild type | 2.9 × 103 | Arrests before metaphase |
| ∷BmlR | qipΔ | 2.4 × 103 | Arrests before metaphase | |
| ∷BmlR | Sad-1Δ | 4008 × 103 | Normal | |
| Round spore | RΔ | Wild type | 0% football | Round ascospores |
| RΔ | qipΔ | 0% football | Round ascospores | |
| RΔ | Sad-1Δ | 100% football | Football-shaped ascospores |
Meiotic silencing of meiotically important genes, such as act+, asm-1+, BmlR, and r+, leads to the reduced production of black (mature), American football (spindle)-shaped ascospores. Crosses carrying Sad-1Δ, not qipΔ, can improve the production of normal ascospores. Strains used in this experiment include F2-29, F2-35, F2-36, F3-23, P3-08, P3-25, and P9-39.
Figure 3.—
Meiotic silencing of the unpaired histone H1-gfp (his-3+∷hH1-gfp) gene is suppressed in a QIP-deficient cross. (A) qip+ × ∷hH1-gfp (P6-07 × P10-16). Since the unpaired hH1-gfp gene is silenced during meiosis in a developing ascus, no fluorescence is observed. (B) qipΔ × ∷hH1-gfp (P12-28 × P10-16). qipΔ does not dominantly suppress the meiotic silencing of unpaired hH1-gfp. The nuclei remain nonfluorescent. (C) ∷qipfs × ∷hH1-gfp qipΔ (P11-38 × P12-13). Silencing of the unpaired hH1-gfp gene is suppressed in a qip knockdown cross, resulting in the presence of nuclear fluorescence. Bar, 10 μm.
The qip gene product is required for meiotic silencing:
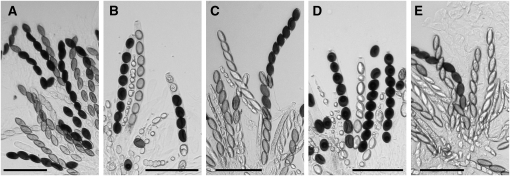
The fact that a qipΔ mutant does not dominantly suppress MSUD suggests either that qip is not involved in the meiotic silencing pathway or that one unpaired copy of the qip gene is not sufficient to silence the silencer. Since qip is required for early sexual development, we cannot examine the expression of unpaired genes in a qipΔ × qipΔ cross (which is completely barren) and unequivocally determine the role of qip in meiotic silencing. To circumvent this technical problem, we utilized a “two unpaired copy knockdown” scheme that was proven successful previously (Alexander et al. 2008). Basically, we constructed a cross heterozygous for qipΔ, heterozygous for an insertion of qipfs (a frameshift null allele), and heterozygous for an insertion of hH1-gfp (an unpaired reporter gene), i.e., his-3+∷qipfs × his-3+∷hH1-gfp qipΔ. In this cross, the presence of a single qip+ gene allows the perithecia to go through early ascus development. However, the single wild-type qip gene is inactivated by meiotic silencing at later stages due to two unpairing events (qip+ unpaired with qipΔ∷hph at the native qip locus and qipfs unpaired with hH1-gfp at the his-3 locus). Results from our cytological examination indicate that the unpaired hH1-gfp reporter gene is expressed throughout meiosis, suggesting that meiotic silencing is indeed deficient in a low QIP background (Figure 3C). We repeated the experiment using r+ as the reporter gene (in a his-3+∷qipfs × his-3+∷r+ qipΔ cross). Our results indicate that the unpaired r+ gene is expressed and that the progeny are of wild-type spindle-shape in the qip knockdown cross (Figure 4). These results indicate that QIP is a necessary component of the MSUD machinery.
Figure 4.—
Meiotic silencing of the unpaired Round spore (r+) gene is suppressed in a QIP-deficient cross. (A) r+ × r+ (P3-07 × P3-08). Normal spindle-shaped (American football-like) ascospores are observed when the r+ gene is expressed. (B) r+ × ∷r+ (P6-08 × P11-36). Meiotic silencing of the r+ gene leads to the production of round ascospores. (C) Sad-1Δ × ∷r+ (P5-52 × P11-36). Sad-1Δ suppresses the meiotic silencing of r+, resulting in the presence of spindle-shaped ascospores. (D) qipΔ × ∷r+ (P11-21 × P11-36). qipΔ does not act as a dominant suppressor of MSUD, resulting in predominantly round ascospores. (E) ∷qipfs × ∷r+ qipΔ (P11-38 × P11-41). The silencing of the unpaired r+ gene is suppressed in a qip knockdown cross, resulting in the presence of spindle-shaped ascospores. Bars, 100 μm.
qip is localized in the perinuclear region:
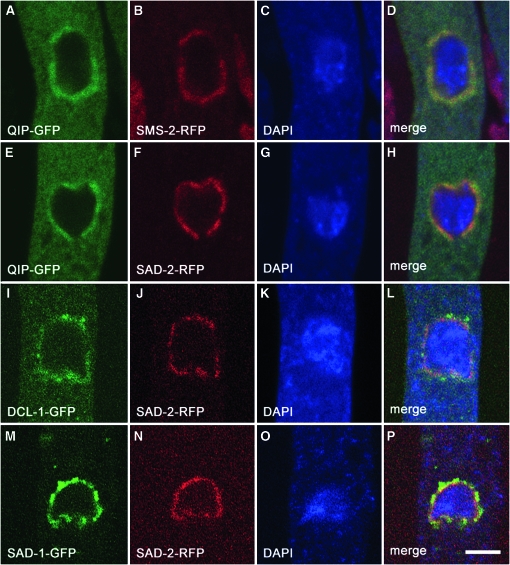
QIP interacts with the QDE-2 Argonaute during the Quelling process (Maiti et al. 2007). If Quelling and MSUD function in a similar manner, at least for the RNA-degradation portion of the pathway, one would expect QIP also to associate with the MSUD Argonaute protein (SMS-2). Previously, we have shown that components of the MSUD machinery, including SAD-1, SAD-2, DCL-1, and SMS-2, are localized in the perinuclear region (Alexander et al. 2008). To determine the subcellular localization of QIP as well as its possible association with other MSUD proteins, we have constructed vectors expressing various green and red fluorescent fusion proteins. Our results indicate that QIP colocalizes with the other MSUD proteins, including the SMS-2 Argonaute, in the perinuclear region (Figure 5). The colocalization pattern is in agreement with the notion that these MSUD proteins are related functionally and spatially and that they may form an RNA-processing complex.
Figure 5.—
Colocalization of MSUD proteins in the perinuclear region. Micrographs illustrate prophase asci expressing (A–D) qip-gfp and sms-2-rfp (P15-02 × P15-03). (E–H) qip-gfp and sad-2-rfp (P14-05 × P6-62). (I–L) dcl-1-gfp and sad-2-rfp (P10-18 × P6-62). (M–P) sad-1-gfp and sad-2-rfp (P13-15 × P6-62). The chromatin was stained with DAPI. Bar, 5 μm.
DISCUSSION
The vegetative silencing machinery in N. crassa is important for the preservation of genome integrity (Chicas et al. 2004; Nolan et al. 2005), the maintenance of ribosomal DNA copies (Cecere and Cogoni 2009), and DNA damage response (Lee et al. 2009). The QIP exonuclease, first identified as a QDE-2-interacting protein, functions to degrade the passenger strand of an siRNA duplex and to activate the RISC during Quelling (Maiti et al. 2007). QIP is of special importance to the delineation of the RNA interference (RNAi) pathway, as the mechanism for passenger strand removal was not obvious before its identification. In this work, we have demonstrated that QIP is also required for MSUD. Thus far, we have shown that at least two proteins, DCL-1 and QIP, have dual functions in the genome surveillance of N. crassa. These results suggest that the crosstalk between the vegetative and meiotic silencing mechanisms is more prevalent than once thought. All the MSUD proteins reported previously, including SAD-1, SAD-2, SMS-2, and DCL-1, are important for sexual development (Shiu et al. 2001, 2006; Lee et al. 2003; Alexander et al. 2008). Homozygous crosses for sad-1 or sad-2 are arrested in prophase, suggesting that some degree of meiotic silencing may be a required checkpoint for cell cycle progression (Shiu et al. 2001, 2006). Unlike sad-1 and sad-2, a cross homozygous for qipΔ does not produce any asci. This observation suggests that qip is important for early ascus development, much like dcl-1 (Alexander et al. 2008). Since QIP and Dicers are essential to the biogenesis of certain microRNA-like RNA (Lee et al. 2010), the lack of these proteins may affect the expression of genes that regulate sexual development. Alternatively, they could regulate endogenous genes that are naturally transcribed in both sense and antisense orientations (Fulci and Macino 2007).
All known components of the MSUD machinery (SAD-1, SAD-2, SMS-2, DCL-1, and QIP), with the exception of Sk-2 and Sk-3 (which have not been molecularly characterized; Raju et al. 2007), are localized in the perinuclear region (Shiu et al. 2001; Alexander et al. 2008). This observation is in contrast with the one made in the Quelling mechanism, in which QDE-1 (and hypothetically, QDE-3) has affinity for repetitive transgenic loci (Nolan et al. 2008). In mammalian cells, siRNA have been shown to accumulate in the perinuclear region, and their proper localization is correlated with the efficiency of RNAi (Grünweller et al. 2003; Chiu et al. 2004). Furthermore, some RNAi proteins have been shown to localize in this region in Drosophila and mouse germ cells (Kotaja and Sassone-Corsi 2007; Lim and Kai 2007; Pane et al. 2007). These observations suggest that the perinuclear region may be an RNAi center for meiotic silencing. It is possible the MSUD machinery examines each RNA molecule as it exits the nucleus, processing any aberrant RNA before it has a chance to reach the exonucleases or the translational machinery. Colocalization of related MSUD proteins may allow the coupling of consecutive reactions and therefore increase the efficiency of the silencing process.
Meiotic silencing can be a useful tool in determining gene functions during meiosis and sexual development. A wide variety of genes, including those encoding actin and β-tubulin, have been silenced using MSUD (Shiu et al. 2001). Although some MSUD mutants, such as Sad-1Δ, Sad-2Δ, Sk-2, and Sk-3, behave as strong dominant MSUD suppressors via the “silencing the silencer” negative feedback system (Shiu et al. 2001, 2006; Raju et al. 2007), others may have difficulties in achieving similar effectiveness. Our use of two unpaired copies in a cross may prove to be useful in silencing genes that are especially hard to silence via the standard “wild type × Δ” scheme, such as those that are highly expressed or those that need few transcripts for normal operation.
The silencing of unpaired chromosomal regions during meiosis is not restricted to fungi. Some form of meiotic silencing is also found in worms, mice, and humans (Bean et al. 2004; Turner et al. 2005; Ferguson et al. 2008). In mammals, the phenomenon is known as meiotic silencing of unsynapsed chromatin, and it is responsible for meiotic sex chromosome inactivation (Burgoyne et al. 2009). Further identification of the genetic factors controlling these phenomena should shed light on the mechanisms involved in targeting unpaired DNA and on whatever similarity or difference there might be among various eukaryotes.
Acknowledgments
The authors thank Namboori Raju for the cytological examination of some of the crosses, the members of the Shiu Laboratory for their technical assistance, and Michael Freitag and Eric Selker for various plasmids. T.M.H. was supported by a University of Missouri Life Sciences Fellowship and a National Institute of General Medical Sciences fellowship. W.G.A. was supported by a Graduate Assistance in Areas of National Need fellowship. This work was supported by National Science Foundation grants EF0412016 (to P.J.P.) and MCB0544237/0918937 (to P.K.T.S.).
Note added in proof: See Lee et al. in this issue (pp. 127–133) for a related work.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.118273/DC1.
References
- Alexander, W. G., N. B. Raju, H. Xiao, T. M. Hammond, T. D. Perdue et al., 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45 719–727. [DOI] [PubMed] [Google Scholar]
- Bean, C. J., C. E. Schaner and W. G. Kelly, 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne, P. S., S. K. Mahadevaiah and J. M. A. Turner, 2009. The consequences of asynapsis for mammalian meiosis. Nat. Rev. Genet. 10 207–216. [DOI] [PubMed] [Google Scholar]
- Cambareri, E. B., B. C. Jensen, E. Schabtach and E. U. Selker, 1989. Repeat-induced G-C to A-T mutations in Neurospora. Science 244 1571–1575. [DOI] [PubMed] [Google Scholar]
- Catalanotto, C., G. Azzalin, G. Macino and C. Cogoni, 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., M. Pallotta, P. Refalo, M. S. Sachs, L. Vayssie et al., 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., T. Nolan and C. Cogoni, 2006. Homology effects in Neurospora crassa. FEMS Microbiol. Lett. 254 182–189. [DOI] [PubMed] [Google Scholar]
- Cecere, G., and C. Cogoni, 2009. Quelling targets the rDNA locus and functions in rDNA copy number control. BMC Microbiol. 9 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas, A., C. Cogoni and G. Macino, 2004. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 32 4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y. L., A. Ali, C. Y. Chu, H. Cao and T. M. Rana, 2004. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem. Biol. 11 1165–1175. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. a Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. b Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286 2342–2344. [DOI] [PubMed] [Google Scholar]
- Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew et al., 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. H., and F. J. de Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17A 79–143. [Google Scholar]
- Ebbole, D., and M. S. Sachs, 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37 17–18. [Google Scholar]
- Ferguson, K. A., V. Chow and S. Ma, 2008. Silencing of unpaired meiotic chromosomes and altered recombination patterns in an azoospermic carrier of a t(8;13) reciprocal translocation. Hum. Reprod. 23 988–995. [DOI] [PubMed] [Google Scholar]
- Freitag, M., and E. U. Selker, 2005. Expression and visualization of red fluorescent protein (RFP) in Neurospora crassa. Fungal Genet. Newsl. 52 14–17. [Google Scholar]
- Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker and N. D. Read, 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41 897–910. [DOI] [PubMed] [Google Scholar]
- Fulci, V., and G. Macino, 2007. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr. Opin. Microbiol. 10 199–203. [DOI] [PubMed] [Google Scholar]
- Glass, N. L., D. J. Jacobson and P. K. T. Shiu, 2000. The genetics of hyphal fusion and vegetative incompatibility in filamentous ascomycete fungi. Annu. Rev. Genet. 34 165–186. [DOI] [PubMed] [Google Scholar]
- Grünweller, A., C. Gillen, V. A. Erdmann and J. Kurreck, 2003. Cellular uptake and localization of a Cy3-labeled siRNA specific for the serine/threonine kinase Pim-1. Oligonucleotides 13 345–352. [DOI] [PubMed] [Google Scholar]
- Kotaja, N., and P. Sassone-Corsi, 2007. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 8 85–90. [DOI] [PubMed] [Google Scholar]
- Lee, D. W., R. J. Pratt, M. McLaughlin and R. Aramayo, 2003. An argonaute-like protein is required for meiotic silencing. Genetics 164 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. C., S. S. Chang, S. Choudhary, A. P. Aalto, M. Maiti et al., 2009. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature 459 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. W., R. Millimaki and R. Aramayo, 2010. QIP, a component of the vegetative RNA silencing pathway, is essential for meiosis and suppresses meiotic silencing in Neurospora crassa. Genetics 186 127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. C., L. Li, W. Gu, Z. Xue, S. K. Crosthwaite et al., 2010. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol. Cell 38 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, A. K., and T. Kai, 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104 6714–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, M., H. C. Lee and Y. Liu, 2007. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 21 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, B. S., M. Freitag and E. U. Selker, 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44 34–36. [Google Scholar]
- McCluskey, K., 2003. The Fungal Genetics Stock Center: from molds to molecules. Adv. Appl. Microbiol. 52 245–262. [DOI] [PubMed] [Google Scholar]
- Nolan, T., L. Braccini, G. Azzalin, A. De Toni, G. Macino et al., 2005. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 33 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, T., G. Cecere, C. Mancone, T. Alonzi, M. Tripodi et al., 2008. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 36 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane, A., K. Wehr and T. Schüpbach, 2007. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 12 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, D. D., A. Radford and M. S. Sachs, 2001. The Neurospora Compendium: Chromosomal Loci. Academic Press, San Diego.
- Raju, N. B., R. L. Metzenberg and P. K. T. Shiu, 2007. Neurospora spore killers Sk-2 and Sk-3 suppress meiotic silencing by unpaired DNA. Genetics 176 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, N., and G. Macino, 1992. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6 3343–3353. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shiu, P. K. T., and N. L. Glass, 1999. Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa. Genetics 151 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K. T., and N. L. Glass, 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr. Opin. Microbiol. 3 183–188. [DOI] [PubMed] [Google Scholar]
- Shiu, P. K. T., and R. L. Metzenberg, 2002. Meiotic silencing by unpaired DNA: properties, regulation, and suppression. Genetics 161 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K. T., N. B. Raju, D. Zickler and R. L. Metzenberg, 2001. Meiotic silencing by unpaired DNA. Cell 107 905–916. [DOI] [PubMed] [Google Scholar]
- Shiu, P. K. T., D. Zickler, N. B. Raju, G. Ruprich-Robert and R. L. Metzenberg, 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 103 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J. M. A., S. K. Mahadevaiah, O. Fernandez-Capetillo, A. Nussenzweig, X. Xu et al., 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37 41–47. [DOI] [PubMed] [Google Scholar]
- Vogel, H. J., 1964. Distribution of lysine pathways among fungi: evolutionary implications. Am. Nat. 98 435–446. [Google Scholar]
- Westergaard, M., and H. K. Mitchell, 1947. Neurospora V: a synthetic medium favoring sexual reproduction. Am. J. Bot. 34 573–577. [Google Scholar]
- Yang, L., L. Ukil, A. Osmani, F. Nahm, J. Davies et al., 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. H., Z. Hamari, K. H. Han, J. A. Seo, Y. Reyes-Domínguez et al., 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41 973–981. [DOI] [PubMed] [Google Scholar]