Abstract
The patterning of initiating organs along specific axes of polarity is critical for the proper development of all higher organisms. Plant lateral organs, such as leaves, are derived from the shoot apical meristems located at the growing tips. After initiation, the leaf primordia of species such as Arabidopsis thaliana differentiate into a polarized structure consisting of a proximal petiole and a distal blade, but the molecular mechanisms that control proximal–distal pattern formation are poorly understood. The transcriptional activators BLADE-ON-PETIOLE1 (BOP1) and BOP2 are known to control Arabidopsis lateral organ differentiation by regulating gene expression along the adaxial–abaxial (dorsal–ventral) and proximal–distal polarity axes. Here, we demonstrate that the development of ectopic blade tissue along bop1 bop2 leaf petioles is strongly suppressed in a dosage-dependant manner by mutations in either of two closely related YABBY (YAB) genes, FILAMENTOUS FLOWER (FIL) and YAB3. Three KNOTTED-LIKE HOMEOBOX (KNOX1) genes also make lesser, and partially redundant, contributions to ectopic blade development in bop1 bop2 leaves. Mutation of these YAB and KNOX1 genes together causes nearly complete suppression of bop1 bop2 ectopic organ outgrowth at the morphological and cellular levels. Our data demonstrate that BOP1 and BOP2 regulate leaf patterning by controlling YAB and KNOX1 gene activity in the developing petiole.
IN higher plants, lateral organs initiate from the flanks of the shoot apical meristem (SAM), which acts as a pluripotent stem cell reservoir throughout the plant life span. Through molecular and genetic studies in model plants such as Arabidopsis and maize, the regulatory mechanisms that control the organ differentiation and patterning process are beginning to be understood. Still, much remains to be learned about the pathways that distinguish lateral organ identity from SAM identity and how these pathways intersect with each other during morphogenesis.
The four related Arabidopsis class 1 KNOTTED-LIKE HOMEOBOX (KNOX1) genes SHOOTMERISTEMLESS (STM), BREVIPEDICELLUS (BP), KNOTTED-like from Arabidopsis thaliana2 (KNAT2), and KNAT6 encode homeodomain transcription factors with overlapping expression and function (Hake et al. 2004). STM is expressed throughout the SAM, and loss-of-function stm mutations result in loss of the SAM (Barton and Poethig 1993). BP has slightly different expression pattern from STM in the SAM and is required for stem and internode morphogenesis (Mele et al. 2003), yet can act redundantly with STM in certain genetic backgrounds (Byrne et al. 2002). KNAT2 plays a role in carpel development (Pautot et al. 2001), whereas KNAT6, which is most similar to KNAT2, acts in root development (Dean et al. 2004). KNAT6, but not KNAT2, functions redundantly with STM in embryonic SAM maintenance and boundary establishment (Belles-Boix et al. 2006). During inflorescence growth, BP restricts KNAT2 and KNAT6 expression to achieve normal shoot morphogenesis, and KNAT2 and KNAT6 reveal their functional redundancy during inflorescence and pedicel development in the absence of BP (Ragni et al. 2008).
In simple-leaved species such as Arabidopsis, ectopic KNOX1 expression causes lobed and rumpled leaf formation (Sinha et al. 1993; Lincoln et al. 1994; Nishimura et al. 2000; Pautot et al. 2001; Dean et al. 2004; Cole et al. 2006), indicating that continuous suppression of KNOX1 activity is required for proper organ differentiation. KNOX1 repression during Arabidopsis organ development is mediated by ASYMMETRIC LEAVES1 (AS1), AS2, BLADE-ON-PETIOLE1 (BOP1), and BOP2 (Byrne et al. 2000; Ori et al. 2000; Semiarti et al. 2001; Ha et al. 2003; Hepworth et al. 2005; Norberg et al. 2005; Ha et al. 2007). AS1 encodes a myb-domain transcription factor that forms a protein complex with the LATERAL-ORGAN-BOUNDARIES (LBD) domain-containing protein AS2 (Byrne et al. 2000; Xu et al. 2003). The BOP1 and BOP2 genes encode BTB/POZ domain and ankyrin repeat-containing proteins (Ha et al. 2004; Hepworth et al. 2005; Norberg et al. 2005). All of these genes are expressed in leaf primordia and when mutated condition a range of leaf developmental phenotypes similar to those of KNOX1 overexpressing plants (Byrne et al. 2000; Ori et al. 2000; Semiarti et al. 2001; Ha et al. 2003; Hepworth et al. 2005; Norberg et al. 2005).
KNOX1 expression is also controlled by members of the YABBY (YAB) family of putative transcription factor genes. KNOX1 transcription is upregulated in the leaves of plants carrying loss-of-function mutations in the YAB genes FILAMENTOUS FLOWER (FIL) and YAB3 (Kumaran et al. 2002), and fil yab3 plants display ectopic shoot development on their blades similar to KNOX1 overexpressing plants (Siegfried et al. 1999; Kumaran et al. 2002). FIL and YAB3 are expressed in the abaxial domain of developing lateral organs and are implicated in the specification of abaxial organ identity (Eshed et al. 1999; Sawa et al. 1999; Siegfried et al. 1999). In addition, YAB activity is required for leaf lamina expansion (Eshed et al. 2004).
Here we show that the bop1 bop2 leaf ectopic outgrowth phenotypes are gradually suppressed by the addition of single, double, and triple bp, knat2, and knat6 allele combinations, revealing functional redundancy of these KNOX1 genes during bop1 bop2 leaf formation. We also find that bop1 bop2 ectopic outgrowth formation requires the activity of the YAB genes FIL and YAB3, with YAB3 playing a more important role than FIL. Furthermore, residual ectopic outgrowth in bop1 bop2 fil yab3 leaves was almost completely abolished by the introduction of the bp, knat2, and knat6 alleles. These results show that KNOX1 genes and also YAB genes contribute to bop1 bop2 ectopic organ outgrowth formation. We conclude that BOP1 and BOP2 play a key role in organ morphogenesis by suppressing both KNOX1 and YAB activity at the leaf base, thereby maintaining the proper cellular environment for normal leaf differentiation.
MATERIALS AND METHODS
Plant materials and genetics:
A. thaliana plants were grown as described (Ha et al. 2004). All mutant alleles were in the Landsberg erecta (Ler) accession except for yab3-1 (Wassilewskija), bop1-4 bop2-11, and knat6-5, which were introgressed from Columbia-0 into Ler three times before analysis. knat2-1 (GT 7953) seeds were obtained from the Cold Spring Harbor Laboratory and knat6-5 (SALK_149322) seeds from the Arabidopsis Biological Resource Center. fil-8 and yab3-2 seeds were kindly provided by Venkatesan Sundaresan (University of California, Davis, CA), fil-5 and yab3-1 seeds by John Bowman (Monash University, Australia), and BP∷GUS seeds by Sarah Hake (University of California, Berkeley, CA).
bp-1 knat2 and bp-1 knat6 plants were generated by crossing bp-1 with knat2-1 or knat6-5, respectively. bp-1 F2 plants were then genotyped for the knat2-1 or knat6-5 allele, respectively. To generate bp-1 knat2-1 knat6-5 plants, F1 plants from a cross between bp-1 knat2-1 and bp-1 knat6-5 were self-pollinated. bp-1 F2 plants were grown on plates containing kanamycin to select for the knat2-1 allele and were genotyped for the bp-1, knat2-1, and knat6-5 alleles. To generate bop1-4 bop2-11 bp-1 knat2-1 knat6-5 plants, F1 plants from a cross between bop1-4 bop2-11 bp-1 knat2-1 and bop1-4 bop2-11 knat2-1 knat6-5 were self-fertilized and the F2 plants genotyped for the bp-1, knat2-1, and knat6-5 alleles. bop1-4 bop2-11 fil-8 and bop1-4 bop2-11 yab3-2 plants were generated by crossing bop1-4 bop2-11 with fil-8 or yab3-2, respectively. bop1-4 bop2-11 F2 plants were then genotyped for the fil-8 and yab3-2 alleles. bop1-4 bop2-11 fil-8 yab3-2 plants were generated by crossing bop1-4 bop2-11 fil-8 plants with bop1-4 bop2-11 yab3-2 plants. bop1-4 bop2-11 F2 plants were genotyped for the fil-8 and yab3-2 alleles. Phenotypic segregation of fil-8 yab3-2 plants in a 1:3 ratio was observed in the F3 generation. To generate bp-1 knat2-1 knat6-5 fil-8 yab3-2 plants, F1 plants from a cross between bp-1 knat2-1 knat6-5 and fil-8 yab3-2 were self-pollinated. bp-1 F2 plants were then genotyped for the knat2-1, knat6-5, fil-8, and yab3-2 alleles. Phenotypic segregation of fil-8 yab3-2 plants in a 1:3 ratio was observed in the F3 generation. To generate bop1-4 bop2-11 bp-1 knat2-1 knat6-5 fil-8 yab3-2 plants, bop1-4 bop2-11 bp-1 knat2-1 knat6-5 plants were crossed to bp-1 knat2-1 knat6-5 fil-8 yab3-2 plants. bop1-4 bop2-11 F2 plants were then genotyped for the fil-8 and yab3-2 alleles. Phenotypic segregation of fil-8 yab3-2 plants in a 1:3 ratio was observed in the F3 generation. Primer sequences for genotyping are listed in supporting information, Table S1.
Histological analysis:
Tissue preparation and sectioning were carried out as described (Ha et al. 2003).
Expression analysis:
RNA was isolated from developing young leaves of 21-day-old plants. Quantification by real-time PCR was performed using the SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) and the MyIQ system (Bio-Rad, Hercules, CA). Primer sequences for mRNA amplification are listed in Table S1. Real-time PCR experiments were repeated using three biological replicates.
GUS analysis:
Thirteen-day-old seedlings were fixed in 90% acetone at −20° for 30 min and washed briefly with 100 mm phosphate buffer. Samples were vacuum infiltrated for 10 min in fresh 100 mm sodium phosphate buffer (pH 7.0) containing 0.5 mm potassium ferrocyanide (Sigma, St. Louis), 0.5 mm potassium ferricyanide (Sigma), 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (Sigma), 1% DMSO, 1% triton X-100, and 10 mm EDTA, and incubated at 37° for 4 hr. The reactions were terminated with 50% ethanol and the samples cleared in a graded ethanol series to 100% ethanol. Cleared samples in 50% ethanol were fixed in FAA solution (50% ethanol, 5% glacial acetic acid, and 3.7% formaldehyde), vacuum infiltrated for 10 min, and mounted in 50% glycerol. Leaves were dissected and viewed using dark-field microscopy.
RESULTS
KNOX1 gene activity conditions the bop1 bop2 leaf phenotype:
Wild-type Arabidopsis rosette leaves are characterized by an ovate blade attached to the main plant stem by a narrow petiole (Figure 1A). bop1 bop2 rosette leaves form extensive outgrowths of blade tissue along the petioles (Figure 1B), indicating that BOP1 and BOP2 are required for proper leaf morphogenesis (Ha et al. 2003, 2004). Misexpression of the KNOX1 genes BP, KNAT2, or KNAT6 in leaves causes lobed and dissected leaf formation, and in extreme cases ectopic shoot initiation on the leaf (Sinha et al. 1993; Lincoln et al. 1994; Chuck et al. 1996; Pautot et al. 2001; Dean et al. 2004; Cole et al. 2006). The phenotypes of KNOX1 misexpressing plants are reminiscent of the bop phenotypes, and indeed KNOX1 genes are misexpressed in bop1 bop2 mutants (Ha et al. 2007). Therefore we used genetic analysis to investigate whether the bop leaf phenotype is caused by KNOX1 gene misexpression during organogenesis.
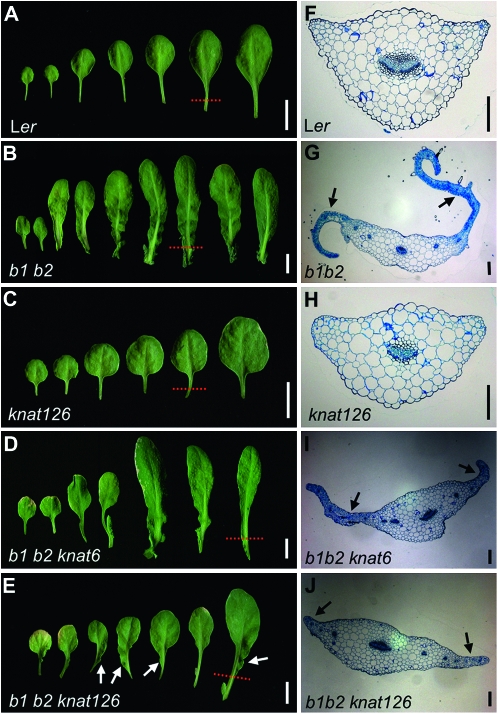
Figure 1.—
Genetic interactions between bop1 bop2 and knox1 alleles. Photographs of dissected rosettes from 38-day-old (A) wild-type Ler, (B) bop1-4 bop2-11 (b1 b2), (C) bp-1 knat2-1 knat6-5 (knat126), (D) bop1-4 bop2-11 knat6-5, and (E) bop1-4 bop2-11 bp-1 knat2-1 knat6-5 plants. Transverse sections of (F) Ler, (G) bop1 bop2, (H) bp-1 knat2-1 knat6-5, (I) bop1-4 bop2-11 knat6-5, and (J) bop1-4 bop2-11 bp-1 knat2-1 knat6-5 leaf petioles. Dotted lines in A–E indicate the sectioned regions in F–J. Bars, 10 mm (for A–E) and 200 μm (for F–J).
We first obtained loss-of-function alleles of BP, KNAT2, and KNAT6 and examined their phenotypes. We used the bp-1 and knat2-1 alleles that had previously been shown to be null alleles (Byrne et al. 2002; Venglat et al. 2002). We were unable to detect KNAT6 transcripts from homozygous knat6-5 plants, indicating that it is also a null allele (Figure S1). To determine the genetic relationship between these KNOX1 genes we constructed bp knat2 knat6 plants. These plants showed partial rescue of the downward-oriented fruit phenotype caused by the bp-1 mutation (Figure S2; Ragni et al. 2008), but had normal leaf development (Figure 1C).
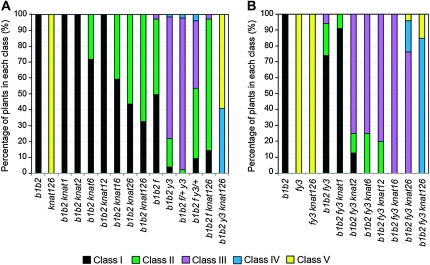
Next we introduced the knox null alleles, singly and in combination, into the bop1 bop2 background. Because rosette leaf morphology varied substantially between the different genetic backgrounds, we quantified the phenotypes by classifying them into five classes on the basis of the extent of suppression of the bop1 bop2 phenotype (Figure S3). Compared to wild-type leaves (Figure S3A), class I plants had leaves with extensive ectopic outgrowths characteristic of bop1 bop2 leaves (Figure S3B). Class II plants had leaves with a slight reduction in ectopic outgrowths compared to class I plants (Figure S3C), and class III plants had serrated leaves with weak petiole outgrowths (Figure S3D). Class IV plants had nearly normal leaves that displayed occasional ectopic outgrowths (Figure S3E), and class V plants had leaves that resembled those of wild-type plants (Figure S3F). Thus, class I plants showed the least suppression of the ectopic outgrowth phenotype, whereas class V plants displayed complete suppression.
Introduction of the bp-1, knat2-1, or bp knat2 alleles into the bop1 bop2 background had no effect on the bop leaf phenotype. In contrast, bop1 bop2 knat6 leaves developed reduced ectopic blade outgrowths in their petiole region (Figure 1D) compared to bop1 bop2 leaves. Whereas all bop1 bop2 bp, bop1 bop2 knat2, and bop1 bop2 bp knat2 plants displayed a class I leaf phenotype, ∼28% of bop1 bop2 knat6 plants displayed a class II leaf phenotype (Figure 3A), showing that mutating KNAT6 slightly suppresses the bop1 bop2 phenotype. bop1 bop2 bp knat6 and bop1 bop2 knat2 knat6 leaves had slightly milder phenotypes than bop1 bop2 knat6 leaves. Class II leaf phenotypes appeared in 41% of bop1 bop2 bp knat6 plants and in 57% of bop1 bop2 knat2 knat6 plants (Figure 3A). In bop1 bop2 bp knat2 knat6 leaves the extent of ectopic outgrowth was further reduced (Figure 1E), with 67% of bop1 bop2 bp knat2 knat6 leaves displaying class II phenotypes (Figure 3A) and some developing ectopic outgrowths along only one side of the petiole (Figure 1E, arrows).
Figure 3.—
Comparision of leaf ectopic outgrowth phenotypes. (A) Graphical representation of the percentage of plants in the bop1 bop2 background displaying rosette and cauline leaf phenotypes of differing severities, grouped from class I (most severe) to class V (least severe). (B) Graphical representation of the percentage of plants in the fil yab3 background displaying rosette and cauline leaf phenotypes of differing severities, grouped from class I (most severe) to class V (least severe). n ≥ 85 plants per genotype.
We also characterized the effects of the KNOX1 alleles on bop1 bop2 ectopic blade organogenesis at the cellular level. Developing wild-type rosette leaf petioles displayed a fully differentiated cellular morphology (Figure 1F), as did bp knat2 knat6 petioles (Figure 1H). In contrast, bop1 bop2 petioles had small and undifferentiated cells at the margins, where the ectopic outgrowths formed (Figure 1G, arrows). The introduction of the knat6 allele into the bop1 bop2 background caused a slight suppression of ectopic outgrowth at the cellular level (Figure 1I, arrow), whereas the introduction of the bp knat2 knat6 allele combination caused a more dramatic suppression of the bop1 bop2 leaf phenotype (Figure 1J, arrows). Thus BP, KNAT2, and KNAT6 are combinatorially responsible for conditioning some of the ectopic blade outgrowth that occurs in bop1 bop2 rosette leaves, with KNAT6 playing a more important role in conditioning the bop phenotype than either BP or KNAT2. These data indicate that an important function for BOP1 and BOP2 during organogenesis is to repress KNOX1 gene activity at the leaf base to ensure proper leaf formation.
YAB gene activity conditions the bop1 bop2 leaf phenotypes:
Because moderate ectopic blade outgrowth still occurred along bop1 bop2 bp knat2 knat6 petioles, we hypothesized that other factors in addition to the KNOX1 genes also contribute to outgrowth production. Two candidates were FIL and YAB3, which promote leaf lamina expansion (Eshed et al. 2004) and are required to repress KNOX1 expression in leaves (Siegfried et al. 1999; Kumaran et al. 2002). Previously we had shown that FIL is ectopically expressed in the adaxial domain of bop1 bop2 leaves, and we proposed that the juxtaposition of ectopic FIL expression with normal class III HD-ZIP expression might lead to ectopic blade outgrowth along new adaxial–abaxial boundaries in bop1 bop2 plants (Ha et al. 2007). We therefore predicted that mutation of FIL, and potentially of YAB3, which has overlapping expression and function (Siegfried et al. 1999), would suppress the bop1 bop2 ectopic outgrowth phenotype.
We tested this hypothesis by performing a genetic analysis between the bop alleles and loss-of-function yab alleles. Because compared to wild-type rosette and cauline leaves (Figure S3A and Figure S4A) fil-8 yab3-2 leaves are small and narrow (Figure S3G and Figure S4B), we separately grouped the variable leaf phenotypes of plants carrying fil and/or yab3 alleles into five classes. Class I plants had four or five leaves that each developed five to eight proximal ectopic organs (Figure S3H). Class II plants had four or five leaves that each developed three to four proximal ectopic organs (Figure S3I). Class III plants had two or three leaves with one or two proximal ectopic organs (Figure S3J). Class IV plants had one rosette and/or cauline leaf with a single ectopic organ (Figure S3K), whereas the leaves of class V plants lacked ectopic outgrowths (Figure S3L).
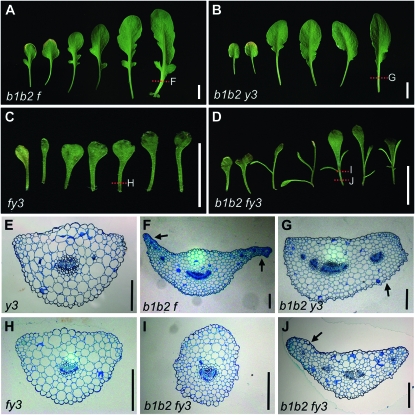
fil-8 and yab3-2 plants displayed almost wild-type rosette leaf morphology, and at the anatomical level the cellular morphology of fil and yab3 petioles was undistinguishable from that of wild-type petioles (Figure 2E). Both bop1 bop2 fil and bop1 bop2 yab3 leaves showed significantly decreased ectopic outgrowth compared to bop1 bop2 leaves (Figure 2, A and B). The bop1 bop2 fil phenotypes ranged from class I to class III, with 3% of bop1 bop2 fil plants forming leaves with class III character (Figure 3A). bop1 bop2 yab3 leaves displayed even stronger suppression of the ectopic outgrowth phenotype. These plants developed leaves with phenotypes ranging from class I to class IV, with 78% of plants displaying either class III or class IV leaf morphology (Figure 3A). However, undifferentiated marginal cells were still evident in bop1 bop2 fil and bop1 bop2 yab3 petioles (Figure 2, F and G, arrows), although they formed reduced ectopic outgrowths compared to bop1 bop2 petioles. These data indicate that FIL and YAB3 each play an important role in conditioning the bop1 bop2 leaf phenotypes, with YAB3 making a more significant contribution.
Figure 2.—
Genetic interactions between bop1 bop2 and yab alleles. Photographs of dissected rosettes from 38-day-old (A) bop1-4 bop2-11 fil-8 (b1b2f), (B) bop1-4 bop2-11 yab3-2 (b1b2y3), (C) fil-8 yab3-2 (fy3), and (D) bop1-4 bop2-11 fil-8 yab3-2 (b1b2fy3) plants. Transverse sections of (E) yab3-2, (F) bop1-4 bop2-11 fil-8, (G) bop1-4 bop2-11 yab3-2, (H) fil-8 yab3-2, and (I and J) bop1-4 bop2-11 fil-8 yab3-2 leaf petioles. Dotted lines in A–D indicate the sectioned regions in F–J. Bars, 10 mm (for A–D) and 200 μm (for E–J).
In addition, we noted a dosage-dependent effect of the fil and yab alleles on the bop1 bop2 ectopic outgrowth phenotype. Compared to 3% of bop1 bop2 fil plants that displayed class III leaf morphology, 46% of bop1 bop2 fil yab3/+ plants displayed class III or class IV leaf morphology (Figure 3A). Similarly, compared to 78% of bop1 bop2 yab3 plants, 98% of bop1 bop2 fil/+ yab3 plants displayed class III or class IV leaf morphology.
fil-8 yab3-2 plants were small and formed narrow leaves with reduced blades (Figure 2C and Figure S4B) as previously reported (Siegfried et al. 1999; Kumaran et al. 2002). Like bop1 bop2 leaves, fil yab3 leaves misexpress KNOX1 genes at high levels and infrequently display ectopic meristem formation on their blades characteristic of KNOX1 overexpressing plants (Kumaran et al. 2002). Thus we tested whether the absence of both FIL and YAB3 expression would affect the bop ectopic outgrowth phenotype. bop1 bop2 fil yab3 plants developed two to four organ outgrowths discontinuously along their rosette leaf petioles (Figure 2D) and four to seven organ outgrowths at the base of their cauline leaves (Figure S4C). The outgrowths were longer and narrower than those produced by bop1 bop2 leaves. The main blade region was also narrow (Figure 2D). The bop1 bop2 fil yab3 phenotypes ranged from class I to class III, with 74% of bop1 bop2 fil yab3 plants forming leaves with class I character (Figure 3B).
We also examined the effects of the fil yab3 alleles on the cellular morphology of rosette leaf petioles. fil yab3 leaves had small petioles with normal cellular morphology (Figure 2H). In contrast, bop1 bop2 fil yab3 petioles exhibited two types of anatomy depending on the region analyzed: petiole regions lacking ectopic outgrowths were nearly radialized, whereas other regions clearly showed ectopic outgrowths at the margins (Figure 2, I and J, arrow). Thus, although mutations in either FIL or YAB3 partially suppressed the bop leaf phenotype, bop1 bop2 fil yab3 leaves still developed substantial ectopic outgrowths potentially because of high-level misexpression of KNOX1 genes through either a common or a converging genetic pathway.
KNOX1 expression analysis:
Our study shows that KNOX1 gene activity is partially responsible for conditioning the bop1 bop2 leaf phenotypes, and both the BOP and YAB genes repress KNOX1 expression during leaf morphogenesis (Kumaran et al. 2002; Ha et al. 2007). To determine whether the BOP and YAB genes regulate KNOX1 transcription via the same or different pathways, we analyzed KNOX1 expression in the young, developing leaves of plants carrying various combinations of bop and yab alleles.
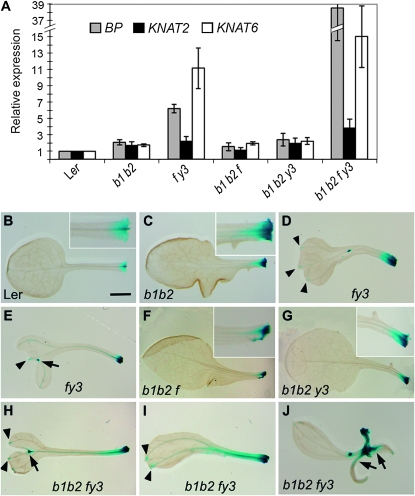
We first confirmed that the steady-state expression levels of BP, KNAT2, and KNAT6 were higher in bop1 bop2 leaves than in wild-type leaves (Figure 4A), as previously reported. KNOX1 expression levels were also higher in fil yab3 leaves than in wild type and bop1 bop2 leaves. KNOX1 expression levels in bop1 bop2 fil and bop1 bop2 yab3 leaves were similar to those in bop1 bop2 leaves (Figure 4A), demonstrating that suppression of the bop1 bop2 ectopic outgrowth phenotype by fil or yab3 is not caused by a reduction in ectopic KNOX1 expression. In contrast, the expression of all three KNOX1 genes, in particular BP, was much higher in bop1 bop2 fil yab3 leaves than either bop1 bop2 or fil yab3 leaves (Figure 4A). This result indicates that the BOP and YAB genes act separately to repress KNOX1 transcription in leaf primordia.
Figure 4.—
Analysis of class 1 KNOX gene expression. (A) BP, KNAT2, and KNAT6 expression in the developing young leaves of 21-day-old seedlings. Mean transcript levels were determined by real-time quantitative PCR analyses and normalized to EF1α. Error bars represent SD. b1 b2, bop1-4 bop2-11; f y3, fil-8 yab3-2. pBP∷GUS activity in (B) Ler, (C) bop1 bop2, (D and E) fil-5 yab3-1, (F) bop1 bop2 fil-5, (G) bop1 bop2 yab3-1, and (H–J) bop1 bop2 fil-8 yab3-2 rosette leaves. Insets in B, C, F, and G are magnified views of the leaf base. Arrowheads indicate BP∷GUS activity in hydathodes. Arrows in E and H indicate BP∷GUS activity in bifurcated leaves. Arrows in J indicate BP∷GUS misexpression in developing ectopic organs.
We next characterized the spatial aspect of KNOX1 misexpression in bop and yab leaves by examining the expression pattern of a BP∷GUS reporter gene. Here we used the fil-5 and yab3-1 alleles rather than the fil-8 and yab3-2 alleles, because both fil-8 and yab3-2 plants already carry a GUS reporter cassette in their genomes (Kumaran et al. 2002). The phenotypes of fil-5 yab3-1 plants were indistinguishable from those of fil-8 yab3-2 plants (Figure S5A; Siegfried et al. 1999). Moreover, the morphological phenotypes of bop1 bop2 fil-5, bop1 bop2 yab3-1, and bop1 bop2 fil-5 yab3-1 plants were indistinguishable from those of bop1 bop2 fil-8, bop1 bop2 yab3-2, and bop1 bop2 fil-8 yab3-2 plants, respectively (Figure S5, B–D).
It has been reported that in wild-type plants BP is expressed in the shoot apex but not in leaf primordia or developing leaves (Ori et al. 2000). However, we found that the base of wild-type leaves showed weak BP∷GUS activity (Figure 4B), likely due either to direct transcription of BP in these cells or to GUS diffusion from the neighboring SAM. Compared to wild-type leaves, bop1 bop2 leaves exhibited stronger and more distally expanded BP∷GUS activity in the petiole (Figure 4C). In fil yab3 leaves, BP∷GUS activity was detected strongly in the petiole and more weakly in the vasculature and hydathodes (Figure 4, D and E, arrowheads), and in the distal region of bifurcated leaves (Figure 4E, arrow). bop1 bop2 fil and bop1 bop2 yab3 leaves had strong BP∷GUS activity restricted to the proximal region of the petiole (Figure 4, F and G), similar to bop1 bop2 leaves. In bop1 bop2 fil yab3 quadruple mutant leaves, BP∷GUS activity was detected in a broader domain than in double- and triple-mutant leaves. In addition to the proximal petiole, the petiole and blade vasculature, the hydathodes and the distal region of bifurcated leaves (Figure 4, H and I), bop1 bop2 fil yab3 leaves also showed strong BP∷GUS activity in the ectopic organs along the petiole (Figure 4J, arrows). Thus KNOX1 expression is both strongly elevated and spatially expanded in bop1 bop2 fil yab3 leaves, and thus may play an important role in conditioning the extensive ectopic organ development seen in bop1 bop2 fil yab3 leaves.
KNOX1 and YAB genes together condition the bop1 bop2 leaf phenotypes:
Although both KNOX1 and YAB mutations were able to partially suppress ectopic organogenesis in bop1 bop2 leaves, the bop phenotypes were not completely rescued by either KNOX1 or YAB mutations. To examine the combinatorial effect of both KNOX1 and YAB genes on leaf morphogenesis, we first combined various knox1 and yab alleles. Despite the fact that KNOX1 gene expression was highly elevated in fil yab3 leaves (Kumaran et al. 2002), the fil yab3 leaf patterning phenotypes were unaffected by the introduction of any combination of the three knox1 alleles (Figure S4D), as was the ectopic shoot formation phenotype (data not shown). Our results demonstrate that derepression of KNOX1 expression does not condition the fil yab3 leaf phenotypes.
Introducing combinations of KNOX1 and YAB mutations into the bop1 bop2 background led to gradually increased suppression of the bop leaf phenotype. The addition of either the knat2 or the knat6 allele into the bop1 bop2 fil yab3 background slightly suppressed the leaf phenotype (Figure 5, A and B, and Figure S4F), with 87% of bop1 bop2 fil yab3 knat2 plants and 100% of bop1 bop2 fil yab3 knat6 plants displaying more suppressed class II or class III leaf phenotypes compared to 5% of bop1 bop2 fil yab3 plants (Figure 3B). In contrast, the addition of the bp allele had little effect, with bop1 bop2 fil yab3 bp leaves displaying phenotypes similar to those of bop1 bop2 fil yab3 leaves (Figure 3B, Figure 5C, and Figure S4E).
Figure 5.—
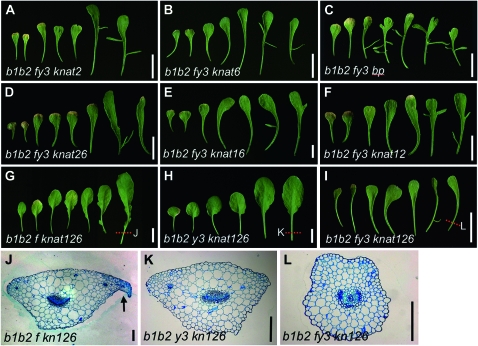
Genetic interactions between yab and knox1 alleles. Photographs of dissected rosette leaves from 38-day-old (A) bop1-4 bop2-11 fil-8 yab3-2 knat2-1, (B) bop1-4 bop2-11 fil-8 yab3-2 knat6-5, (C) bop1-4 bop2-11 fil-8 yab3-2 bp-1, (D) bop1-4 bop2-11 fil-8 yab3-2 knat2-1 knat6-5, (E) bop1-4 bop2-11 fil-8 yab3-2 bp-1 knat6-5, (F) bop1-4 bop2-11 fil-8 yab3-2 bp-1 knat2-1, (G) bop1-4 bop2-11 fil-8 bp-1 knat2-1 knat6-5, (H) bop1-4 bop2-11 yab3-2 bp-1 knat2-1 knat6-5, and (I) bop1-4 bop2-11 fil-8 yab3-2 bp-1 knat2-1 knat6-5 plants. Transverse sections of (J) bop1-4 bop2-11 fil-8 bp-1 knat2-1 knat6-5, (K) bop1-4 bop2-11 yab3-2 bp-1 knat2-1 knat6-5, and (L) bop1-4 bop2-11 fil-8 yab3-2 bp-1 knat2-1 knat6-5 leaf petioles. Dotted lines in G–I indicate the sectioned regions in J–L. Bars, 10 mm (for A–I) and 200 μm (for J–L).
We then introduced the knat2 knat6, bp knat2, and bp knat6 alleles into the bop1 bop2 fil yab3 background. All hextuple mutants showed suppression of the bop1 bop2 fil yab3 leaf phenotypes (Figure 3B, Figure 5, D–F, and Figure S4G). Whereas 75% of bop1 bop2 fil yab3 knat6 plants exhibited class III character, 80% of bop1 bop2 fil yab3 bp knat2 and 100% of bop1 bop2 fil yab3 bp knat6 plants had class III phenotypes (Figure 3B). The bop1 bop2 fil yab3 knat2 knat6 allelic combination generated the greatest degree of suppression, with all plants displaying class III, class IV, or even class V leaf character (Figure 3B).
Introduction of the fil or yab3 alleles into bop1 bop2 bp knat2 knat6 plants induced further suppression of the leaf phenotypes (Figure 5, G and H). bop1 bop2 fil bp knat2 knat6 plants exhibited phenotypes ranging from class I to class III, indicating a reduction in the severity of the class I and class II bop1 bop2 bp knat2 knat6 phenotypes (Figure 3A). bop1 bop2 yab3 bp knat2 knat6 plants developed almost wild-type leaves displaying either class IV or class V morphology (Figure 3A). These data confirm that YAB3 plays a more important role than FIL in conditioning the bop1 bop2 leaf phenotypes. Finally, the bop1 bop2 fil yab3 bp knat2 knat6 septuple mutant combination conditioned nearly complete suppression of the bop1 bop2 leaf phenotype (Figure 5I and Figure S4H). bop1 bop2 fil yab3 bp knat2 knat6 leaves exhibited only class IV or class V character (Figure 3B), with an occasional single ectopic outgrowth observed along the petiole or proximal blade region.
At the cellular level, the extent of ectopic outgrowth reduction was striking in bop1 bop2 fil bp knat2 knat6 petioles, which developed far smaller ectopic organs than bop1 bop2 petioles (Figure 5J). The anatomy of bop1 bop2 yab3 bp knat2 knat6 petioles was very similar to wild-type petioles (Figure 5K). bop1 bop2 fil yab3 bp knat2 knat6 petioles were radialized in shape, although approximately 80% (33/41) showed normal polarity in their central vasculature (Figure 5L). The remaining 20% (8/41) of bop1 bop2 fil yab3 bp knat2 knat6 petioles had vasculature consisting of xylem surrounded by phloem, indicating abaxialization. These histological data clearly show at the cellular level that the extent of ectopic outgrowth gradually decreases with the stepwise addition of mutations in KNOX1 and YAB genes. In sum, our data demonstrate that three KNOX1 genes, BP, KNAT2, and KNAT6, and two YAB genes, FIL and YAB3, are together responsible for ectopic lamina development in bop1 bop2 leaves.
DISCUSSION
Leaf morphogenesis in Arabidopsis to form a narrow proximal petiole and a wide distal blade requires carefully regulated patterning coupled with controlled cell proliferation. BOP1 and BOP2 play key roles in the patterning process because bop1 bop2 leaves display ectopic outgrowth of blade tissue along the petiole (Ha et al. 2004). This phenotype is associated with ectopic KNOX1 and YAB expression (Ha et al. 2003, 2007), yet whether these genes mediate bop outgrowth formation had not previously been determined.
Here we demonstrate that BP, KNAT2, and KNAT6 make a modest contribution to the bop ectopic outgrowth phenotype. The expression of all three genes is elevated in bop1 bop2 leaves, and BP∷GUS activity expands more distally in bop1 bop2 than wild-type leaves (Figure 4). Removal of KNAT6, but not BP or KNAT2, activity alone slightly rescued the bop leaf phenotype (Figure 1D). However, introduction of the bp or knat2 allele into the bop1 bop2 knat6 background further attenuated the bop phenotype, and inactivation of all three KNOX1 genes produced the greatest extent of rescue (Figure 1E and Figure 3A). Thus BP and KNAT2 function redundantly with each other, and overlap with KNAT6, to condition the bop ectopic outgrowth phenotype. These data confirm that an important function for BOP1 and BOP2 during leaf morphogenesis is to repress KNOX1 activity to permit proper petiole differentiation.
FIL and YAB3 also make a significant contribution to ectopic outgrowth formation in bop1 bop2 leaves. We found that compromising either FIL or YAB3 activity reduced bop ectopic outgrowth, with YAB3 playing a more important role than FIL (Figure 2, D and E). These genes also suppressed the bop phenotype in a dose-dependent fashion, with bop1 bop2 fil yab3/+ and bop1 bop2 yab3 fil/+ plants showing more complete suppression than bop1 bop2 fil and bop1 bop2 yab3 plants, respectively (Figure 3A). Thus another key role for the BOP genes during leaf development is to repress YAB expression in the adaxial leaf domain.
Interestingly, the complete loss of both FIL and YAB3 did not cause the disappearance of bop1 bop2 leaf outgrowths. Instead, bop1 bop2 fil yab3 leaves developed extensive ectopic outgrowths compared to bop1 bop2 fil and bop1 bop2 yab3 leaves (Figure 2). This result is explained by strong ectopic expression of KNOX1 genes in bop1 bop2 fil yab3 plants (Figure 4), because the removal of BP, KNAT2, and KNAT6 function strongly suppressed the bop1 bop2 fil yab3 phenotype, such that the leaves of all bop1 bop2 fil yab3 bp knat2 knat6 plants either formed a single ectopic outgrowth or lacked them entirely (Figure 3B and Figure 5I). We conclude that BOP1/BOP2 and FIL/YAB3 act through separate pathways to suppress KNOX1 transcription during leaf morphogenesis.
Several lines of evidence suggest that the roles of the YAB genes in repressing KNOX1 expression and promoting lamina outgrowth are separable. First, removal of BP, KNAT2, and KNAT6 activity in fil yab3 leaves did not restore blade growth (Figure S4D). Second, we did not detect a direct correlation between the level of KNOX1 expression and the extent of ectopic organ outgrowth in the various bop yab backgrounds. bop1 bop2, bop1 bop2 fil, and bop1 bop2 yab3 plants all exhibited similar KNOX1 expression levels and patterns (Figure 4), yet the bop1 bop2 fil and bop1 bop2 yab3 ectopic outgrowth phenotypes were weaker than the bop1 bop2 phenotype (Figure 3A). In addition, KNOX1 mRNA levels were much higher in bop1 bop2 fil yab3 plants than in bop1 bop2 plants (Figure 4A), although the bop1 bop2 fil yab3 phenotype was slightly weaker than the bop1 bop2 phenotype (Figure 3, A and B). We interpret these data in the following way. In bop1 bop2 plants, ectopic KNOX1 and YAB expression produces ectopic outgrowths along the petiole by sustaining the cells in a proliferative state, while simultaneously the juxtaposition of normal PHV and ectopic KAN1 expression domains (Ha et al. 2007) forms new adaxial–abaxial boundaries that trigger lamina outgrowth. In bop1 bop2 fil and bop1 bop2 yab3 plants, ectopic KNOX1 expression is unaltered but the capacity for YAB-mediated lamina outgrowth is reduced, and thus the phenotype is attenuated. In bop1 bop2 fil yab3 plants, the capacity for lamina outgrowth is further compromised, but is largely compensated for in the proximal leaf region by the dramatic increase in ectopic KNOX1 expression (Figure 4, A and H–J) that sustains the cells in an excessively proliferative capacity.
A slight amount of residual ectopic organ outgrowth was observed in bop1 bop2 fil yab3 bp knat2 knat6 leaves, and a higher proportion of bop1 bop2 yab3 bp knat2 knat6 plants than bop1 bop2 fil yab3 bp knat2 knat6 plants displayed fully wild-type leaf morphology (Figure 3). One explanation for these observations is that the residual ectopic organ outgrowth could be conditioned by misregulation of other YAB genes, such as YAB2 and YAB5, which show molecular and functional redundancy with FIL and YAB3 (Siegfried et al. 1999; Izhaki and Bowman 2007). Alternatively, STM, the remaining KNOX1 gene, might function in this process. STM expression is restricted to the SAM and is not detected in developing leaves except infrequently in the sinus (Kawamura et al. 2010). Ectopic expression of a nuclear-localizable STM construct produced plants with lobed and rumpled leaves similar to the phenotypes of BP, KNAT2, and KNAT6 overexpressing plants (Lincoln et al. 1994; Pautot et al. 2001; Dean et al. 2004; Cole et al. 2006). We found that ectopic STM transcripts were not detectable in the developing young leaves of bop1 bop2 yab3 bp knat2 knat6 plants, but were highly elevated in those of bop1 bop2 fil yab3 bp knat2 knat6 plants (Figure S6). These data suggest that STM misexpression is likely responsible for the residual ectopic leaf outgrowth observed in the bop1 bop2 fil yab3 bp knat2 knat6 plants. Unfortunately, it is not feasible to genetically test the contribution of STM to the ectopic organ outgrowth phenotype because stm null mutants lack leaves (Barton and Poethig 1993) and weak stm mutants produce fused leaves with abnormal morphology (Endrizzi et al. 1996).
Arabidopsis as1 and as2 plants in which KNOX1 genes are misexpressed also infrequently develop leaflet-like lobed organs along the petioles (Byrne et al. 2000; Ori et al. 2000; Serrano-Cartagena et al. 2000; Semiarti et al. 2001). Interestingly, unlike bop1 bop2 ectopic outgrowth formation, which is partially suppressed by the bp knat2 knat6 allelic combination, as1 and as2-lobed leaf morphogenesis was not affected by the bp knat2 knat6 allelic combination (Ikezaki et al. 2010). Furthermore, as1 fil yab3 and as2 fil yab3 plants showed additive rather than suppressed phenotypic effects (Fu et al. 2007).
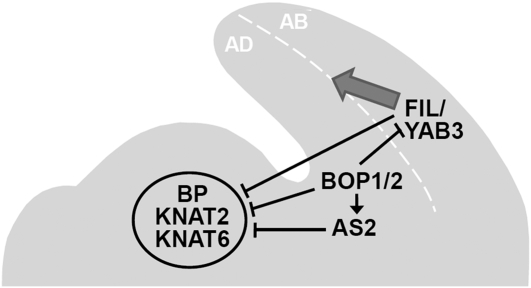
Our data point toward a complex network of interactions between transcriptional regulators during leaf patterning (Figure 6). We have shown that BOP1 is a transcriptional activator that directly associates with specific sites in the AS2 promoter to induce its expression at the base of developing leaf primordia (Jun et al. 2010). AS2 acts with AS1 to maintain KNOX1 repression in leaf primordia (Guo et al. 2008). We demonstrate that BOP1/2 also repress KNOX1 expression, but several observations indicate that this effect is not mediated entirely via AS2. First, bop as2 plants have synergistic phenotypes (Ha et al. 2003, 2007), showing that these genes are not part of a strictly linear genetic pathway. Second, knox1 mutations partially rescue the bop but not the as2 leaf phenotypes (Ikezaki et al. 2010). The FIL and YAB3 genes represent a third set of transcriptional regulators that suppress KNOX1 gene expression in developing leaves (Kumaran et al. 2002). Although FIL is misregulated in bop1 bop2 leaves (Ha et al. 2007) there is no epistatic relationship between bop and yab mutations, implying that BOP1 and BOP2 negatively regulate KNOX1 expression through a separate pathway from the YAB genes. BOP1 and BOP2 also negatively regulate YAB expression in proximal, adaxial leaf cells (Ha et al. 2007), preventing lamina expansion along the petiole. We conclude that ectopic blade outgrowth along Arabidopsis leaf petioles is prevented by BOP1/2 repression both of ectopic YAB gene expression in the adaxial, proximal domain that would promote lamina expansion and of ectopic KNOX1 gene expression that would sustain the leaf cells in an excessively proliferative state.
Figure 6.—
Model for Arabidopsis gene function during lateral organ development. The BOP1 and BOP2 genes directly induce AS2 expression and also negatively regulate FIL and YAB3 activity in developing leaves. In addition, the BOP genes repress KNOX1 (BP, KNAT2, KNAT6) expression via an AS2-independent pathway. AS2 and FIL/YAB3 play separate roles in negatively regulating KNOX1 transcription during lateral organ growth. FIL/YAB3 also promote lamina outgrowth along the adaxial–abaxial boundary (shaded arrow) through a KNOX1 independent pathway. AD, adaxial; AB, abaxial.
Acknowledgments
We thank Venkatesan Sundaresan, John Bowman, Sarah Hake, the Cold Spring Harbor Laboratory, and the Arabidopsis Biological Resource Center for supplying seeds. This work was supported by a U.S. Department of Agriculture Current Research Information System grant to J.C.F.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.118703/DC1.
References
- Barton, M. K., and R. S. Poethig, 1993. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Belles-Boix, E., O. Hamant, S. M. Witiak, H. Morin, J. Traas et al., 2006. KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18 1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M. E., R. Barley, M. Curtis, J. M. Arroyo, M. Dunham et al., 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M. E., J. Simorowski and R. A. Martienssen, 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chuck, G., C. Lincoln and S. Hake, 1996. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, M., C. Nolte and W. Werr, 2006. Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete sub-domains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 34 1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, G., S. Casson and K. Lindsey, 2004. KNAT6 gene of Arabidopsis is expressed in roots and is required for correct lateral root formation. Plant Mol. Biol. 54 71–84. [DOI] [PubMed] [Google Scholar]
- Endrizzi, K., B. Moussian, A. Haecker, J. Z. Levin and T. Laux, 1996. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., S. F. Baum and J. L. Bowman, 1999. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., A. Izhaki, S. F. Baum, S. K. Floyd and J. L. Bowman, 2004. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Fu, Y., L. Xu, B. Xu, L. Yang, Q. Ling et al., 2007. Genetic interactions between leaf polarity-controlling genes and ASYMMETRIC LEAVES1 and 2 in Arabidopsis leaf patterning. Plant Cell Physiol. 48 724–735. [DOI] [PubMed] [Google Scholar]
- Guo, M., J. Thomas, G. Collins and M. C. Timmermans, 2008. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C. M., G.-T. Kim, B. C. Kim, J. H. Jun, M. S. Soh et al., 2003. The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130 161–172. [DOI] [PubMed] [Google Scholar]
- Ha, C. M., J. H. Jun, H. G. Nam and J. C. Fletcher, 2004. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45 1361–1370. [DOI] [PubMed] [Google Scholar]
- Ha, C. M., J. H. Jun, H. G. Nam and J. C. Fletcher, 2007. BLADE-ON-PETIOLE1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19 1809–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake, S., H. M. S. Smith, H. Holtan, E. Magnani, G. Mele et al., 2004. The role of knox genes in plant development. Annu. Rev. Cell. Dev. Biol. 20 125–151. [DOI] [PubMed] [Google Scholar]
- Hepworth, S. R., Y. Zhang, S. McKim, X. Li and G. W. Haughn, 2005. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezaki, M., M. Kojima, H. Sakakibara, S. Kojima, Y. Ueno et al., 2010. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 61 70–82. [DOI] [PubMed] [Google Scholar]
- Izhaki, A., and J. L. Bowman, 2007. KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun, J. H., C. M. Ha and J. C. Fletcher, 2010. BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura, E., G. Horiguchi and H. Tsukaya, 2010. Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 62 429–441. [DOI] [PubMed] [Google Scholar]
- Kumaran, M. K., J. L. Bowman and V. Sundaresan, 2002. YABBY polarity genes mediate the repression of KNOX homeobox genes in Arabidopsis. Plant Cell 14 2761–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., J. Long, J. Yamaguchi, K. Serikawa and S. Hake, 1994. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele, G., N. Ori, S. Sato and S. Hake, 2003. The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17 2088–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A., M. Tamaoki, T. Sakamoto and M. Matsuoka, 2000. Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol. 41 583–590. [DOI] [PubMed] [Google Scholar]
- Norberg, M., M. Holmlund and O. Nilsson, 2005. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132 2203–2213. [DOI] [PubMed] [Google Scholar]
- Ori, N., Y. Eshed, G. Chuck, J. L. Bowman and S. Hake, 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Pautot, V., J. Dockx, O. Hamant, J. Kronenberger, O. Grandjean et al., 2001. KNAT2: evidence for a link between Knotted-like genes and carpel development. Plant Cell 13 1719–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni, L., E. Belles-Boix, M. Gunl and V. Pautot, 2008. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., T. Ito, Y. Shimura and K. Okada, 1999. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell 11 69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Y. Ueno, H. Tsukaya, H. Iwakawa, C. Machida et al., 2001. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena, J., H. Candela, P. Robles, M. R. Ponce, J. M. Perez-Perez et al., 2000. Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K. R., Y. Eshed, S. F. Baum, D. Otsuga, G. N. Drews et al., 1999. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 4117–4128. [DOI] [PubMed] [Google Scholar]
- Sinha, N., R. E. Williams and S. Hake, 1993. Overexpression of the maize homeobox gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7 787–795. [DOI] [PubMed] [Google Scholar]
- Venglat, S. P., T. Dumonceaux, K. Rozwadowski, L. Parnell, V. Babic et al., 2002. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Y. Xu, A. Dong, Y. Sun, L. Pi et al., 2003. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]