Abstract
Many arthropod species are infected with maternally inherited endosymbionts that induce a shift in the sex ratio of their hosts by feminizing or killing males (cytoplasmic sex-ratio distorters, or SRDs). These endosymbionts can have profound impacts on evolutionary processes of their hosts. Here, I derive analytical expressions for the coalescent effective size Ne of populations that are infected with SRDs. Irrespective of the type of SRD, Ne for mitochondrial genes is given by the number of infected females. For nuclear genes, the effective population size generally decreases with increasing prevalence of the SRD and can be considerably lower than the actual size of the population. For example, with male-killing bacteria that have near perfect maternal transmission, Ne is reduced by a factor that is given to a good approximation by the proportion of uninfected individuals in the population. The formulae derived here also yield the effective size of populations infected with mutualistic endosymbionts or maternally inherited bacteria that induce cytoplasmic incompatibility, although in these cases, the reduction in Ne is expected to be less severe than for cytoplasmic SRDs.
SIMPLE null models are essential in science. In population genetics, this role is filled by the Wright–Fisher model and its retrospective counterpart, the Kingman coalescent. Both of these models have proven to be immensely useful in spite of the fact that natural populations usually violate the assumptions made in these models. The reason for this is that often, the Wright–Fisher model can be rescaled so that it behaves in many important respects like a more complex population model. This rescaling is achieved through the concept of the effective population size, Ne. Roughly speaking, a complex population model is said to have a certain Ne if the haploid Wright–Fisher model with population size Ne experiences the same amount of random genetic drift as the complex model. Reflecting the different ways in which drift can be measured, Ne can be defined in different ways, e.g., as the inbreeding, the variance, or the coalescent effective population size. Different definitions often produce the same value for Ne, but may also yield drastically different numbers (Kimura and Crow 1963).
The coalescent effective population size is defined through the factor by which time needs to be rescaled in a complex population model to produce the standard coalescent with time scale given by the population size N (Nordborg and Krone 2002). It has been argued that this is the most useful definition for Ne because “the coalescent essentially embodies all of the information that can be found in sampled genetic data” (Sjödin et al. 2005). More recently, Wakeley and Sargsyan (2009) have proposed two extensions of the coalescent effective population size in which they advocate including a mutation parameter in the definition and also allowing for a nonlinear relationship between Ne and N.
One frequently encountered feature in natural populations that complicates population genetics is infection with maternally inherited endosymbionts. In particular, many arthropod species harbor a great number of phylogenetically diverse microorganisms that influence their hosts' biology in different ways (Bourtzis and Miller 2003; Bourtzis and Miller 2006; Bourtzis and Miller 2009). Because of their maternal transmission, many of these microorganisms—for example, the bacteria Wolbachia pipientis and Cardinium hertigii—have evolved intricate manipulations of their hosts' reproductive system that allows them to spread in a host population through exploitation of male hosts (reproductive parasitism, reviewed in Engelstädter and Hurst 2009a). Most manipulations involve a shift in the sex ratio of their hosts (both primary and at the population level), and the inducing endosymbionts are consequently referred to as cytoplasmic sex-ratio distorters (SRDs). In some species, genetic males develop into females if they are infected (“feminization”: Martin et al. 1973; Rigaud 1997; Bouchon et al. 1998; Hiroki et al. 2002). More commonly, infected males are killed by the endosymbionts early in their development (male killing: reviewed in Hurst et al. 2003). The adaptive advantage of this strategy is seen in an early fitness boost in the surviving females in a brood, for example, through reduced sibling competition or cannibalism of the dead brothers (Hurst 1991; Hurst and Majerus 1993; Jaenike et al. 2003). Some examples for species infected with male-killing or feminizing SRDs are given in Table 1. Finally, in haplodiploid species, unfertilized eggs that would normally develop into males can be diploidized by different bacteria so that they develop into females. The spread of such bacteria may lead to completely asexually reproducing populations (parthenogenesis induction: reviewed in Huigens and Stouthamer 2003).
TABLE 1.
Empirical examples for cytoplasmic SRDs with parameter estimates
| Host | SRD | 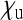 |
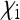 |
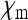 |
References |
|---|---|---|---|---|---|
| Acrea encedona (butterfly) | Wolbachia | ≈ 0 | ≈ 0.5 | ≈ 0 | Jiggins et al. (2002) |
| Adalia bipunctatab (Ladybird beetle) | Rickettsia | 0.076 | 0.506 | 0.076 | Hurst et al. (1993) |
| Drosophila innubilab | Wolbachia | 0.013 | 0.509 | 0.037 | Dyer and Jaenike (2004) |
| Gammarus duebenic (freshwater shrimp) | microsporidium | 0.127 | 0.706 | 0.167 | Dunn et al. (1993) |
| Hypolimnas bolinaa (butterfly) | Wolbachia | ≈ 0 | ≈ 0.5 | ≈ 0 | Dyson et al. (2002) |
With the exception of the feminizing microsporidia in G. duebeni, all SRDs are male-killing bacteria.
In these two butterfly species, maternal transmission and male-killing penetrance is very close to perfect, so that virtually no sons or uninfected daughters are produced by infected mothers. The fact that the infection has not spread to fixation in these species suggests that the fitness benefit of surviving siblings in a brood is absent or very low, leading to the prediction that infected females produce about as many daughters as uninfected females (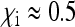 ).
).
In these two species, parameter estimates were obtained from the transmission rate and the prevalence (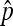 ) reported in the respective references (see Equation 2). In the case of D. innubila, I used the data from the 2002 sampling in Dyer and Jaenike (2004), as this was the largest sample and lay in between the other two samples with regard to infection prevalence.
) reported in the respective references (see Equation 2). In the case of D. innubila, I used the data from the 2002 sampling in Dyer and Jaenike (2004), as this was the largest sample and lay in between the other two samples with regard to infection prevalence.
To calculate the parameter values given for this species in Dunn et al. (1993, Table 1), I assumed equal overall offspring production of infected and uninfected females (as indicated in Dunn et al. 1993) and discarded occasionally produced intersexes. Also note that the primary sex ratio in uninfected G. duebeni is determined by environmental cues and can therefore deviate from 1:1.
Previous theoretical studies indicate that cytoplasmic SRDs will have a strong impact on evolutionary processes for both mitochondrial and nuclear host genes. This is essentially because the host population consists of different classes of individuals (male/female, infected/uninfected) with different reproductive success. Johnstone and Hurst (1996) showed that genetic variation in mtDNA is expected to be strongly reduced during the spread of male-killing bacteria. After the male killers have reached a stable equilibrium in the population, mtDNA variation will recover, but will still be permanently reduced to a value that approximately corresponds to the expected variation if the population consisted only of infected females. In other words, the equilibrium Ne equals approximately the number of infected females in this case. Conversely, for nuclear host genes, Engelstädter and Hurst (2007) showed through computer simulations that to a good approximation, a male-killer infected population behaves as if only uninfected individuals were present.
Here, I derive analytical expressions for the coalescent effective size of host populations infected with cytoplasmic SRDs at equilibrium frequency. This is done for both mitochondrial and nuclear genes. The approach is considerably more general than in the two above-mentioned previous studies in that not only male killing, but also feminizing and even endosymbionts without sex-ratio distorting activity, are covered. However, diploid hosts are assumed throughout this article, so that the derivation for Ne in populations infected with parthenogenesis-inducing bacteria is left for future investigations.
MODEL AND RESULTS
Infection dynamics:
Let us assume a randomly mating diploid population of size N. Females can be either infected by the cytoplasmic SRD or uninfected, and transmission of the SRDs is strictly maternal (no paternal or horizontal transmission). Uninfected females are assumed to produce offspring in a 1:1 sex ratio. Infected females can produce uninfected daughters, infected daughters, and sons, and the relative numbers of offspring of these three classes are denoted by 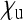 ,
, 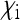 , and
, and 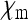 , respectively. These numbers are relative to the offspring number of uninfected females; i.e., infected females may have the same fecundity as uninfected females (
, respectively. These numbers are relative to the offspring number of uninfected females; i.e., infected females may have the same fecundity as uninfected females (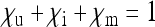 ), but they may also produce fewer (
), but they may also produce fewer (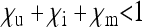 ) or more (
) or more (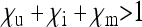 ) offspring than uninfected females. This parameterization covers a broad spectrum of different types of cytoplasmic SRDs, and some examples for parameters estimated in natural systems are given in Table 1.
) offspring than uninfected females. This parameterization covers a broad spectrum of different types of cytoplasmic SRDs, and some examples for parameters estimated in natural systems are given in Table 1.
Let us denote by p the proportion of infected females among all females in the population. Assuming that the host population is large enough relative to selection on SRD spread, so that random genetic drift of the SRD can be ignored, the infection dynamics of the SRD are approximated by a deterministic model with infinite population size and follow the equation
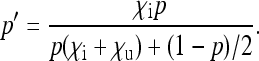 |
(1) |
Solving p′=p yields the nontrivial equilibrium frequency of the SRD
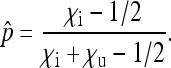 |
(2) |
This equilibrium exists and is stable if and only if 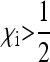 , i.e., if infected females produce more infected daughters than uninfected females produce daughters. This fundamental result goes back to Fine (1975) and has been obtained in a variety of models with different parametrizations (e.g., Bull 1983; Taylor 1990; Hurst 1991; Lipsitch et al. 1995; Engelstädter and Hurst 2009b). Note that the number of males produced by infected females (
, i.e., if infected females produce more infected daughters than uninfected females produce daughters. This fundamental result goes back to Fine (1975) and has been obtained in a variety of models with different parametrizations (e.g., Bull 1983; Taylor 1990; Hurst 1991; Lipsitch et al. 1995; Engelstädter and Hurst 2009b). Note that the number of males produced by infected females (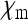 ) does not feature in Equation 2, implying that sex-ratio distortion per se is relevant for the spread of the SRD only insofar as it increases the number
) does not feature in Equation 2, implying that sex-ratio distortion per se is relevant for the spread of the SRD only insofar as it increases the number  of infected daughters or decreases the number
of infected daughters or decreases the number 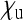 of uninfected daughters that infected females produce.
of uninfected daughters that infected females produce.
Based on 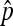 , we can now derive the equilibrium frequencies
, we can now derive the equilibrium frequencies 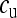 ,
, 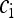 , and
, and 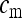 of uninfected females, infected females and males among all individuals in the population as
of uninfected females, infected females and males among all individuals in the population as
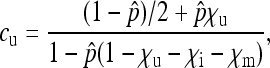 |
(3a) |
 |
(3b) |
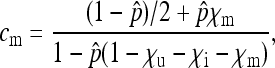 |
(3c) |
with 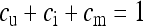 . Finally, these equilibrium frequencies can be used to calculate probabilities of descent, which will be needed in the following sections to calculate
. Finally, these equilibrium frequencies can be used to calculate probabilities of descent, which will be needed in the following sections to calculate 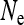 . Let
. Let 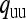 be the probability that the mother of an uninfected female is also uninfected, and
be the probability that the mother of an uninfected female is also uninfected, and 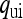 the probability that the mother of an uninfected female is infected. Similarly, let
the probability that the mother of an uninfected female is infected. Similarly, let 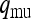 and
and 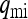 be the probabilities that the mother of a male is uninfected and infected, respectively. We then have
be the probabilities that the mother of a male is uninfected and infected, respectively. We then have
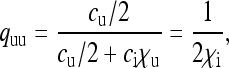 |
(4a) |
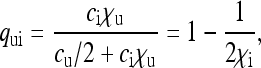 |
(4b) |
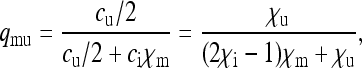 |
(4c) |
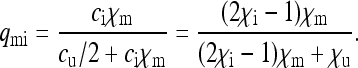 |
(4d) |
Since we assume that the population has reached the equilibrium state with regard to SRD infection, these probabilities will remain constant in time. Table 2 summarizes all parameters and variables of the model.
TABLE 2.
Parameters and auxiliary variables of the model
| Parameters | ||
| N | Actual population size | |
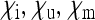
|
Numbers of infected daughters, uninfected daughters and sons, respectively, that an infected female produces relative to a total offspring production of 1 of uninfected females | |
| Auxiliary variables | ||
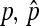
|
Frequency and equilibrium frequency, respectively, of the SRD, i.e., proportion of infected females among all females in the population | |
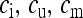
|
Equilibrium proportion of infected females, uninfected females, and males, respectively, among all individuals in the population | |
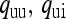
|
Probability that the mother of an uninfected female is uninfected or infected, respectively ( ) ) |
|
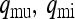
|
Probability that the mother of a male is uninfected or infected, respectively (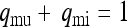 ) ) |
|
Ne for mitochondrial genes:
To derive the effective population size for mitochondrial genes, we follow the ancestry of two gene copies backward in time and determine the average time until coalescence of these gene copies occurs. We assume that all mitochondrial genomes within a single host are identical (no heteroplasmy). Since males do not transmit mitochondria, we need only to follow the coalescence process in females. To this end, we construct a Markov process with the following four states: (1) both gene copies are in different uninfected females, (2) both gene copies are in different infected females, (3) one gene copy is in an infected and the other copy in an uninfected female, and (4) the two gene copies have coalesced in a single (infected or uninfected) female. Table 3 illustrates these four states and gives the transition probability matrix Π between these states (i.e., 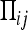 is the probability of switching from state i in the current to state j in the previous generation).
is the probability of switching from state i in the current to state j in the previous generation).
TABLE 3.
Probability transition matrix Π for the ancestral process of mitochondrial genes
Applying Möhle's method of separation of time scales (Möhle 1998), we can partition the matrix Π as 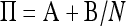 , where
, where
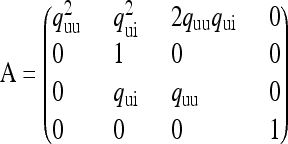 |
(5a) |
and
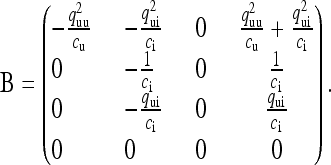 |
(5b) |
The matrix A is again a probability transition matrix and can be interpreted as describing the backward dynamics on a short time scale. The equilibrium of this short-term process is given by the limit
 |
(6) |
This means that when starting with two gene copies in two different females, the probability that these two gene copies will be found in two infected females (state 2) will converge to one on this short time scale.
As proven by Möhle (1998), the coalescence process converges to the continuous-time process  . Here, the rate matrix PBP is given by
. Here, the rate matrix PBP is given by
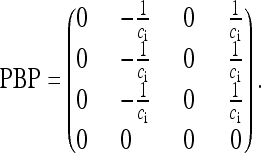 |
(7) |
This shows that by rescaling the coalescence process by a factor 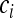 , the Kingman coalescent is obtained if N is sufficiently large. Hence, the effective population size for mitochondrial genes is given by
, the Kingman coalescent is obtained if N is sufficiently large. Hence, the effective population size for mitochondrial genes is given by 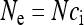 in this case, i.e., the number of infected females. This result has been previously obtained for male-killing bacteria (Johnstone and Hurst 1996) and has important consequences for the interpretation of mitochondrial nucleotide polymorphism data (Hurst and Jiggins 2005). Figure 1 shows how the scaling factor
in this case, i.e., the number of infected females. This result has been previously obtained for male-killing bacteria (Johnstone and Hurst 1996) and has important consequences for the interpretation of mitochondrial nucleotide polymorphism data (Hurst and Jiggins 2005). Figure 1 shows how the scaling factor 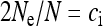 , giving the change in
, giving the change in 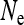 relative to an uninfected population with N/2 females, depends on the parameters of the model.
relative to an uninfected population with N/2 females, depends on the parameters of the model. 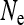 is substantially reduced when infected mothers produce only little more infected daughters than uninfected females produce daughters (
is substantially reduced when infected mothers produce only little more infected daughters than uninfected females produce daughters (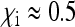 ) and have a low rate of maternal transmission (relatively large
) and have a low rate of maternal transmission (relatively large 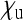 ), a situation observed in many male-killing bacteria that are at low infection frequency within a population. On the other hand, when
), a situation observed in many male-killing bacteria that are at low infection frequency within a population. On the other hand, when 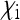 is large or
is large or 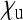 is low—as occurs in feminizing microbes or male-killers with high maternal transmission rate—
is low—as occurs in feminizing microbes or male-killers with high maternal transmission rate—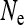 is increased relative to an uninfected population.
is increased relative to an uninfected population.
Figure 1.—
Scaling factor 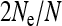 for the effective population size of mitochondrial genes in an SRD-infected population, depending on the relative numbers
for the effective population size of mitochondrial genes in an SRD-infected population, depending on the relative numbers 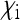 and
and 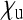 of infected and uninfected daughters that an infected female produces. In this contour plot, the factor by which
of infected and uninfected daughters that an infected female produces. In this contour plot, the factor by which 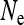 is altered through SRD infection is indicated by areas of different color, where the small numbers in the plot give
is altered through SRD infection is indicated by areas of different color, where the small numbers in the plot give 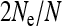 at the borders of these areas. The relative number of sons,
at the borders of these areas. The relative number of sons,  , that an infected female produces, is assumed to be equal to the number of uninfected daughters
, that an infected female produces, is assumed to be equal to the number of uninfected daughters 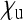 . The dashed red line marks the situation where infected females produce as many offspring as uninfected females.
. The dashed red line marks the situation where infected females produce as many offspring as uninfected females.
It should be noted that although the above derivation is based on a sample size of two, convergence to the Kingman coalescent would also occur for larger sample sizes. This was demonstrated by Nordborg and Krone (2002) for the general case of a structured population where the rates of “migration” of ancestral lineages between different “patches” do not depend on the population size N (this is the case of α = 0 in Nordborg and Krone (2002)). The present model is a special case of this more general model: patches correspond to infection states (infected or uninfected), and migration corresponds to movement of lineages from the infected state to the uninfected state through imperfect maternal transmission of the endosymbionts. Thus, the effective population size of 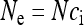 obtained here is a generally applicable one, because the structure of the population—given by the infection states—collapses to the Kingman coalescent as N tends to infinity.
obtained here is a generally applicable one, because the structure of the population—given by the infection states—collapses to the Kingman coalescent as N tends to infinity.
Ne for nuclear genes:
The effective population size for nuclear genes will now be derived with the same method as in the previous section. In contrast to the case of mitochondrial genes, we now have to also consider inheritance through males as well as diploidy. Again, we consider the ancestral process only for a sample of size two, but on the basis of previous results (Möhle 1998; Nordborg and Krone 2002), the same process for larger samples is also expected to converge to the Kingman coalescent as the population size tends to infinity.
The Markov chain for the ancestral process of two gene copies now has the following 10 states:
Both genes are in a single uninfected female (but have not coalesced)
Both genes are in a single infected female
Both genes are in a single male
The genes are in different uninfected females
The genes are in different infected females
The genes are in different males
One gene is in an uninfected female and the other in an infected female
One gene is in an uninfected female and the other in a male
One gene is in an infected female and the other in a male
The two genes have coalesced (in an uninfected or infected female or in a male)
Table 4 illustrates these 10 states and gives the probability transition matrix Π of this Markov process.
TABLE 4.
Probability transition matrix Π for the ancestral process of nuclear genes
Again, we can separate time scales by partitioning Π into 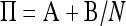 . The matrices A and B (not shown) are readily obtained from Π.
. The matrices A and B (not shown) are readily obtained from Π. 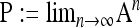 takes the form
takes the form
 |
(8) |
where the formulae for the placeholders 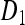 ,
, 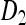 ,
, 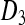 , and Q are given in the appendix. The matrix P can be interpreted as the short-term equilibrium probability distribution. Thus, the probability that the two gene copies will be in two different males (state 6), and the probability that the two genes will be in two different females (states 4, 5, and 7) will converge to
, and Q are given in the appendix. The matrix P can be interpreted as the short-term equilibrium probability distribution. Thus, the probability that the two gene copies will be in two different males (state 6), and the probability that the two genes will be in two different females (states 4, 5, and 7) will converge to 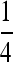 on this time scale, whereas the probability that one gene will be in a female and the other in a male (states 8 and 9) will converge to
on this time scale, whereas the probability that one gene will be in a female and the other in a male (states 8 and 9) will converge to 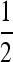 . These probabilities follow from the simple fact that in spite of the potentially complicated transmission dynamics and the sex-ratio distortion, every individual has one maternally and one paternally derived allele.
. These probabilities follow from the simple fact that in spite of the potentially complicated transmission dynamics and the sex-ratio distortion, every individual has one maternally and one paternally derived allele.
The scaling factor determining the effective population size is given by the entries in the last column of the rate matrix PBP. The resulting formula for Ne is given in the appendix. This rather unwieldy formula simplifies considerably in a few important special cases that will now be considered.
Special case 1: Infected females produce equal numbers of uninfected daughters and sons ( ):
):
This situation arises when there is perfect penetrance of the induced sex-ratio distortion, i.e., if all infected male offspring are killed or feminized and hence no infected males occur in the population. Perfect penetrance seems to be the rule in both male-killing and feminizing Wolbachia and is therefore a very realistic assumption. With 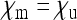 , the scaling factor by which the effective population size is reduced is given by
, the scaling factor by which the effective population size is reduced is given by
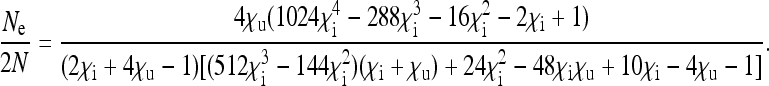 |
(9) |
Figure 2 shows how this scaling factor depends on the two parameters 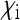 and
and 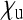 (
(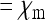 ). The strongest reduction of Ne is obtained when
). The strongest reduction of Ne is obtained when 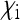 is large and
is large and 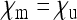 are small. This corresponds to a high prevalence
are small. This corresponds to a high prevalence 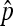 of the SRD in the population and to a strongly female-biased population sex ratio (low cm). Indeed, it can be seen from Equation 9 that
of the SRD in the population and to a strongly female-biased population sex ratio (low cm). Indeed, it can be seen from Equation 9 that 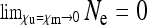 for any fixed
for any fixed 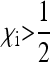 ; i.e., as the number of uninfected daughters and males produced by infected females approaches zero, the effective population size converges to zero.
; i.e., as the number of uninfected daughters and males produced by infected females approaches zero, the effective population size converges to zero.
Figure 2.—
Scaling factor 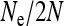 for the effective population size of nuclear genes in an SRD-infected population, depending on the relative numbers
for the effective population size of nuclear genes in an SRD-infected population, depending on the relative numbers 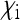 of infected daughters and
of infected daughters and 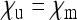 of uninfected daughters and sons that an infected female produces. The dashed red line marks the situation where infected females produce as many offspring as uninfected females.
of uninfected daughters and sons that an infected female produces. The dashed red line marks the situation where infected females produce as many offspring as uninfected females.
We can also ask what the effective population size is with a male-killing SRD that has a very high transmission rate (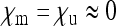 ), but that is selectively close to neutral (
), but that is selectively close to neutral (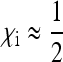 ). This latter assumption is probably very realistic for many male-killer systems in which the death of male offspring does only very weakly enhance the fitness of the surviving siblings. To obtain the effective population size for this case, we can derive an approximation of Equation 9 assuming that
). This latter assumption is probably very realistic for many male-killer systems in which the death of male offspring does only very weakly enhance the fitness of the surviving siblings. To obtain the effective population size for this case, we can derive an approximation of Equation 9 assuming that  is very small, but the fraction ci of infected females in the population takes a constant value. For given parameters
is very small, but the fraction ci of infected females in the population takes a constant value. For given parameters 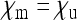 , the parameter
, the parameter 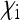 that is required to produce a certain equilibrium proportion ci of infected females in the population is given from Equation 3b by
that is required to produce a certain equilibrium proportion ci of infected females in the population is given from Equation 3b by
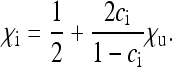 |
(10) |
Inserting this into Equation 9 and assuming  yields
yields
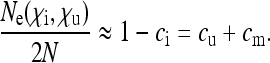 |
(11) |
Thus, the effective size of a population infected with a “nearly neutral male killer” as defined above is given simply by twice the number of uninfected individuals in the population. This result has been anticipated through verbal arguments (Sullivan and Jaenike 2006; Engelstädter and Hurst 2007) and was also confirmed in computer simulations (Engelstädter and Hurst 2007).
In Figure 3, the scaling factor for the effective population size is shown in relation to the infection frequency 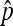 in the population (see Equation 2) and the maternal transmission rate of the endosymbionts,
in the population (see Equation 2) and the maternal transmission rate of the endosymbionts,  . This plot was obtained by a straightforward reparametrization in which
. This plot was obtained by a straightforward reparametrization in which 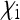 and
and 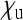 in Equation 9 are replaced by expressions involving
in Equation 9 are replaced by expressions involving 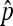 and the transmission rate.
and the transmission rate. 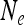 decreases with increasing prevalence of the SRD and with increasing rate of maternal transmission. The plot also indicates that for high rates of maternal transmission, Equation 11 provides a good approximation for
decreases with increasing prevalence of the SRD and with increasing rate of maternal transmission. The plot also indicates that for high rates of maternal transmission, Equation 11 provides a good approximation for 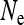 .
.
Figure 3.—
Scaling factor 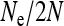 for the effective population size of nuclear genes in an SRD-infected population, with
for the effective population size of nuclear genes in an SRD-infected population, with 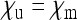 .
. 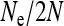 is expressed as a function of the equilibrium frequency
is expressed as a function of the equilibrium frequency 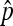 of the SRD and in relation to the following rates of maternal transmission
of the SRD and in relation to the following rates of maternal transmission 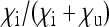 : 0.9 (solid line), 0.95 (dotted line), and 0.99 (dashed line). Note that
: 0.9 (solid line), 0.95 (dotted line), and 0.99 (dashed line). Note that  can never be greater than any given transmission rate. The thick line gives the value for
can never be greater than any given transmission rate. The thick line gives the value for 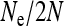 that is obtained with the approximation
that is obtained with the approximation 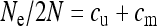 (see Equation 11).
(see Equation 11).
Special case 2: SRDs with perfect transmission but incomplete penetrance (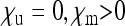 ):
):
In this scenario, all offspring from infected females will also be infected, and a certain fraction of sons will be feminized or killed by the bacteria. Provided that 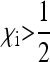 (either trough feminization or male killing with fitness compensation), Equation 2 shows that the SRD will spread to fixation in the population. Thus, there remain only infected females in the population that produce offspring with a male:female sex ratio of
(either trough feminization or male killing with fitness compensation), Equation 2 shows that the SRD will spread to fixation in the population. Thus, there remain only infected females in the population that produce offspring with a male:female sex ratio of 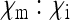 . It is not surprising therefore that the scaling factor for the effective population size simplifies to
. It is not surprising therefore that the scaling factor for the effective population size simplifies to
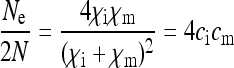 |
(12) |
in this special case, which is Wright's formula for the effective size of a population with arbitrary sex ratio (Wright 1931). Note that with feminizing SRDs, this special case of 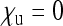 and
and 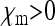 may also ensue when maternal transmission is not complete (see the discussion).
may also ensue when maternal transmission is not complete (see the discussion).
Special case 3: Endosymbionts that do not induce sex-ratio distortion (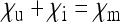 ):
):
Although not the primary focus of this study, the model presented here also covers maternally inherited endosymbionts that do not induce any sex-ratio distortion, but spread by enhancing the fitness of their hosts in some way. It is clear from the condition 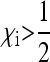 that for this type of maternally inherited endosymbiont to spread and be stably maintained in a population,
that for this type of maternally inherited endosymbiont to spread and be stably maintained in a population, 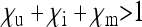 needs to be fulfilled. This means that infected females must have an increased overall survival rate and/or fecundity compared to uninfected females, as is the case for many endosymbionts that provide their hosts with nutrients or protect them from parasites (e.g., Aksoy 2003; Douglas 2003; Oliver et al. 2003; Scarborough et al. 2005; Hosokawa et al. 2010).
needs to be fulfilled. This means that infected females must have an increased overall survival rate and/or fecundity compared to uninfected females, as is the case for many endosymbionts that provide their hosts with nutrients or protect them from parasites (e.g., Aksoy 2003; Douglas 2003; Oliver et al. 2003; Scarborough et al. 2005; Hosokawa et al. 2010).
No simple formula for Ne was obtained for this special case. Figure 4 shows that for realistic parameter values of 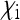 and
and 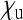 (with
(with 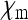 given by the sum of these two), the reduction in Ne is only very weak. The strongest reduction is expected when
given by the sum of these two), the reduction in Ne is only very weak. The strongest reduction is expected when 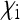 is relatively small, but
is relatively small, but 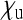 is large. This will be the case when maternal transmission is very inefficient, but when the endosymbionts still produce a benefit for their hosts that is large enough to ensure stable persistence in the population.
is large. This will be the case when maternal transmission is very inefficient, but when the endosymbionts still produce a benefit for their hosts that is large enough to ensure stable persistence in the population.
Figure 4.—
Scaling factor  for the effective population size of nuclear genes in endosymbiont harboring populations that do not induce sex-ratio distortion (i.e.,
for the effective population size of nuclear genes in endosymbiont harboring populations that do not induce sex-ratio distortion (i.e., 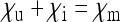 ). The dashed green line marks endosymbionts with a transmission rate
). The dashed green line marks endosymbionts with a transmission rate 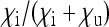 of 0.95.
of 0.95.
Aside from endosymbionts that provide benefits (e.g., nutritional or protective) to their hosts, this special case also covers endosymbionts that induce cytoplasmic incompatibility (CI), a reproductive incompatibility between uninfected females and infected males (reviewed in Engelstädter and Telschow 2009). Here, it is essentially the offspring production of uninfected females that is altered by the endosymbionts, but through rescaling, the parameters 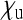 ,
, 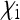 , and
, and 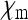 relative to a total offspring production 1 in uninfected females can be obtained. Although these parameters will depend on the frequency of the endosymbionts, at equilibrium they are constant and therefore can be used to obtain Ne as for other types endosymbionts. In most natural systems where this has been studied,
relative to a total offspring production 1 in uninfected females can be obtained. Although these parameters will depend on the frequency of the endosymbionts, at equilibrium they are constant and therefore can be used to obtain Ne as for other types endosymbionts. In most natural systems where this has been studied, 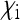 will be very large and
will be very large and 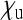 will be rather small due maternal transmission rates close to one and high offspring mortality in incompatible crosses. Hence, only minor reductions in Ne are expected with CI-inducing microbes.
will be rather small due maternal transmission rates close to one and high offspring mortality in incompatible crosses. Hence, only minor reductions in Ne are expected with CI-inducing microbes.
DISCUSSION
The coalescent effective size of populations infected with maternally inherited endosymbionts—in particular those inducing a sex-ratio distortion in their hosts—was derived for both mitochondrial and nuclear host genes. This was done using the method of the separation of time scales, a powerful tool that can be used to simplify various types of structured population, including geographical, age, or class structure (Möhle 1998; Nordborg and Krone 2002; Ramachandran et al. 2008; see also Wakeley 2009, Chap. 6, for an accessible account of this method). The model and parametrization used here was kept deliberately simple in that the endosymbionts are characterized by only three parameters (the relative numbers of infected daughters, uninfected daughters, and sons that infected females produce). Despite this simplicity, the model can be applied to a large variety of situations, including both male-killing and feminizing microbes, arbitrary rates of maternal transmission, arbitrary penetrance of the sex-ratio distortion, and arbitrary reduction in overall fecundity of infected females. In addition, the model also produces predictions on the effective size of populations infected with maternally inherited symbionts that do not induce sex-ratio distortion, but spread in the population by providing hosts with some form of benefit or inducing cytoplasmic incompatibility.
For mitochondrial genes, the effective population size is given simply by the number of infected females in the population, independent of the type of sex-ratio distortion or other means of spread of the endosymbionts. This means that the strongest reduction in Ne is caused by endosymbionts that are stably maintained at a low prevalence in the population. Many male-killing bacteria are indeed present at a low prevalence (reviewed in Hurst et al. 2003), and endosymbionts where the induced phenotype is unknown are also often characterized by a low infection frequency (e.g., Duron et al. 2008; Hilgenboecker et al. 2008). Because low prevalence endosymbionts have a strong impact on mitochondrial Ne and are common but easy to miss, these endosymbionts can easily confound interpretations of mtDNA polymorphism data (Hurst and Jiggins 2005).
For nuclear genes, maternally inherited SRDs influence effective population size in the opposite direction: the higher the SRD prevalence, the lower Ne. The general formula for nuclear Ne with endosymbiont infections is rather complicated, but can nevertheless be calculated explicitly without computer simulations. In some special cases, this formula can be reduced considerably. In particular, the nuclear effective size of a male-killer infected population is often given to a good approximation by twice the number of uninfected individuals, a simple result obtained previously through verbal arguments and computer simulations (Sullivan and Jaenike 2006; Engelstädter and Hurst 2007). In situations where feminizing microbes have become fixed in the population, Ne is given by Wright's well-known formula for sex-biased populations (Wright 1931). For endosymbionts that do not induce any sex-ratio distortion—including facultative mutualistic endosymbionts or bacteria inducing cytoplasmic incompatibility—Ne is also reduced, but only weakly.
Limitations of the model:
Several simplifying assumptions have been made to obtain analytical expression of the effective population size. Most importantly, I assumed that the frequency of the endosymbionts in the population is at a constant equilibrium. For mitochondrial genes, it has been demonstrated that the effective population size can also be strongly reduced during the spread of the SRD (Johnstone and Hurst 1996). This is because the mitochondrial genotype that happens to be associated with the ancestral bacterial infection in the population will hitchhike along with the SRD in the population, a process not covered by the current model. Moreover, even if the infection is at equilibrium in the population, fluctuations around this equilibrium frequency due to random genetic drift can be expected. The magnitude of these fluctuations will depend on the size of the population and the type and penetrance of sex-ratio distortion (i.e., the strength of selection for the SRD). For example, in many male-killing bacteria showing close to perfect maternal transmission but weak levels of fitness compensation for females in a brood, we expect substantial fluctuations in frequency around the equilibrium value. Such fluctuations are likely to further reduce the effective population size below the values predicted by this model, an expectation stemming from previous results on populations with changing size. [With fast fluctuations in population size, Ne is given by the harmonic mean of the actual sizes of the population, which is smaller than the arithmetic mean (Kimura and Crow 1963; Sjödin et al. 2005)]. For the case of male-killing bacteria, this reasoning is in accord with simulation results by Engelstädter and Hurst (2007).
Another important assumption of the model presented here is that the proportions of offspring that females produce depend only on their infection state. This assumption is violated in species with a ZW sex chromosome system (i.e., where ZW individuals develop into females and ZZ individuals normally develop into males) that are infected with feminizing endosymbionts. This situation has been reported in the woodlouse Armadillidium vulgare as well as in the butterfly Eurema hecabe, both infected with feminizing Wolbachia (Martin et al. 1973; Hiroki et al. 2002). There can be two types of females in such a population: ZW females (i.e., “real” genetic females) and ZZ females (genetically male, but feminized). Clearly, infected ZW females may produce all possible types of offspring if transmission is imperfect, whereas ZZ females cannot produce uninfected daughters. However, modeling has shown that the spread of the feminizing endosymbionts will eventually lead to extinction of the W chromosome (Taylor 1990). Thus, at equilibrium all individuals will have a ZZ constitution and their sex is determined solely by the presence or absence of the endosymbionts, a condition observed—albeit complicated by an additional feminizing factor—in some populations of A. vulgare (reviewed in Rigaud et al. 1997). At equilibrium, this complicated situation is therefore covered by the model presented here, more precisely by special case 2, where 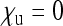 and Wright's (1931) formula for Ne in populations with biased population sex ratio is valid.
and Wright's (1931) formula for Ne in populations with biased population sex ratio is valid.
Next, the method of separation of time scales used here assumes that a within-population equilibrium of the distribution of gene copies is reached quickly relative to coalescent events. This assumption may be violated for mitochondrial genes when maternal transmission is very efficient. In this case, coalescence of two gene copies in uninfected females may occur faster than transition of these copies into the class of infected females. In the extreme case of perfect maternal transmission—which requires selective neutrality of the SRD if it is not to become fixed in the population—no such transitions will occur, so that both classes can be regarded as isolated subpopulations. However, for large populations, infection frequencies that are not extremely small and with at least some low level of imperfect maternal transmission, the approximation through Möhle's (1998) theorem is expected to hold. Furthermore, no such problems are expected for nuclear genes, because here, there will necessarily be ample “gene flow” between infected and uninfected individuals through males.
Finally, I assumed that the effective size of the population in the absence of infection is given by the census population size (i.e., 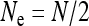 for mitochondrial and
for mitochondrial and 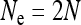 for nuclear genes). In reality, many factors are known in addition to cytoplasmic SRDs that also lead to reduced or increased effective population sizes. For example, if there is strong sexual selection in the population so that males have a high variance in reproductive success, this will decrease Ne (Kimura and Crow 1963). On the other hand, when bottlenecks during mitochondrial transmission are large so that intra-individual mitochondrial variation (heteroplasmy) can build up, the effective population size for mitochondrial genes can also be larger than the number of females in the population (Birky et al. 1983). In many situations, modifications in Ne caused by cytoplasmic SRDs may combine with other such modifications in a simple way (i.e., the factors quantifying these modifications in Ne are multiplied), but it is not clear that this should always be so.
for nuclear genes). In reality, many factors are known in addition to cytoplasmic SRDs that also lead to reduced or increased effective population sizes. For example, if there is strong sexual selection in the population so that males have a high variance in reproductive success, this will decrease Ne (Kimura and Crow 1963). On the other hand, when bottlenecks during mitochondrial transmission are large so that intra-individual mitochondrial variation (heteroplasmy) can build up, the effective population size for mitochondrial genes can also be larger than the number of females in the population (Birky et al. 1983). In many situations, modifications in Ne caused by cytoplasmic SRDs may combine with other such modifications in a simple way (i.e., the factors quantifying these modifications in Ne are multiplied), but it is not clear that this should always be so.
Implications for genetic variation:
The effective population size is a measure for the magnitude of random genetic drift in a population and therefore determines many population genetic quantities. Genetic variation is the most directly observable of these quantities. The results obtained here imply that studies investigating and interpreting genetic variation in arthropod populations should take potential infections with maternally inherited endosymbionts into account. For mutualistic infections as well as for bacteria inducing cytoplasmic incompatibility that are at equilibrium in the population, reductions in Ne for nuclear genes caused by these microbes can probably safely be ignored in most cases. The change in nuclear Ne due to cytoplasmic SRDs, on the other hand, will often strongly affect the expected level of genetic variation in a population.
It is important to stress in this respect that even in the absence of an SRD infection in a population, a past infection that has been cleared from the population may still have left a population genetic trace of reduced genetic variation. Given that cytoplasmic SRDs are common in arthropods and that because of their parasitic nature, host suppressor genes may often drive them extinct, such footprints of past infections may be very widespread. A case in point is an Independent Samoan population of the butterfly Hypolimnas bolina. For at least about 100 years until 2001, this population was infected with male-killing Wolbachia at a very high prevalence (99% infected females: Dyson and Hurst 2004), but within only 10 generations, a host suppressor gene effecting survival of infected males spread to fixation in this population, restoring the population sex ratio to 1:1 (Charlat et al. 2007). Consequently, this population experienced a prolonged and drastic reduction in its effective size for nuclear genes (to approximately 1% of the Ne without the infection), but then recovered quickly to its uninfected Ne. The reduced level of genetic variation that this reduction in Ne is likely to have caused is expected to be still strong in this population. Moreover, the selective sweep that occurred during the spread of the suppressor gene will have caused further reduction in genetic variation at loci linked to this suppressor gene. Without knowledge of the infection history of this population, the strong reduction in genetic variation might easily be mistaken for signs of demographic events like population bottlenecks. Thus, this example shows that cytoplasmic SRDs can severely confound interpretations not only of mitochondrial (Hurst and Jiggins 2005), but also of nuclear DNA diversity.
Acknowledgments
I thank Tanja Stadler, John Wakeley, Jon Wilkins, and an anonymous reviewer for helpful comments on the manuscript and acknowledge support from the Swiss National Science Foundation.
APPENDIX
Here, the formulae for the expressions specifying the matrix P in Equation 8 as well as for Ne for nuclear genes in the general model are given. To simplify the notation, let us define the following placeholders:
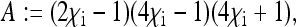 |
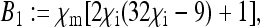 |
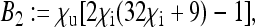 |
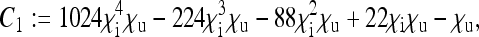 |
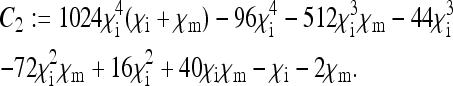 |
The placeholders in matrix P in Equation 8 are then given by
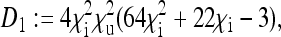 |
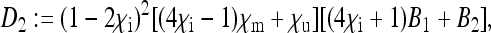 |
 |
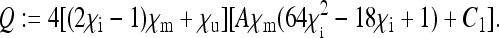 |
The effective population size for nuclear genes in the most general scenario is given by
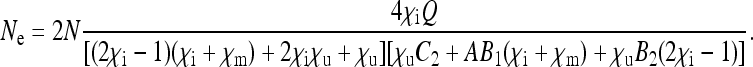 |
References
- Aksoy, S., 2003. Symbiosis in tsetse, pp. 53–65 in Insect Symbiosis, edited by K. Bourtzis and T. A. Miller. CRC Press, Boca Raton, FL.
- Birky, C. W., T. Maruyama and P. Fuerst, 1983. An approach to population and evolutionary genetic theory for genes in mitochondria and chloroplasts, and some results. Genetics 103 513–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon, D., T. Rigaud and P. Juchault, 1998. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc. R. Soc. Lond. B Biol. Sci. 265 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis, K., and T. A. Miller (Editors), 2003. Insect Symbiosis. CRC Press, Boca Raton, FL.
- Bourtzis, K., and T. A. Miller (Editors), 2006. Insect Symbiosis, Vol. 2. CRC Press, Boca Raton, FL.
- Bourtzis, K., and T. A. Miller (Editors), 2009. Insect Symbiosis, Vol. 3. CRC Press, Boca Raton, FL.
- Bull, J. J., 1983. The Evolution of Sex Determination Mechanisms. Benjamin/Cummings Publishing, Menlo Park, CA.
- Charlat, S., E. A. Hornett, J. H. Fullard, J. Davies, G. K. Roderick et al., 2007. Extraordinary flux in sex-ratio. Science 317 214. [DOI] [PubMed] [Google Scholar]
- Douglas, A. E., 2003. Buchnera bacteria and other symbionts of aphids, pp. 23–28 in Insect Symbiosis, edited by K. Bourtzis and T. A. Miller. CRC Press, Boca Raton, FL.
- Dunn, A. M., J. Adams and J. E. Smith, 1993. Transovarial transmission and sex-ratio distortion by a microsporidian parasite in a shrimp. J. Invert. Pathol. 61 248–252. [Google Scholar]
- Duron, O., D. Bouchon, S. Boutin, L. Bellamy, L. Zhou et al., 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, K. A., and J. Jaenike, 2004. Evolutionarily stable infection by a male-killing endosymbiont in Drosophila innubila. Genetics 168 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, E. A., and G. D. D. Hurst, 2004. Persistence of an extreme sex-ratio bias in a natural population. Proc. Natl. Acad. Sci. USA 101 6520–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, E. A., M. K. Kamath and G. D. D. Hurst, 2002. Wolbachia infection associated with all-female broods in Hypolimnas bolina (Lepidoptera: Nymphalidae): evidence for horizontal transmission of a butterfly male killer. Heredity 88 166–171. [DOI] [PubMed] [Google Scholar]
- Engelstädter, J., and G. D. D. Hurst, 2007. The impact of male-killing bacteria on host evolutionary processes. Genetics 175 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstädter, J., and G. D. D. Hurst, 2009. a The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. System. 40 127–149. [Google Scholar]
- Engelstädter, J., and G. D. D. Hurst, 2009. b What use are male hosts?: the dynamics of maternally inherited bacteria showing sexual transmission or male-killing. Am. Nat. 173 E159–E170. [DOI] [PubMed] [Google Scholar]
- Engelstädter, J., and A. Telschow, 2009. Cytoplasmic incompatibility and host population structure. Heredity 103 196–207. [DOI] [PubMed] [Google Scholar]
- Fine, P. E. M., 1975. Vectors and vertical transmission: an epidemiologic perspective. Ann. NY Acad. Sci. 266 173–194. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker, K., P. Hammerstein, P. Schlattmann, A. Telschow and J. H. Werren, 2008. How many species are infected with Wolbachia?: a statistical analysis of current data. FEMS Microbiol. Lett. 281 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroki, M., Y. Kato, T. Kamito and K. Miura, 2002. Feminization of genetic males by a symbiotic bacterium in a butterfly, Eurema hecabe (Lepidoptera:Pieridae). Naturwissenschaften 89 167–170. [DOI] [PubMed] [Google Scholar]
- Hosokawa, T., K. Koga, Y. Kikuchi, X. Y. Meng and T. Fukatsu, 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 107 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huigens, M. E., and R. Stouthamer, 2003. Parthenogenesis associated with Wolbachia, pp. 247–266 in Insect Symbiosis, edited by K. Bourtzis and T. A. Miller. CRC Press, Boca Raton, FL.
- Hurst, G. D. D., and F. M. Jiggins, 2005. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc. R. Soc. Lond. B Biol. Sci. 272 1525–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, G. D. D., and M. E. N. Majerus, 1993. Why do maternally inherited microorganisms kill males? Heredity 71 81–95. [Google Scholar]
- Hurst, G. D. D., M. E. N. Majerus and L. E. Walker, 1993. The importance of cytoplasmic male-killing elelments in the natural populations of the two spot ladybird, Adalia bipunctata (Linnaeus) (Coleoptera; Coccinellidae). Biol. J. Linnean Soc. 49 195–202. [Google Scholar]
- Hurst, G. D. D., F. M. Jiggins and M. E. N. Majerus, 2003. Inherited microorganisms that selectively kill male hosts: the hidden players of insect evolution?, pp. 177–198 in Insect Symbiosis, edited by K. Bourtzis and T. A. Miller. CRC Press, Boca Raton, FL.
- Hurst, L. D., 1991. The incidences and evolution of cytoplasmic male killers. Proc. R. Soc. Lond. B Biol. Sci. 244 91–99. [Google Scholar]
- Jaenike, J., K. A. Dyer and L. K. Reed, 2003. Within-population structure of competition and the dynamics of male-killing Wolbachia. Evol. Ecol. Res. 5 1023–1036. [Google Scholar]
- Jiggins, F. M., J. P. Randerson, G. D. D. Hurst and M. E. N. Majerus, 2002. How can sex ratio distorters reach extreme prevalences? Male-killing Wolbachia are not suppressed and have near-perfect vertical transmission efficiency in Acraea encedon. Evolution 56 2290–2295. [DOI] [PubMed] [Google Scholar]
- Johnstone, R. A., and G. D. D. Hurst, 1996. Maternally inherited male-killing microorganisms may confound interpretation of mitochondrial DNA variability. Biol. J. Linnean Soc. 58 453–470. [Google Scholar]
- Kimura, M., and J. F. Crow, 1963. The measurement of effective population number. Evolution 17 279–288. [Google Scholar]
- Lipsitch, M., M. A. Nowak, D. Ebert and R. M. May, 1995. The population-dynamics of vertically and horizontally transmitted parasites. Proc. R. Soc. Lond. B Biol. Sci. 260 321–327. [DOI] [PubMed] [Google Scholar]
- Martin, G., P. Juchault and J. J. Legrand, 1973. Mise en évidence d'un micro-organisme intracytoplasmique symbiote de I'Oniscoide Armadillidium vulgare L. dont la présence accompagne I'intersexualité ou la féminisation totale des mâles génétiques de la lignée thélygène. (Evidence for an intracytoplasmic symbiotic microorganism of the oniscoid Armadillidium vulgare L. whose presence is associated with intersexes or complete feminization of genetic males in thelygene lines.) C. R. Acad. Sci. Paris 276 2313–2316. [Google Scholar]
- Möhle, M., 1998. A convergence theorem for Markov chains arising in population genetics and the coalescent with selfing. Adv. Appl. Probab. 30 493–512. [Google Scholar]
- Nordborg, M., and S. M. Krone, 2002. Separation of time scales and convergence to the coalescent in structured populations, pp. 194–232 in Modern Developments in Theoretical Population Genetics: The Legacy of Gustave Malécot, edited by M. Slatkin and M. Veuille. Oxford University Press, Oxford.
- Oliver, K. M., J. A. Russell, N. A. Moran and M. S. Hunter, 2003. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. USA 100 1803–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, S., N. A. Rosenberg, M. W. Feldman and J. Wakeley, 2008. Population differentiation and migration: coalescence times in a two-sex island model for autosomal and X-linked loci. Theor. Popul. Biol. 74 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaud, T., 1997. Inherited microorganisms and sex determination of arthropod hosts, pp. 81–102 in Influential Passengers: Inherited Microorganisms and Invertebrate Reproduction, edited by S. L. O'Neill, A. A. Hoffmann and J. H. Werren. Oxford University Press, New York.
- Rigaud, T., P. Juchault and J. P. Mocquard, 1997. The evolution of sex determination in isopod crustaceans. BioEssays 19 409–416. [Google Scholar]
- Scarborough, C. L., J. Ferrari and H. C. J. Godfray, 2005. Aphid protected from pathogen by endosymbiont. Science 310 1781. [DOI] [PubMed] [Google Scholar]
- Sjödin, P., I. Kaj, S. Krone, M. Lascoux and M. Nordborg, 2005. On the meaning and existence of an effective population size. Genetics 169 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J., and J. Jaenike, 2006. Male-killing Wolbachia and male mate choice: a test with Drosophila innubila. Evol. Ecol. Res. 8 91–102. [Google Scholar]
- Taylor, D. R., 1990. Evolutionary consequences of cytoplasmic sex ratio distorters. Evol. Ecol. 4 235–248. [Google Scholar]
- Wakeley, J., 2009. Coalescent Theory: An Introduction. Roberts and Company Publishers, Greenwood Village, CO.
- Wakeley, J., and O. Sargsyan, 2009. Extensions of the coalescent effective population size. Genetics 181 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1931. Evolution in Mendelian populations. Genetics 16 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]








