Abstract
Mutations in maternally transmitted symbionts can affect host fitness. In this study we investigate a mutation in an obligate bacterial symbiont (Buchnera), which has dramatic effects on the heat tolerance of pea aphid hosts (Acyrthosiphon pisum). The heat-sensitive allele arises through a single base deletion in a homopolymer within the promoter of ibpA, which encodes a universal small heat-shock protein. In laboratory cultures reared under cool conditions (20°), the rate of fixation (1.4 × 10−3 substitutions per Buchnera replication) is much higher than the previously estimated mutation rate for single base deletions in homopolymers in the Buchnera genome, implying a strong selective benefit. This mutation recurs in natural populations, but seldom reaches high frequencies, implying that it is only rarely favored by selection. Another potential source of physiological stress in pea aphids is infection by other microorganisms, including facultative bacterial symbionts, which occur in a majority of pea aphids in field populations. Frequency of the heat-sensitive Buchnera allele is negatively correlated with presence of facultative symbionts in both laboratory colonies and field populations, suggesting that these infections impose stress that is ameliorated by ibpA expression. This single base polymorphism in Buchnera has the potential to allow aphid populations to adapt quickly to prevailing conditions.
APHIDS have engaged in symbiosis with the maternally transmitted bacterial symbiont Buchnera aphidicola for >160 million years in a mutually obligate relationship, providing aphids with a source of essential amino acids lacking in nutrient-poor plant phloem (Buchner 1965; Sandström and Moran 1999; Shigenobu et al. 2000). A major determinant of a species' ecology and geographic distribution is tolerance to thermal stress; for aphids this is constrained by tolerances of Buchnera. Pea aphids (Acyrthosiphon pisum) are sensitive to heat (Chen et al. 2000; Russell and Moran 2006), reflecting in part, the heat sensitivity of Buchnera cells (Montllor et al. 2002; Ohtaka and Ishikawa 1991; Dunbar et al. 2007; Burke et al. 2009). Laboratory and field experiments have shown that a naturally occurring Buchnera mutation, preventing heat induction of ibpA, which encodes a small heat-shock protein, has severe effects upon aphid fitness following heat stress in both the laboratory (Dunbar et al. 2007) and the field (Harmon et al. 2009).
Buchnera exhibits constitutively high expression of several heat-shock genes, presumably assisting folding of proteins that are destabilized due to degenerative evolution in the Buchnera genome (Wilcox et al. 2003; Wilson et al. 2006). However, ibpA is not constitutively upregulated in Buchnera (Wilcox et al. 2003); furthermore, under cool conditions (20°), decreased expression of ibpA, due to a single base deletion inactivating the promoter, has been shown to increase aphid fitness (Dunbar et al. 2007). The IbpA heat-shock protein is detrimental in cool conditions, perhaps due to its binding with other Buchnera proteins; in other organisms, it binds promiscuously with many cellular proteins (Kuczynska-Wisnik et al. 2002). Therefore, the ibpA promoter mutation of Buchnera is expected to undergo either positive or negative selection at the level of aphid hosts, depending on the thermal conditions encountered by a population.
Buchnera is clonal and inherited strictly maternally within aphids (Peccoud et al. 2009), so the occurrence of this mutation can be mapped on matriline phylogenies derived from other regions of the Buchnera genome. In this study, we explore the occurrence of the ibpA promoter mutation of Buchnera both in laboratory clones (genetically identical asexually reproducing aphids that originated from a single female) and in worldwide samples. We estimate the laboratory substitution rate and use a matriline phylogeny of pea aphids as a basis for identifying independent origins of the Buchnera ibpA allele. In addition, we test whether ibpA promoter type is correlated with presence of facultative symbionts, which are widespread in pea aphids and which are a potential source of cellular stress for Buchnera and aphid hosts.
MATERIALS AND METHODS
Field collections:
We examined the ibpA promoter region for Buchnera from a worldwide collection of pea aphids, including 193 individuals from North America, Europe, Japan, and western Asia. Most samples corresponded to a subset of those used in a recent study of the matriline phylogeny of pea aphids (Peccoud et al. 2009), based on two Buchnera genomic regions (corresponding to positions 20408–21459 (groEL–efp) and 30176–31218 (cof–metE) in the published genome for Buchnera from the APS lineage of pea aphids (NC_002528; Shigenobu et al. 2000). We used the tree, pruned of branches corresponding to collections for which the ibpA promoter was not sequenced, as a phylogenetic framework to examine the derivation of ibpA promoter alleles. To explore the frequency of the alternative ibpA promoter alleles within populations, we examined an additional 196 pea aphids sampled at random within target populations in North America.
Laboratory clones:
Laboratory pea aphid clones are derived from a single field collected female and then maintained asexually on Vicia faba at constant 20°. Two clones were subdivided into two or more subclones, derived from the same founding female. These were 5A, originating from a female collected in Madison, WI in 1999, and Tucson, originating from a female collected in Tucson, AZ in 1999. These clones contain Buchnera, representing distant matrilines within pea aphids (Peccoud et al. 2009). Lab clones were monitored with molecular markers to ensure that they were not cross-contaminated. Samples were frozen every 3 months at −80° during routine culture maintenance. For this study, all clones were sampled and the ibpA promoter sequenced. For any samples containing the mutation, the promoter was resequenced for verification, and the approximate time of origin of the allele was determined using the frozen samples.
PCR and sequencing of the ibpA allele:
The ibpA promoter alleles differ by the number of adenines within the spacer between −10 and −35 binding sites: the common allele has 11 A's (the spacer is 13 nucleotides in total) and the heat sensitive allele has 10 A's (spacer length, 12 nucleotides). This difference can only be reliably determined by sequencing. The region was amplified using polymerase chain reaction (PCR) with the primers ibpA (NM2) F: 5′-CAAATCAAACTATGTAGAAGAACATTTAGATC-3′ and ibpA (NM2) R: 5′-CTAATTTCAATAGACCTAAAGATAACTCTGC-3′ to give a 659-bp amplicon, including 385 bp (81%) of the ibpA coding sequence. Reaction volumes were 10 μl, with 1 μL of reaction buffer (including 1.5 mm MgCl2), 1 mm MgCl2, 2.5 mm of each dNTP, 0.4 μmol of each primer, 0.25 units of Eppendorf taq polymerase, and ∼15 ng of DNA. Reactions were incubated at 94° for 2 min, then cycled 35 times as follows: 94° for 30 sec, 58° for 30 sec, and 72° for 45 sec. Amplification products underwent final extension at 72° for 6 min. Samples were sequenced using an ABI 3730 at the University of Arizona Genetics Core.
Diagnostic facultative symbiont infection screens:
To determine presence of facultative symbionts, screens were performed using PCR to specifically amplify gyrB, using the primers gyrBR-type-402R:
5′-CGCTGAACAGCTACATGGAA-3′ and gyrBR-type-158F:
5′-GCCGACCACAATTTTAGCAT-3′ for Serratia symbiotica and gyrBT-type484F: 5′-TTCCTGAAATCCATCGTTCC-3′ and gyrBT-type685R: 5′-CAAACGCAACGATCAAGAAA-3′ for Hamiltonella defensa (Oliver et al. 2006).
RESULTS AND DISCUSSION
Evolution of the ibpA mutation in laboratory conditions:
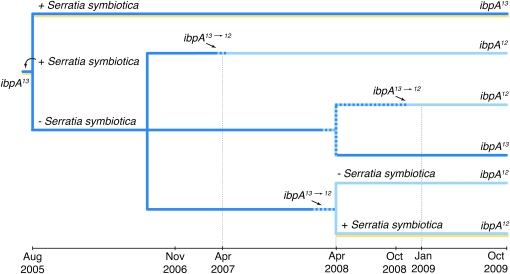
Previously, two subclones of 5A were identified that had undergone fixation of the heat-sensitive allele (ibpA12) from the common allele (ibpA13) in Buchnera (Dunbar et al. 2007). Here, we present evidence that the ibpA13→12 mutation (the deletion of one nucleotide in a homopolymeric tract of the ibpA promoter region) occurred and was fixed multiple times in a second lab clone (Tucson). This mutation was discovered in three Tucson subclones, ∼600, 960, and 1230 days after establishment of the facultative symbiont-cured Tucson subclone in August 2005 (Figure 1). We assume that mutations are fixed or lost quickly after their origin, due to bottlenecks both during progeny inoculation and during maintenance of lab clones that are repeatedly reduced to a few individuals. The approximate substitution rate per Buchnera replication in lab clones can be estimated on the basis of the time until appearance of the mutation, the average aphid generation time, and average number of Buchnera cell divisions per aphid generation (Mira and Moran 2002, Table 1). On the basis of five occurrences of the ibpA13→12 mutation in our laboratory clones, we estimate a substitution rate of 1.4 × 10−3 per Buchnera replication. In comparison, the estimated mutation rate for all homopolymeric tracts of similar size in the Buchnera genome is 7.5 × 10−8 insertion/deletion (indel) events per homopolymeric tract per replication; this estimate is based on intergenic spacers in which insertions or deletions are approximately neutral, and almost all indels involve a single nucleotide base pair (Moran et al. 2009). For nucleotide base substitutions, the genome-wide mutation rate of Buchnera is estimated at 4 × 10−9 substitutions per site per replication.
Figure 1.—
Chronology of ibpA promoter mutations in laboratory subclones of the Tucson pea aphid genotype. In August 2005, a Tucson subclone was cured of its facultative symbiont, Serratia symbiotica. Three subclones were established from the cured aphids, and all underwent ibpA13→12 mutations before their discovery in April 2008. Hatched bars indicate polymorphism in the ibpA promoter sequence among a population.
TABLE 1.
Estimation of substitution rate of ibpA13→12 in laboratory aphid populations, reflecting both mutation rate and selection in two aphid clones
| Genotype | Time until mutation appearance | Pea aphid generationsa | Buchnera cell divisions | Substitutions in lab colonies |
|---|---|---|---|---|
| 5A | June 1999—January 2001, 570 days | 47.5 | 380 | |
| June 1999—September 2005, 1890 days | 157.5 | 1260 | ||
| Tucson | August 2005—April 2007, 600 days | 50 | 400 | |
| August 2005—April 2008, 960 days | 80 | 640 | ||
| August 2005—January 2009, 1230 days | 103 | 824 | ||
| Mean | 701 | 1.4 × 10−3 |
Pea aphids have a generation time of 12 days, and Buchnera cells divide approximately eight times per aphid generation (Mira and Moran 2002).
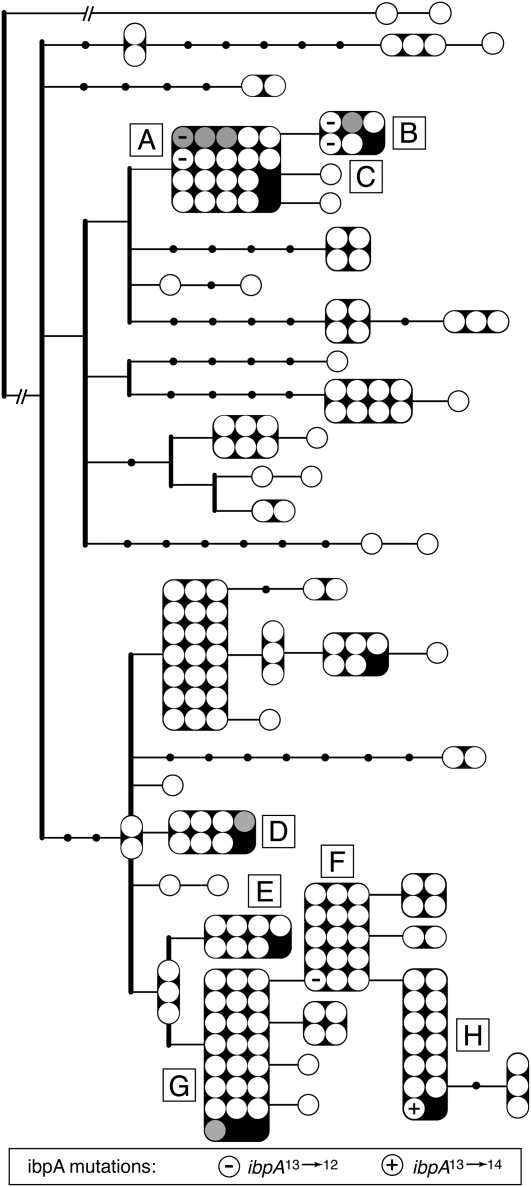
The Buchnera ibpA promoter undergoes recurrent mutation, as demonstrated by the fact that the shortened, inactive promoter (ibpA12) evolved multiple times in the laboratory, in two different pea aphid genetic backgrounds representing distant matrilines (C and E on the Buchnera haplotype tree, Figure 2). The estimated rate of ibpA13→12 substitution in laboratory colonies is >104 times higher than the mutation rate previously estimated for all homopolymeric tracts in intergenic spacers 10–12 bp in size in the Buchnera genome, and 105 times higher than the mutation rate for single base substitutions, based on neutral sites in the full genome data for Buchnera strains (Moran et al. 2009). If the ibpA13→12 mutation were neutral, its rate of substitution in lab clones would correspond to the genome-wide mutation rate for homopolymeric tracts (Moran et al. 2009) and we would not expect to sample the mutation at all during the limited duration of the lab colonies. Furthermore, single nucleotide insertions are equivalent in number to deletions based on genome-wide mutational patterns for homopolymeric tracts (Moran et al. 2009). We have not observed any insertions in Buchnera ibpA promoters, including any reversions (ibpA12→13) in the laboratory, presumably due to the lack of selection for ibpA13 or ibpA14.
Figure 2.—
ibpA promoter mutations mapped onto a haplotype map of pea aphid Buchnera lineages. Discs represent an aphid individual or population. Gray discs indicate that multiple aphids were sampled; a “population.” Letters in boxes indicate haplotypes referred to in text.
Therefore, ibpA12 must be positively selected under the lab environment. This supposition is consistent with previous demonstration of a fitness benefit for aphids bearing Buchnera with the ibpA12 allele under constant 20° (Dunbar et al. 2007). Selection on Buchnera mutations acts on two levels: among the symbionts within a host insect and among hosts within the insect population. Under cool conditions, we presume that a mutant Buchnera cell with ibpA12 gains an advantage over cells with ibpA13 through improved cellular function, resulting in increased frequency and fixation of ibpA12 within the individual aphid and matriline. Between aphids, hosts with more functional nutritional symbionts are favored by selection (Dunbar et al. 2007). Thus, in cool conditions we expect selection to act in the same direction (favoring ibpA12) at both symbiont and host population levels. As drift impairs the efficacy of selection at both the symbiont and host levels due to bottlenecks in symbiont transmission and within small laboratory populations, respectively, its evolution in laboratory populations on such a short time scale indicates strong selection for ibpA12.
Calculation of s and N corresponding to the observed rate of substitution is complex because selection coefficients and relevant effective population sizes are different for each level at which selection acts. Efficacies of within-host and between-host selection are most affected by transmission bottlenecks and by host population size, respectively (Rispe and Moran 2000). Although it is not feasible to calculate exact values of s or N, we note that N must exceed 37,333 (1.4 × 10−3/3.75 × 10−8 or the estimated substitution rate divided by the estimated unidirectional mutation rate for indels). This value corresponds to N when probability of fixation of each mutation is one (since substitution rate = Nμ × probability of fixation of a new mutation; Graur and LI 2000), and thus is a minimum. Although it seems large, this value of N is feasible since it refers to the population size of Buchnera, which number ∼4 × 104 per pea aphid embryo (Mira and Moran 2002). Our laboratory cultures are bottlenecked to 10 aphids at each culture change, so ∼4 × 105 Buchnera cells are present in the transferred individuals during their early development, giving an upper bound for the effective population size and corresponding to a lower bound of s = 0.05 to yield the observed rate of substitution. Almost certainly N is considerably smaller and s larger; for example, with N of 5 × 104, s is 0.7. Daily fecundity of aphids grown at 20° was 1.17 times higher for ibpA12 than for ibpA13 aphids (Dunbar et al. 2007), and, assuming this is an index for fitness, s is 0.17, corresponding to a required N of ∼1.3 × 105. These estimates are compatible with the range of possible values calculated above. These rough calculations show that the overall selective benefit of ibpA12 must be substantial to generate the observed rate of substitution.
Distribution of the Buchnera ibpA promoter mutation in field populations:
To investigate the prevalence of this mutation in field populations, the ibpA promoter region of Buchnera was sequenced for 193 pea aphids taken from diverse host plants and localities (supporting information, Table S1 and Table S2). Previous sampling of field populations for ibpA promoter mutations was limited to the United States and included some of the same sites with fewer individuals per site (Dunbar et al. 2007). The 545-bp region sequenced contained 10 polymorphic sites. In these worldwide samples, we found six alleles with unusual ibpA promoter length. Five were the same heat-sensitive allele (ibpA12), which eliminates ibpA transcriptional response to heat (Dunbar et al. 2007). The sixth unusual allele lengthened the promoter sequence by one adenine (ibpA14). No loss-of-function mutations were found in the ibpA coding sequence, and only synonymous changes in the ibpA coding region were present in eight fully sequenced genomes of Buchnera from distinct pea aphid strains (Moran et al. 2009).
Mutations in the Buchnera ibpA promoter were mapped onto the pea aphid matriline genealogy from Peccoud et al. (2009) (Figure 2). The six ibpA promoter mutations represent four Buchnera haplotypes and thus at least four independent mutational events. In haplotype B, the genotypes with ibpA12 are 7a and 7-2-1, collected in Cayuga County, NY, in 2000 and 2001. Haplotype A also exhibited two ibpA12 alleles, found in genotypes A2A and WI1999B. Haplotype F has one individual collection, Colmar, a long-term laboratory clone with ibpA12. In haplotype H, the aphid individual LL01_VF (also a long-term laboratory clone) had an ibpA14 allele. Whether these two mutations occurred before or after establishment as laboratory colonies is unknown.
Despite the high rate of the ibpA13→12 mutation, ibpA12 is rare in most field populations. Only five ibpA12 alleles were found from 193 individuals from diverse field collections. Thus, although ibpA13→12 recurs and is advantageous in our laboratory conditions, ibpA12 remains rare in field conditions. This is probably due to selection for IbpA production under the warm or stressful conditions that are common for natural populations. We note that even brief exposure to higher temperatures can favor the ibpA13 allele and that aphids that fall from a plant can experience radiant heat that exceeds local air temperatures; thus, even in cooler climates, temperature stress may often favor ibpA13.
We sequenced more individuals from three populations (aphids found at the same location and date) that showed evidence for promoter mutations (Table S2). All 49 individuals sampled from a population in Wisconsin in 1999 (called WI1999) belong to the same Buchnera haplotype (haplotype A), but they include distinct aphid clones, including two different color morphs (color polymorphism is a Mendelian trait in pea aphids so morphs represent different clones). Of 33 red pea aphids screened, all had ibpA12. The 16 green pea aphids (which represent one or more clones distinct from the red aphids) collected at the same location and date had ibpA13. Thus, ibpA12 was observed at high frequency in the WI1999 population (33/49 or 67%).
In some conditions, ibpA12 may be advantageous (Dunbar et al. 2007), due to the cost of making IbpA protein or to negative effects of this protein (Kuczynska-Wisnik et al. 2002). The ibpA12 aphids possibly represent a single clone, as they shared the same Buchnera haplotype and body color. Their high frequency could be attributable either to an advantage of ibpA12 under recent field conditions or to other genetic differences affecting fitness of particular clones. Two populations, NY2000 and NY2003, were sampled from Cayuga County, NY in 2000 and 2003, respectively. In both years, two aphid haplotypes were present, but none had ibpA12. The populations collected in 2000 consisted of Buchnera haplotypes A (NY2000B, July 2000, N = 8 and October 2000, N = 18) and D (NY2000A, July 2000, N = 16 and October 2000, N = 28). The 2003 populations contained Buchnera haplotypes B (NY2003B, N = 24) and G (NY2003A, N = 53).
Although the 7a and 7-2-1 clones (haplotype B) collected in Cayuga County, NY in May 2000 and August 2001 bore ibpA12, ibpA12 alleles were absent from 147 individuals from populations of the same or closely related haplotypes collected from the same region in July 2000, October 2000, and July 2003. Thus, the ibpA12 allele can change prevalence in pea aphid populations on the scale of months (χ2 likelihood ratio = 21.2, P = 0.0003).
Within a haplotype, ibpA13→12 mutations occur repeatedly, as observed in two clones representing distant Buchnera genotypes monitored in the lab. In Buchnera haplotype B, the 7a and 7-2-1 aphids are distinct aphid clones, based on color morph and year of collection. Thus, either ibpA12 evolved twice in this haplotype or persisted across a sexual generation involving reassortment at the aphid locus determining color. The two ibpA12 alleles found for Buchnera haplotype A likely represent independent mutations, as one was collected in Wisconsin and one in Utah (1800 km apart in different years). Together, these data illustrate a recurrent Buchnera mutation that is highly dynamic in pea aphid populations, possibly with significant effects upon aphid fitness and adaptation to environmental fluctuations.
One haplotype displayed ibpA14, a mutation that increased the promoter spacer length from 13 to 14 bp. In Escherichia coli, variants with 14-bp spacers display the highest level of transcriptional induction upon heat shock, ∼2.5-fold higher than promoters with 13-bp spacers (Wang and Dehaseth 2003). Since the consensus promoter and the Sigma factor binding sites are identical for E. coli and Buchnera (Wilcox et al. 2003), the longer spacer length may increase ibpA expression and thermal tolerance.
Correlation of ibpA alleles and facultative symbionts:
Mutations in the Buchnera ibpA promoter are negatively correlated with infection by facultative symbionts, which infect a majority of individuals in most pea aphid populations (references within Oliver et al. 2010). In four of five origins of ibpA12 in laboratory colonies, mutations have occurred in subclones naturally uninfected or uninfected following curing of a facultative symbiont (Table 2, two-sided P = 0.0027, Fisher's exact test). The exception occurred in a subclone of 5A infected with S. symbiotica, but the facultative symbiont infection was lost 1 month after the mutation was established, resulting in an uninfected ibpA12 5A subclone (Dunbar et al. 2007). Most of our laboratory clones are infected with facultative symbionts, and no others have undergone promoter mutations, despite being grown under the same conditions. This trend is also observed for aphids sampled from the field. Arizona-collected aphids, which belong to haplotype E, are invariably infected with S. symbiotica, and none have been found to contain Buchnera with ibpA12 (table 1 of Dunbar et al. 2007). All five aphids with ibpA12 from the worldwide samples lacked infection with any of the three facultative symbionts common in pea aphids (S. symbiotica, H. defensa, and Regiella insecticola). Finally, in the WI1999 population, facultative symbiont infection frequency was higher in ibpA13 aphids (14/15 or 93%) compared to ibpA12 aphids (13/26 or 46%) (two-sided P = 0.0061, Fisher's exact test). Twelve ibpA12 aphids were infected with H. defensa and one with S. symbiotica, and 14 ibpA13 aphids were infected with H. defensa.
TABLE 2.
Buchnera ibpA promoter substitutions in long-term laboratory stocks with differing facultative symbiont infections
| Facultative symbiont infection | Number of aphid subclones | Total years of subclone evolution | Number of ibpA13→12 substitutions |
|---|---|---|---|
| Regiella insecticola | 3 | 22 | 0 |
| Hamiltonella defensa | 3 | 20 | 0 |
| Serratia symbiotica | 6 | 35 | 1a |
| None (starting as ibpA13) | 4 | 10 | 4 |
The probability of substitution is different, depending upon facultative symbiont infection (two-tailed P = 0.0027, Fisher's exact test).
S. symbiotica infection was lost 1 month after Buchnera ibpA13→12 substitution (figure 3 in Dunbar et al. 2007).
Thus, Buchnera ibpA12 is correlated with absence of a facultative symbiont, in both field and laboratory aphids. This is most readily explained by a fitness cost to ibpA12 in aphids harboring a facultative symbiont. Under such a cost, ibpA12 may spread only when it arises in aphids lacking facultative symbionts (a minority of individuals in most populations). Facultative symbionts have a large effect upon the metabolism of aphids (Burke et al. 2009), possibly resulting in oxidative or other stress for Buchnera cells. The combination of facultative symbiont-induced stress and low expression of this heat-shock protein (due to the Buchnera ibpA12 allele) may confer low fitness and could lead these lineages to extinction, resulting in their absence from field collections.
The ibpA13→12 mutation must arise in every field population, based on its recurrence in lab colonies. Although we did not observe reversions (ibpA12→13) in laboratory colonies, they are likely common, as homopolymers in noncoding regions show roughly equal numbers of insertions and deletions (Moran et al. 2009). The ibpA12/ibpA13 polymorphism may allow aphid populations to adapt quickly to prevailing conditions.
Acknowledgments
We thank Kim Hammond for maintaining aphid colonies and Laurel Johnstone for work on DNA sequencing. Aphid samples were kindly provided by Saghar Aleosfoor, Paul Baumann, Emilie Bilodeau, René Bournoville, Marina Caillaud, Steve Clement, Conrad Cloutier, Angela Douglas, Sanford Eigenbrode, Owain Edwards, Christian Figueroa, Takema Fukatsu, Nicole Gerardo, Hanene Harbaoui, Anthony Ives, Blas Lavandero, Weinong Lu, Hanem Makni, Susan Masta, Kerry Oliver, Yannick Outreman, Jean Peccoud, Servane Penvern, Manuel Plantegenest, Glen Powell, Claudio Ramirez, Jacob Russell, Beatriz Sabater-Munoz, Tsutomu Tsuchida, Wolfgang Weisser, Alex Wilson, and Steve Wratten.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.117440/DC1.
References
- Buchner, P., 1965. Endosymbiosis in animals with plant microorganisms. John Wiley & Sons, New York.
- Burke, G. R., O. Fiehn and N. A. Moran, 2009. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J. 4 242–252. [DOI] [PubMed] [Google Scholar]
- Chen, D.-Q., C. B. Montllor and A. H. Purcell, 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, the blue alfalfa aphid, A. kondoi. Entomol. Exp. Appl. 95 315–323. [Google Scholar]
- Dunbar, H. E., A. C. Wilson, N. R. Ferguson and N. A. Moran, 2007. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 5 e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur, D. and W.-H. Li, 2000. Fundamentals of Molecular Evolution. Sinauer, Sunderland, MA.
- Harmon, J. P., N. A. Moran and A. R. Ives, 2009. Species response to environmental change: impacts of food web interactions and evolution. Science 323 1347–1350. [DOI] [PubMed] [Google Scholar]
- Kuczynska-Wisnik, D., S. Kcdzierska, E. Matuszewska, P. Lund, A. Taylor et al., 2002. The Escherichia coli small heat-shock proteins IbpA and IbpB prevent the aggregation of endogenous proteins denatured in vivo during extreme heat shock. Microbiology 148 1757–1765. [DOI] [PubMed] [Google Scholar]
- Montllor, C., A. Maxmen and A. H. Purcell, 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27 189–195. [Google Scholar]
- Moran, N. A., H. J. McLaughlin and R. Sorek, 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323 379–382. [DOI] [PubMed] [Google Scholar]
- Mira, A., and N. A. Moran, 2002. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microbiol. Ecol. 44 137–143. [DOI] [PubMed] [Google Scholar]
- Ohtaka, C., and H. Ishikawa, 1991. Effects of heat treatments on the symbiotic systems of an aphid mycetocyte. Symbiosis 11 19–30. [Google Scholar]
- Oliver, K. M., Moran, N. A., and M. S. Hunter, 2006. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. B Biol. Sci. 273 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, K. M., P. H. Degnan, G. R. Burke and N. A. Moran, 2010. Facultative symbionts of aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55 247–266. [DOI] [PubMed] [Google Scholar]
- Peccoud, J., J.-C. Simon, H. J. McLaughlin and N. A. Moran, 2009. Post-Pleistocene radiation of the pea aphids complex revealed by rapidly evolving endosymbionts. Proc. Natl. Acad. Sci. USA 106 16315–16320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispe, C., and N. A. Moran, 2000. Accumulation of deleterious mutations in endosymbionts: Muller's ratchet with two levels of selection. Am. Nat. 156 425–441. [DOI] [PubMed] [Google Scholar]
- Russell, J. A. and N. A. Moran, 2006. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc. R. Soc. B Biol. Sci. 273 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström, J. P., and N. A. Moran, 1999. How nutritionally imbalanced in phloem sap for aphids? Entomol. Exp. Appl. 91 203–210. [Google Scholar]
- Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki and H. Ishikawa, 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407 81–86. [DOI] [PubMed] [Google Scholar]
- Wilcox, J. L., H. E. Dunbar, R. D. Wolfinger and N. A. Moran, 2003. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol. Microbiol. 48 1491–1500. [DOI] [PubMed] [Google Scholar]
- Wilson, A. C., H. E. Dunbar, G. K. Davis, W. B. Hunter, D. L Stern et al., 2006. A dual-genome microarray for the pea aphid, Acyrthosiphon pisum, and its obligate bacterial symbiont, Buchnera aphidicola. BMC Genomics 7 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., and P. L. deHaseth, 2003. Sigma 32-dependent promoter activity in vivo: sequence determinants of the groE promoter. J. Bacteriol. 185 5800–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]