Abstract
Mutant screens have proven powerful for genetic dissection of a myriad of biological processes, but subsequent identification and isolation of the causative mutations are usually complex and time consuming. We have made the process easier by establishing a novel strategy that employs whole-genome sequencing to simultaneously map and identify mutations without the need for any prior genetic mapping.
THE challenges posed by the identification of a causal mutation in a mutant of interest have in effect restricted the use of forward genetics to those organisms benefiting from a solid genetic toolbox. Whole-genome sequencing (WGS) is promising to revolutionize the way phenotypic traits are assigned to genes. However, current strategies to identify causal mutations using WGS require first the identification of an approximate genomic location containing the mutation of interest (Sarin et al. 2008; Smith et al. 2008; Srivatsan et al. 2008; Blumenstiel et al. 2009; Irvine et al. 2009). This is because genomes contain many natural sequence variations (Denver et al. 2004; Hillier et al. 2008; Sarin et al. 2010), which, along with mutagen-induced ones, complicate the identification of the causal mutation when an approximate genomic location has not been previously identified. Mapping has previously been achieved with time-consuming and laborious techniques that, in addition, rely on an organism's single-nucleotide polymorphism (SNP) map and established variant strains. For example, traditional SNP-based mapping (Wicks et al. 2001; Davis et al. 2005) has previously been used in Caenorhabditis elegans to narrow down the genomic region containing the mutation of interest, prior to conducting WGS (Sarin et al. 2008). In Arabidopsis, simultaneous SNP mapping and mutation identification has been achieved with WGS, but this requires the generation of a mapping population of up to 500 F2 progeny to identify only one allele (Schneeberger et al. 2009). This is a challenging prospect for many model systems. Indeed, if the mutant phenotype is subtle, the isolation of such numbers of recombinants is very tedious. Furthermore, it is not applicable in those organisms where a mapping population cannot be generated, simply because of a lack of intercrossable variants or because of life cycles (parasitic organisms, for example) that would make it extremely difficult to follow and isolate many recombinant individuals.
Here, we describe a strategy to simultaneously and rapidly locate and identify multiple mutations from a mutagenesis screen with WGS that circumvents these limitations. This powerful and straightforward method directly uses mutagen-induced nucleotide changes that are linked to the causal mutation to identify its specific genomic location, thus negating the construction of genetic mapping populations and subsequent mapping.
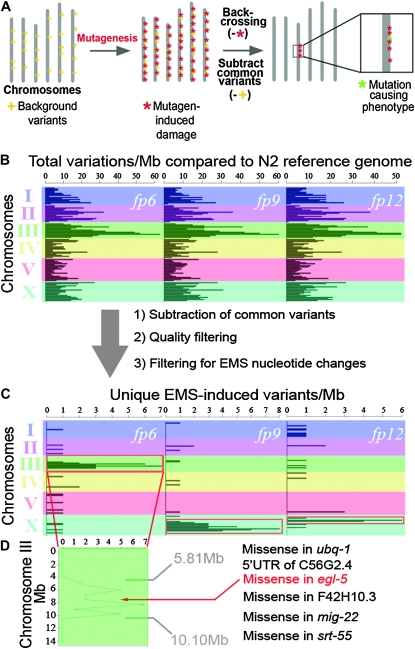
Treatment of organisms with a chemical mutagen induces nucleotide changes throughout the genome. Following mutagenesis, backcrossing or outcrossing of the mutagenized organism to unmutagenized counterparts is performed to eliminate mutagen-induced mutations (Figure 1A; supporting information, File S2). The phenotype-causing mutation remains as only backcrossed individuals showing the phenotype of interest are retained. In addition, mutagen-induced nucleotide changes that are genetically linked to the causal mutation and physically surround it on the chromosome will remain, in contrast to unlinked nucleotide changes (Figure 1A). As a result of this genetic linkage, a high-density cluster of typical mutagen-induced variants is visualized from sequence data obtained by WGS, which is positioned around the causal mutation. By locating such high-density regions, one maps the approximate genomic location of the causal mutation and subsequently identifies the affected gene within this region.
Figure 1.—
Mapping mutations on the basis of density of mutagen-induced DNA damage across the genome. (A) Visual representation of our WGS cloning strategy. Mutagen treatment induces point mutations throughout the genome (red asterisks). Backcrossing to the original unmutated parent strain removes much of the mutagen-induced nucleotide changes except for the causal mutation (green asterisk) and those genetically linked to it. WGS sequencing can be used to detect canonical mutagen-induced point mutations, thus revealing a physical position for the causal mutation. Shared background variants (yellow crosses) are filtered out from WGS data by comparing the sequences of mutants sequenced side-by-side, revealing a high-density variant cluster in only one genomic region. Importantly, genomic sequences of mutants derived from the same starting strain must be compared, to allow subtraction of nucleotide variants that are common to this particular strain, through sequence comparison. (B) Physical map of total nucleotide variations per megabase across the genome compared to the wild-type reference genome for each mutant (fp6, fp9, and fp12) after WGS. (C) After sequence quality filtering, subtraction of common variants between the 3 mutants, and filtering out noncanonical EMS nucleotide changes, high-density variant peaks are obtained in one genomic location for each mutant (red boxes). Steps 1 and 3 are essential for clear visualization of the high-density peaks whereas step 2 improves visualization. (D) Close-up of variants on chromosome III for fp6. Within this peak we identified only 6 candidate mutations that could potentially affect a protein sequence. We confirmed that the missense mutation in egl-5 was the causal mutation (Figure S2). For fp9 and fp12 we identified only 10 (9 missense and 1 3′-UTR) and 4 (2 premature stop and 2 missense) candidate mutations, respectively, within each mutant's EMS-based mapped region. Thus, our method consistently allowed precise mapping in 3 different mutants to a region small enough to contain only a handful of candidate mutations.
As a proof-of-principle, we simultaneously mapped and sequenced the causal mutations of multiple C. elegans mutants isolated from an EMS mutagenesis screen using this strategy. The mutagenesis screen itself was undertaken to identify genes that controlled the reprogramming of a single cell called Y into another cell called PDA during C. elegans development (Jarriault et al. 2008). After EMS treatment, three distinct mutant alleles (fp6, fp9, and fp12) were backcrossed to the original unmutagenized strain 4-6X. It is important to note that a backcrossing or outcrossing step is necessary for the analysis of mutants obtained from all mutagenesis screens, irrespective of the type of mutant identification strategy used or the type of mutagen or organism used (and, as such, does not represent an extra step introduced by our method). The mutants then underwent WGS side-by-side (Table S1, Table S2, Figure S1, and File S2). After alignment to the wild-type N2 reference genome using MAQgene software (Bigelow et al. 2009), the sequencing data obtained for each mutant were compared, and we subtracted common nucleotide variants that were shared between at least two of our three mutants (File S1). These shared variants, which are very unlikely to be either the causal mutation or EMS-induced mutations from the screen itself, represent strain differences between the N2 used to generate the reference genome and the PS3662 strain used here for mutagenesis. Note that this step eliminated ∼2000 point mutations as potential candidates for our causal mutation. This result strongly emphasizes the advantage of conducting WGS on two or more mutants side-by-side, as reference genomes may contain many nucleotide variations when compared to organisms sequenced from the laboratory (Denver et al. 2004; Hillier et al. 2008; Sarin et al. 2010; this study) and as such would confound mutation identification.
To identify EMS-induced changes linked to the causal mutation and expose its location, we looked only at variants that matched the canonical EMS-induced G/C > A/T transitions (Drake and Baltz 1976), revealing localized peaks of high-density variation on a single chromosome for each mutant (Figure 1, B and C). These peaks correspond to regions of high mutagen-induced damage that were not removed during backcrossing and therefore are most likely genetically linked to the causal mutation. We therefore focused our attention on these physical regions to identify candidate mutations within them. We localized fp6 to a 4.29-Mb region on chromosome III, fp9 to a 7.11-Mb region on chromosome X, and fp12 to a 1.28-Mb region on a different part of chromosome X (Figure 1C).
As a proof of principle, we further examined the nucleotide changes present in the interval to which fp6 was linked. Taking into consideration all variant types (point mutations and indels), we identified only six candidate mutations that potentially affected a gene's function (Figure 1D and Table S3). One of these, affecting the egl-5/hox gene, lies almost perfectly in the middle of the predicted EMS-based mapped region. We confirmed the existence of the mutation in egl-5 by manual resequencing. Both egl-5 targeted RNAi and noncomplementation with the egl-5(n945) null allele confirmed that fp6 affected egl-5 and caused the Y-to-PDA reprogramming defect (Figure S2). fp9 and fp12 each map to distinct regions on chromosome X that also contain only a handful of candidate mutations (10 and 4, respectively) (Figure 1C). Thus, our method consistently allowed precise mapping in 3 different mutants to a region small enough to contain only a handful of candidate mutations and subsequent identification of the causal mutation.
We calculated that comparison of WGS data for only two mutants of the same mutagenesis screen is sufficient to localize and sequence the causal mutation (Table 1, Table S4). Thirteen times sequence coverage has been found to be sufficient to identify a mutation in a pre-SNP mapped C. elegans mutant (Shen et al. 2008). Here, we tested the sequence coverage necessary to perform simultaneous mapping and mutant identification using our strategy and found that 13× was more than enough (Table 1, Table S4). In addition, by performing longer reads and/or paired-end sequencing, our method can be scaled up to bigger genomes or allow multiple mutant sequencing on each flow cell lane [for, e.g., using multiplex WGS (Cronn et al. 2008)]. Furthermore, because direct sequence comparison is ultimately made between two mutants sequenced side-by-side, the quality of an organism's reference genome (which is used only for alignment purposes) does not have a bearing on the mapping or mutant identification outcome. Moreover, recent advances in de novo alignment of short reads generated from next generation sequencing platforms (Li et al. 2010; Nowrousian et al. 2010; Webb and Rosenthal 2010; Young et al. 2010) suggest that a reference genome may not even be required to perform mutagen-based mapping and mutant identification with WGS. We predict that technical advances in these areas will make it possible to perform mutagenesis screens on any nonsequenced and genetically uncharacterized organism and use our strategy to quickly identify the causal mutation of an interesting mutant.
TABLE 1.
Summary of WGS cloning strategy
| Conditions used | Minimal requirements tested | |
|---|---|---|
| Backcrossing | 4–6× | 4× enough |
| No. of mutants sequenced | 3 | 2 enough |
| Sequencing of mutant | 2× flow cell lanes, paired-end reads (57mer) | 1× flow cell lane enough, single-end reads (57mer) enough |
| Average sequence coverage | 52.2–55.3× | 13.6× enough |
| Advantages | ||
| Any SNP or genetic map information is not necessary | ||
| No prior wet lab work necessary: generation of a recombinant mapping population is not necessary | ||
| Multiple alleles identified at once | ||
| Amenable to scaling up: can be equally used for bigger genomes | ||
| Fast: 7 days sequencing, 12 hr MAQGene alignment, and 1 hr mapping | ||
| Modest sequence coverage requirements limit cost | ||
| Reference genome sequence quality is not important and may not even be necessary | ||
| Very straightforward without any specialized software | ||
| Requirement | ||
| Species must be amenable to mutagenesis and backcrossing | ||
We found that all of the minimal requirements tested here were more than adequate to use our mapping strategy. Therefore, it is possible that fewer backcrosses and less sequencing coverage may suffice than is shown here. For example, for genomes with a similar size to C. elegans (∼100 Mb), this method can easily be scaled up by sequencing eight mutants per flow cell. As for any WGS experiments, total cost depends on genome size.
By eliminating any prior work except for back/outcrossing, a necessary step for any mutant characterization, our simple and quick strategy provides a significant saving of time and labor as the time needed to map and identify a candidate causal mutation is trimmed down to the sequencing time (currently 7 days) and sequence analysis time (<1 day, see Table 1). In addition, our strategy allows simultaneous discovery of multiple mutant alleles from a mutagenesis screen without any mapping population generation, thus making it conceptually easy to apply to many species. Indeed, our strategy is applicable to any vertebrate or invertebrate organism subjected to mutagenesis and will be particularly useful for those organisms where traditional genetic mapping is tedious and long. The only requirement to carry on this mutant identification strategy is that the organism be amenable to back/outcrossing. Perhaps most importantly, the strategy does not use species-specific SNPs to map the mutation, thus avoiding many constraints of previous methods. Thus, the spiraling-down cost of next generation sequencing technology and the establishment of our strategy open the exciting prospects of performing creative mutagenesis screens in a wide range of organisms.
Acknowledgments
We thank Arnaud Ahier, Irwin Davidson, Maria Doitsidou, and Bernard Jost for discussions and advice; Serge Vicaire from the Institut de Génétique et de Biologie Moléculaire et Cellulaire solexa platform for libraries preparation and sequencing; and Paul Ebert, Jean-Louis Mandel, Jean-Marc Reichhard, and Julien Vermot for a critical reading of the manuscript. This work was supported by a Université de Strasbourg fellowship to S.Z. and grants from the Centre National de la Recherche Scientifique (CNRS) and the Fondation pour la Recherche Médicale to S.J. S.J. is an investigator of the CNRS.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.119230/DC1.
References
- Bigelow, H., M. Doitsidou, S. Sarin and O. Hobert, 2009. MAQGene: software to facilitate C. elegans mutant genome sequence analysis. Nat. Methods 6 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel, J. P., A. C. Noll, J. A. Griffiths, A. G. Perera, K. N. Walton et al., 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn, R., A. Liston, M. Parks, D. S. Gernandt, R. Shen et al., 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36 e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, M. W., M. Hammarlund, T. Harrach, P. Hullett, S. Olsen et al., 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver, D. R., K. Morris, M. Lynch and W. K. Thomas, 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430 679–682. [DOI] [PubMed] [Google Scholar]
- Drake, J. W., and R. H. Baltz, 1976. The biochemistry of mutagenesis. Annu. Rev. Biochem. 45 11–37. [DOI] [PubMed] [Google Scholar]
- Hillier, L. W., G. T. Marth, A. R. Quinlan, D. Dooling, G. Fewell et al., 2008. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5 183–188. [DOI] [PubMed] [Google Scholar]
- Irvine, D. V., D. B. Goto, M. W. Vaughn, Y. Nakaseko, W. R. McCombie et al., 2009. Mapping epigenetic mutations in fission yeast using whole-genome next-generation sequencing. Genome Res. 19 1077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault, S., Y. Schwab and I. Greenwald, 2008. A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. USA 105 3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., W. Fan, G. Tian, H. Zhu, L. He et al., 2010. The sequence and de novo assembly of the giant panda genome. Nature 463 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian, M., J. E. Stajich, M. Chu, I. Engh, E. Espagne et al., 2010. De novo assembly of a 40 Mb eukaryotic genome from short sequence reads: Sordaria macrospora, a model organism for fungal morphogenesis. PLoS Genet. 6 e1000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin, S., S. Prabhu, M. M. O'Meara, I. Pe'er and O. Hobert, 2008. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5 865–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin, S., V. Bertrand, H. Bigelow, A. Boyanov, M. Doitsidou et al., 2010. Analysis of multiple EMS-mutagenized Caenorhabditis elegans strains by whole genome sequencing. Genetics 185 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger, K., S. Ossowski, C. Lanz, T. Juul, A. H. Petersen et al., 2009. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6 550–551. [DOI] [PubMed] [Google Scholar]
- Shen, Y., S. Sarin, Y. Liu, O. Hobert and I. Pe'er, 2008. Comparing platforms for C. elegans mutant identification using high-throughput whole-genome sequencing. PLoS One 3 e4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. R., A. R. Quinlan, H. E. Peckham, K. Makowsky, W. Tao et al., 2008. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 18 1638–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan, A., Y. Han, J. Peng, A. K. Tehranchi, R. Gibbs et al., 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4 e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, K. M., and B. M. Rosenthal, 2010. Deep resequencing of Trichinella spiralis reveals previously un-described single nucleotide polymorphisms and intra-isolate variation within the mitochondrial genome. Infect. Genet. Evol. 10 304–310. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28 160–164. [DOI] [PubMed] [Google Scholar]
- Young, A. L., H. O. Abaan, D. Zerbino, J. C. Mullikin, E. Birney et al., 2010. A new strategy for genome assembly using short sequence reads and reduced representation libraries. Genome Res. 20 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]